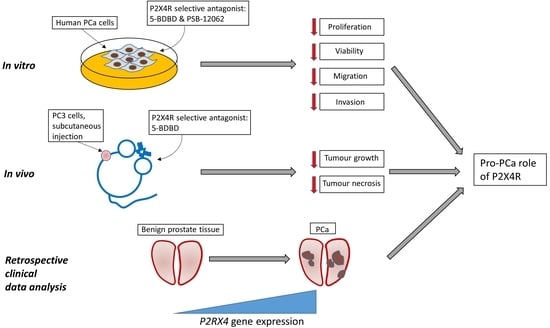
Inhibiting the P2X4 Receptor Suppresses Prostate Cancer Growth In Vitro and In Vivo, Suggesting a Potential Clinical Target
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Quantitative Real-Time PCR
2.3. P2X4R Agonist and Antagonists
2.4. Calcium Influx Assay
2.5. Viability Assay
2.6. Proliferation Assay
2.7. Apoptosis Assay
2.8. Migration Assay by Scratch Test
2.9. Cell Migration Assay by Transwell
2.10. Invasion Assay
2.11. In Vivo Study
2.12. Histology
2.13. Retrospective Analysis of Clinical Datasets
2.14. Statistical Analysis
3. Results
3.1. P2X4R is the Highest Expressed Functional P2 Receptor in PCa Cell Lines
3.2. Inhibiting P2X4R Impairs Proliferation and Viability of PCa Cells but Not Apoptosis
3.3. Inhibiting P2X4R Reduces PCa Cell Mobility
3.4. P2X4R Antagonist Shows Anti-Tumourigenic Effects in a PCa Xenograft Model
3.5. Retrospective Analysis of Clinical Datasets Suggests That P2X4R Associates with PCa Malignancy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swami, U.; McFarland, T.R.; Nussenzveig, R.; Agarwal, N. Advanced Prostate Cancer: Treatment Advances and Future Directions. Trends Cancer 2020, 6, 702–715. [Google Scholar] [CrossRef] [PubMed]
- Grozescu, T.; Popa, F. Prostate cancer between prognosis and adequate/proper therapy. J. Med. Life 2017, 10, 5–12. [Google Scholar] [PubMed]
- Fountain, S.J. Primitive ATP-activated P2X receptors: Discovery, function and pharmacology. Front. Cell. Neurosci. 2013, 7, 247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhuna, K.; Felgate, M.; Bidula, S.M.; Walpole, S.; Bibic, L.; Cromer, B.A.; Angulo, J.; Sanderson, J.; Stebbing, M.J.; Stokes, L. Ginsenosides Act As Positive Modulators of P2X4 Receptors. Mol. Pharmacol. 2019, 95, 210–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Guzman, M.; Soto, F.; Gomez-Hernandez, J.M.; Lund, P.E.; Stuhmer, W. Characterization of recombinant human P2X4 receptor reveals pharmacological differences to the rat homologue. Mol. Pharmacol. 1997, 51, 109–118. [Google Scholar] [CrossRef] [Green Version]
- Soto, F.; Garcia-Guzman, M.; Gomez-Hernandez, J.M.; Hollmann, M.; Karschin, C.; Stuhmer, W. P2X4: An ATP-activated ionotropic receptor cloned from rat brain. Proc. Natl. Acad. Sci. USA 1996, 93, 3684–3688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, R.; Taly, A.; Lemoine, D.; Martz, A.; Cunrath, O.; Grutter, T. Tightening of the ATP-binding sites induces the opening of P2X receptor channels. EMBO J. 2012, 31, 2134–2143. [Google Scholar] [CrossRef] [Green Version]
- Soto, F.; Garcia-Guzman, M.; Karschin, C.; Stuhmer, W. Cloning and tissue distribution of a novel P2X receptor from rat brain. Biochem. Biophys. Res. Commun. 1996, 223, 456–460. [Google Scholar] [CrossRef]
- Suurvali, J.; Boudinot, P.; Kanellopoulos, J.; Ruutel Boudinot, S. P2X4: A fast and sensitive purinergic receptor. Biomed. J. 2017, 40, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Stokes, L.; Layhadi, J.A.; Bibic, L.; Dhuna, K.; Fountain, S.J. P2X4 Receptor Function in the Nervous System and Current Breakthroughs in Pharmacology. Front. Pharmacol. 2017, 8, 291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Chai, H.; Ehinger, K.; Egan, T.M.; Srinivasan, R.; Frick, M.; Khakh, B.S. Imaging P2X4 receptor subcellular distribution, trafficking, and regulation using P2X4-pHluorin. J. Gen. Physiol. 2014, 144, 81–104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, Y.; Yang, B.; Liu, Z.; Yu, Q. Microencapsulated olfactory ensheathing-cell transplantation reduces pain in rats by inhibiting P2X4 receptor overexpression in the dorsal root ganglion. Neuroreport 2019, 30, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Ulmann, L.; Hirbec, H.; Rassendren, F. P2X4 receptors mediate PGE2 release by tissue-resident macrophages and initiate inflammatory pain. EMBO J. 2010, 29, 2290–2300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montilla, A.; Mata, G.P.; Matute, C.; Domercq, M. Contribution of P2X4 Receptors to CNS Function and Pathophysiology. Int. J. Mol. Sci. 2020, 21, 5562. [Google Scholar] [CrossRef] [PubMed]
- Paalme, V.; Rump, A.; Mado, K.; Teras, M.; Truumees, B.; Aitai, H.; Ratas, K.; Bourge, M.; Chiang, C.S.; Ghalali, A.; et al. Human Peripheral Blood Eosinophils Express High Levels of the Purinergic Receptor P2X4. Front. Immunol. 2019, 10, 2074. [Google Scholar] [CrossRef]
- Ralevic, V. P2X receptors in the cardiovascular system and their potential as therapeutic targets in disease. Curr. Med. Chem. 2015, 22, 851–865. [Google Scholar] [CrossRef]
- Yang, A.; Sonin, D.; Jones, L.; Barry, W.H.; Liang, B.T. A beneficial role of cardiac P2X4 receptors in heart failure: Rescue of the calsequestrin overexpression model of cardiomyopathy. Am. J. Physiol. Heart Circ. Physiol. 2004, 287, H1096–H1103. [Google Scholar] [CrossRef]
- Burnstock, G.; Di Virgilio, F. Purinergic signalling and cancer. Purinergic. Signal. 2013, 9, 491–540. [Google Scholar] [CrossRef]
- Huo, J.F.; Chen, X.B. P2X4R silence suppresses glioma cell growth through BDNF/TrkB/ATF4 signaling pathway. J. Cell. Biochem. 2019, 120, 6322–6329. [Google Scholar] [CrossRef]
- Chong, J.H.; Zheng, G.G.; Zhu, X.F.; Guo, Y.; Wang, L.; Ma, C.H.; Liu, S.Y.; Xu, L.L.; Lin, Y.M.; Wu, K.F. Abnormal expression of P2X family receptors in Chinese pediatric acute leukemias. Biochem. Biophys. Res. Commun. 2010, 391, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Asif, A.; Khalid, M.; Manzoor, S.; Ahmad, H.; Rehman, A.U. Role of purinergic receptors in hepatobiliary carcinoma in Pakistani population: An approach towards proinflammatory role of P2X4 and P2X7 receptors. Purinergic. Signal. 2019, 15, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.C.; Kuo, D.; Sheieh, P.; Chen, F.; Kuo, C.; Jan, C. Effect of the antidepressant paroxetine on Ca2+ movement in PC3 human prostate cancer cells. Drug Dev. Res. 2009, 71, 120–126. [Google Scholar] [CrossRef]
- Ardura, J.A.; Alvarez-Carrion, L.; Gutierrez-Rojas, I.; Alonso, V. Role of Calcium Signaling in Prostate Cancer Progression: Effects on Cancer Hallmarks and Bone Metastatic Mechanisms. Cancers 2020, 12, 1071. [Google Scholar] [CrossRef]
- Ghalali, A.; Ye, Z.W.; Hogberg, J.; Stenius, U. PTEN and PHLPP crosstalk in cancer cells and in TGFbeta-activated stem cells. Biomed. Pharmacother. 2020, 127, 110112. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Layhadi, J.A.; Fountain, S.J. P2X4 Receptor-Dependent Ca2+ Influx in Model Human Monocytes and Macrophages. Int. J. Mol. Sci. 2017, 18, 2261. [Google Scholar] [CrossRef] [Green Version]
- Balazs, B.; Danko, T.; Kovacs, G.; Koles, L.; Hediger, M.A.; Zsembery, A. Investigation of the inhibitory effects of the benzodiazepine derivative, 5-BDBD on P2X4 purinergic receptors by two complementary methods. Cell. Physiol. Biochem. 2013, 32, 11–24. [Google Scholar] [CrossRef]
- Zhang, W.J.; Luo, H.L.; Zhu, Z.M. The role of P2X4 receptors in chronic pain: A potential pharmacological target. Biomed. Pharmacother. 2020, 129, 110447. [Google Scholar] [CrossRef]
- Srivastava, P.; Cronin, C.G.; Scranton, V.L.; Jacobson, K.A.; Liang, B.T.; Verma, R. Neuroprotective and neuro-rehabilitative effects of acute purinergic receptor P2X4 (P2X4R) blockade after ischemic stroke. Exp. Neurol. 2020, 329, 113308. [Google Scholar] [CrossRef]
- Long, T.; He, W.; Pan, Q.; Zhang, S.; Zhang, D.; Qin, G.; Chen, L.; Zhou, J. Microglia P2X4R-BDNF signalling contributes to central sensitization in a recurrent nitroglycerin-induced chronic migraine model. J. Headache Pain 2020, 21, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, T.; He, W.; Pan, Q.; Zhang, S.; Zhang, Y.; Liu, C.; Liu, Q.; Qin, G.; Chen, L.; Zhou, J. Microglia P2X4 receptor contributes to central sensitization following recurrent nitroglycerin stimulation. J. Neuroinflamm. 2018, 15, 245. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varambally, S.; Yu, J.; Laxman, B.; Rhodes, D.R.; Mehra, R.; Tomlins, S.A.; Shah, R.B.; Chandran, U.; Monzon, F.A.; Becich, M.J.; et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 2005, 8, 393–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanni, S.; Priolo, C.; Grasselli, A.; D’Eletto, M.; Merola, R.; Moretti, F.; Gallucci, M.; De Carli, P.; Sentinelli, S.; Cianciulli, A.M.; et al. Epithelial-restricted gene profile of primary cultures from human prostate tumors: A molecular approach to predict clinical behavior of prostate cancer. Mol. Cancer Res. 2006, 4, 79–92. [Google Scholar] [CrossRef] [Green Version]
- Tomlins, S.A.; Mehra, R.; Rhodes, D.R.; Cao, X.; Wang, L.; Dhanasekaran, S.M.; Kalyana-Sundaram, S.; Wei, J.T.; Rubin, M.A.; Pienta, K.J.; et al. Integrative molecular concept modeling of prostate cancer progression. Nat. Genet. 2007, 39, 41–51. [Google Scholar] [CrossRef]
- Dhulipala, P.D.; Wang, Y.X.; Kotlikoff, M.I. The human P2X4 receptor gene is alternatively spliced. Gene 1998, 207, 259–266. [Google Scholar] [CrossRef]
- Carpenter, D.; Meadows, H.J.; Brough, S.; Chapman, G.; Clarke, C.; Coldwell, M.; Davis, R.; Harrison, D.; Meakin, J.; McHale, M.; et al. Site-specific splice variation of the human P2X4 receptor. Neurosci. Lett. 1999, 273, 183–186. [Google Scholar] [CrossRef]
- Chen, D.; Rauh, M.; Buchfelder, M.; Eyupoglu, I.Y.; Savaskan, N. The oxido-metabolic driver ATF4 enhances temozolamide chemo-resistance in human gliomas. Oncotarget 2017, 8, 51164–51176. [Google Scholar] [CrossRef] [Green Version]
- Draganov, D.; Gopalakrishna-Pillai, S.; Chen, Y.R.; Zuckerman, N.; Moeller, S.; Wang, C.; Ann, D.; Lee, P.P. Modulation of P2X4/P2X7/Pannexin-1 sensitivity to extracellular ATP via Ivermectin induces a non-apoptotic and inflammatory form of cancer cell death. Sci. Rep. 2015, 5, 16222. [Google Scholar] [CrossRef] [Green Version]
- Ricote, M.; Garcia-Tunon, I.; Bethencourt, F.; Fraile, B.; Onsurbe, P.; Paniagua, R.; Royuela, M. The p38 transduction pathway in prostatic neoplasia. J. Pathol. 2006, 208, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Koul, S.; Huang, M.; Chaturvedi, L.; Meacham, R.B.; Koul, H.K. p42/p44 Mitogen-activated protein kinase signal transduction pathway regulates interleukin-6 expression in PC3 cells, a line of hormone-refractory prostate cancer cells. Ann. N. Y. Acad. Sci. 2004, 1030, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Koul, H.K.; Pal, M.; Koul, S. Role of p38 MAP Kinase Signal Transduction in Solid Tumors. Genes Cancer 2013, 4, 342–359. [Google Scholar] [CrossRef] [PubMed]
- Skjoth, I.H.; Issinger, O.G. Profiling of signaling molecules in four different human prostate carcinoma cell lines before and after induction of apoptosis. Int. J. Oncol. 2006, 28, 217–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu-Lee, L.Y.; Yu, G.; Lee, Y.C.; Lin, S.C.; Pan, J.; Pan, T.; Yu, K.J.; Liu, B.; Creighton, C.J.; Rodriguez-Canales, J.; et al. Osteoblast-Secreted Factors Mediate Dormancy of Metastatic Prostate Cancer in the Bone via Activation of the TGFbetaRIII-p38MAPK-pS249/T252RB Pathway. Cancer Res. 2018, 78, 2911–2924. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Chen, S.; Bergan, R.C. MAPKAPK2 and HSP27 are downstream effectors of p38 MAP kinase-mediated matrix metalloproteinase type 2 activation and cell invasion in human prostate cancer. Oncogene 2006, 25, 2987–2998. [Google Scholar] [CrossRef] [Green Version]
- Acosta, A.M.; Al Rasheed, M.R.H.; Rauscher, G.H.; Vormittag, E.; Mon, K.S.; Sharif, A.; Kajdacsy-Balla, A.; Mohapatra, G. Tumor necrosis in radical prostatectomies with high-grade prostate cancer is associated with multiple poor prognostic features and a high prevalence of residual disease. Hum. Pathol. 2018, 75, 1–9. [Google Scholar] [CrossRef]
- Humphrey, P.A. Histopathology of Prostate Cancer. Cold Spring Harb. Perspect. Med. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Zhang, J.; Zhang, W.; Zhang, J.; Yang, J.; Li, K.; He, Y. ATP-P2X4 signaling mediates NLRP3 inflammasome activation: A novel pathway of diabetic nephropathy. Int. J. Biochem. Cell Biol. 2013, 45, 932–943. [Google Scholar] [CrossRef]
- Han, S.J.; Lovaszi, M.; Kim, M.; D’Agati, V.; Hasko, G.; Lee, H.T. P2X4 receptor exacerbates ischemic AKI and induces renal proximal tubular NLRP3 inflammasome signaling. FASEB J. 2020, 34, 5465–5482. [Google Scholar] [CrossRef] [Green Version]
- Hoorens, A.; Stange, G.; Pavlovic, D.; Pipeleers, D. Distinction between interleukin-1-induced necrosis and apoptosis of islet cells. Diabetes 2001, 50, 551–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vercammen, E.; Staal, J.; Van Den Broeke, A.; Haegman, M.; Vereecke, L.; Schotte, P.; Beyaert, R. Prolonged exposure to IL-1beta and IFNgamma induces necrosis of L929 tumor cells via a p38MAPK/NF-kappaB/NO-dependent mechanism. Oncogene 2008, 27, 3780–3788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- England, H.; Summersgill, H.R.; Edye, M.E.; Rothwell, N.J.; Brough, D. Release of interleukin-1alpha or interleukin-1beta depends on mechanism of cell death. J. Biol. Chem. 2014, 289, 15942–15950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, K.; Sokabe, T.; Matsumoto, T.; Yoshimura, K.; Shibata, M.; Ohura, N.; Fukuda, T.; Sato, T.; Sekine, K.; Kato, S.; et al. Impaired flow-dependent control of vascular tone and remodeling in P2X4-deficient mice. Nat. Med. 2006, 12, 133–137. [Google Scholar] [CrossRef]
- Sim, J.A.; Chaumont, S.; Jo, J.; Ulmann, L.; Young, M.T.; Cho, K.; Buell, G.; North, R.A.; Rassendren, F. Altered hippocampal synaptic potentiation in P2X4 knock-out mice. J. Neurosci. 2006, 26, 9006–9009. [Google Scholar] [CrossRef]
- Khoja, S.; Huynh, N.; Asatryan, L.; Jakowec, M.W.; Davies, D.L. Reduced expression of purinergic P2X4 receptors increases voluntary ethanol intake in C57BL/6J mice. Alcohol 2018, 68, 63–70. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, J.; Zhou, Y.; Arredondo Carrera, H.M.; Sprules, A.; Neagu, R.; Zarkesh, S.A.; Eaton, C.; Luo, J.; Gartland, A.; Wang, N. Inhibiting the P2X4 Receptor Suppresses Prostate Cancer Growth In Vitro and In Vivo, Suggesting a Potential Clinical Target. Cells 2020, 9, 2511. https://doi.org/10.3390/cells9112511
He J, Zhou Y, Arredondo Carrera HM, Sprules A, Neagu R, Zarkesh SA, Eaton C, Luo J, Gartland A, Wang N. Inhibiting the P2X4 Receptor Suppresses Prostate Cancer Growth In Vitro and In Vivo, Suggesting a Potential Clinical Target. Cells. 2020; 9(11):2511. https://doi.org/10.3390/cells9112511
Chicago/Turabian StyleHe, Jiepei, Yuhan Zhou, Hector M. Arredondo Carrera, Alexandria Sprules, Ramona Neagu, Sayyed Amin Zarkesh, Colby Eaton, Jian Luo, Alison Gartland, and Ning Wang. 2020. "Inhibiting the P2X4 Receptor Suppresses Prostate Cancer Growth In Vitro and In Vivo, Suggesting a Potential Clinical Target" Cells 9, no. 11: 2511. https://doi.org/10.3390/cells9112511