ProNGF/p75NTR Axis Drives Fiber Type Specification by Inducing the Fast-Glycolytic Phenotype in Mouse Skeletal Muscle Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Satellite Cell Isolation, Culturing and Treatments
2.2. Animals
2.3. Total Lysate Preparation and Western Blot Analysis
2.4. Cell Fractionation
2.5. Morphological Analysis
2.6. EdU Staining
2.7. RNA Extraction and Real-Time PCR
2.8. Co-Immunoprecipitation
2.9. Statistical Analysis
3. Results
3.1. Satellite Cell-Derived Myoblasts Express proNGF and p75NTR
3.2. Evaluation of proNGF Effects on SCDM Proliferation and Differentiation
3.3. ProNGF Promotes a Fast Phenotype during SCDM Differentiation
3.4. ProNGF Induces Slow-to-Fast Transition in Myotubes
3.5. ProNGF and p75NTR Levels Are Increased in Dystrophic Muscles, and Are Accompanied by the Reduction in Markers of Slow/Oxidative Phenotype
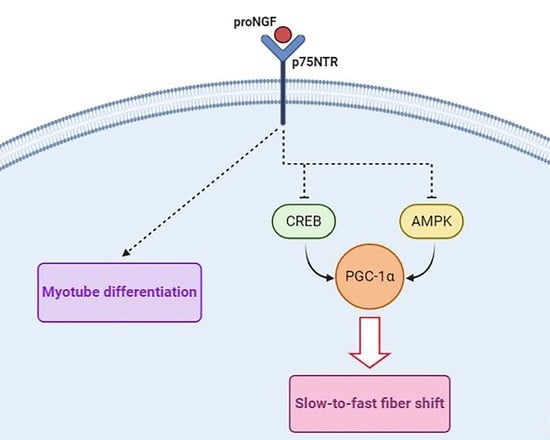
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sakuma, K.; Yamaguchi, A. The recent understanding of the neurotrophin‘s role in skeletal muscle adaptation. J. Biomed. Biotechnol. 2011, 2011, 201696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bibel, M.; Barde, Y.A. Neurotrophins: Key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000, 14, 2919–2937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longo, F.M.; Massa, S.M. Small-molecule modulation of neurotrophin receptors: A strategy for the treatment of neurological disease. Nat. Rev. Drug Discov. 2013, 12, 507–525. [Google Scholar] [CrossRef] [PubMed]
- Segatto, M.; Fico, E.; Gharbiya, M.; Rosso, P.; Carito, V.; Tirassa, P.; Plateroti, R.; Lambiase, A. VEGF inhibition alters neurotrophin signalling pathways and induces caspase-3 activation and autophagy in rabbit retina. J. Cell Physiol. 2019, 234, 18297–18307. [Google Scholar] [CrossRef]
- Chevrel, G.; Hohlfeld, R.; Sendtner, M. The role of neurotrophins in muscle under physiological and pathological conditions. Muscle Nerve 2006, 33, 462–476. [Google Scholar] [CrossRef]
- Klein, R.; Silos-Santiago, I.; Smeyne, R.J.; Lira, S.A.; Brambilla, R.; Bryant, S.; Zhang, L.; Snider, W.D.; Barbacid, M. Disruption of the neurotrophin-3 receptor gene trkC eliminates la muscle afferents and results in abnormal movements. Nature 1994, 368, 249–251. [Google Scholar] [CrossRef]
- Ernfors, P.; Lee, K.F.; Kucera, J.; Jaenisch, R. Lack of neurotrophin-3 leads to deficiencies in the peripheral nervous system and loss of limb proprioceptive afferents. Cell 1994, 77, 503–512. [Google Scholar] [CrossRef]
- Belluardo, N.; Westerblad, H.; Mudó, G.; Casabona, A.; Bruton, J.; Caniglia, G.; Pastoris, O.; Grassi, F.; Ibáñez, C.F. Neuromuscular junction disassembly and muscle fatigue in mice lacking neurotrophin-4. Mol. Cell Neurosci. 2001, 18, 56–67. [Google Scholar] [CrossRef] [Green Version]
- Kulakowski, S.A.; Parker, S.D.; Personius, K.E. Reduced TrkB expression results in precocious age-like changes in neuromuscular structure, neurotransmission, and muscle function. J. Appl. Physiol. (1985) 2011, 111, 844–852. [Google Scholar] [CrossRef] [Green Version]
- Clow, C.; Jasmin, B.J. Brain-derived neurotrophic factor regulates satellite cell differentiation and skeltal muscle regeneration. Mol. Biol. Cell 2010, 21, 2182–2190. [Google Scholar] [CrossRef] [Green Version]
- Colombo, E.; Bedogni, F.; Lorenzetti, I.; Landsberger, N.; Previtali, S.C.; Farina, C. Autocrine and immune cell-derived BDNF in human skeletal muscle: Implications for myogenesis and tissue regeneration. J. Pathol. 2013, 231, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Delezie, J.; Weihrauch, M.; Maier, G.; Tejero, R.; Ham, D.J.; Gill, J.F.; Karrer-Cardel, B.; Rüegg, M.A.; Tabares, L.; Handschin, C. BDNF is a mediator of glycolytic fiber-type specification in mouse skeletal muscle. Proc. Natl. Acad. Sci. USA 2019, 116, 16111–16120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capsoni, S.; Ruberti, F.; Di Daniel, E.; Cattaneo, A. Muscular dystrophy in adult and aged anti-NGF transgenic mice resembles an inclusion body myopathy. J. Neurosci. Res. 2000, 59, 553–560. [Google Scholar] [CrossRef]
- Ruberti, F.; Capsoni, S.; Comparini, A.; Di Daniel, E.; Franzot, J.; Gonfloni, S.; Rossi, G.; Berardi, N.; Cattaneo, A. Phenotypic knockout of nerve growth factor in adult transgenic mice reveals severe deficits in basal forebrain cholinergic neurons, cell death in the spleen, and skeletal muscle dystrophy. J. Neurosci. 2000, 20, 2589–2601. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Sobue, G.; Yamamoto, K.; Terao, S.; Mitsuma, T. Expression of mRNAs for neurotrophic factors (NGF, BDNF, NT-3, and GDNF) and their receptors (p75NGFR, trkA, trkB, and trkC) in the adult human peripheral nervous system and nonneural tissues. Neurochem. Res. 1996, 21, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Deponti, D.; Buono, R.; Catanzaro, G.; De Palma, C.; Longhi, R.; Meneveri, R.; Bresolin, N.; Bassi, M.T.; Cossu, G.; Clementi, E.; et al. The low-affinity receptor for neurotrophins p75NTR plays a key role for satellite cell function in muscle repair acting via RhoA. Mol. Biol. Cell 2009, 20, 3620–3627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Perini, A.; Dimauro, I.; Duranti, G.; Fantini, C.; Mercatelli, N.; Ceci, R.; Di Luigi, L.; Sabatini, S.; Caporossi, D. The p75. BMC Res. Notes 2017, 10, 686. [Google Scholar]
- Colombo, E.; Romaggi, S.; Medico, E.; Menon, R.; Mora, M.; Falcone, C.; Lochmüller, H.; Confalonieri, P.; Mantegazza, R.; Morandi, L.; et al. Human neurotrophin receptor p75NTR defines differentiation-oriented skeletal muscle precursor cells: Implications for muscle regeneration. J. Neuropathol. Exp. Neurol. 2011, 70, 133–142. [Google Scholar] [CrossRef]
- Seidl, K.; Erck, C.; Buchberger, A. Evidence for the participation of nerve growth factor and its low-affinity receptor (p75NTR) in the regulation of the myogenic program. J. Cell Physiol. 1998, 176, 10–21. [Google Scholar] [CrossRef]
- Proserpio, V.; Fittipaldi, R.; Ryall, J.G.; Sartorelli, V.; Caretti, G. The methyltransferase SMYD3 mediates the recruitment of transcriptional cofactors at the myostatin and c-Met genes and regulates skeletal muscle atrophy. Genes Dev. 2013, 27, 1299–1312. [Google Scholar] [CrossRef] [Green Version]
- Trapani, L.; Segatto, M.; La Rosa, P.; Fanelli, F.; Moreno, S.; Marino, M.; Pallottini, V. 3-hydroxy 3-methylglutaryl coenzyme A reductase inhibition impairs muscle regeneration. J. Cell Biochem. 2012, 113, 2057–2063. [Google Scholar] [CrossRef] [PubMed]
- Crescenzo, R.; Mazzoli, A.; Cancelliere, R.; Bucci, A.; Naclerio, G.; Baccigalupi, L.; Cutting, S.M.; Ricca, E.; Iossa, S. Beneficial effects of carotenoid-producing cells of Bacillus indicus HU16 in a rat model of diet-induced metabolic syndrome. Benef. Microbes 2017, 8, 823–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tonini, C.; Colardo, M.; Colella, B.; Di Bartolomeo, S.; Berardinelli, F.; Caretti, G.; Pallottini, V.; Segatto, M. Inhibition of Bromodomain and Extraterminal Domain (BET) Proteins by JQ1 Unravels a Novel Epigenetic Modulation to Control Lipid Homeostasis. Int. J. Mol. Sci. 2020, 21, 1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cartocci, V.; Segatto, M.; Di Tunno, I.; Leone, S.; Pfrieger, F.W.; Pallottini, V. Modulation of the Isoprenoid/Cholesterol Biosynthetic Pathway During Neuronal Differentiation In Vitro. J. Cell Biochem. 2016, 117, 2036–2044. [Google Scholar] [CrossRef]
- Trapani, L.; Segatto, M.; Simeoni, V.; Balducci, V.; Dhawan, A.; Parmar, V.S.; Prasad, A.K.; Saso, L.; Incerpi, S.; Pallottini, V. Short- and long-term regulation of 3-hydroxy 3-methylglutaryl coenzyme A reductase by a 4-methylcoumarin. Biochimie 2011, 93, 1165–1171. [Google Scholar] [CrossRef]
- Pesiri, V.; Totta, P.; Segatto, M.; Bianchi, F.; Pallottini, V.; Marino, M.; Acconcia, F. Estrogen receptor α L429 and A430 regulate 17β-estradiol-induced cell proliferation via CREB1. Cell Signal 2015, 27, 2380–2388. [Google Scholar] [CrossRef]
- Segatto, M.; Fittipaldi, R.; Pin, F.; Sartori, R.; Dae Ko, K.; Zare, H.; Fenizia, C.; Zanchettin, G.; Pierobon, E.S.; Hatakeyama, S.; et al. Epigenetic targeting of bromodomain protein BRD4 counteracts cancer cachexia and prolongs survival. Nat. Commun. 2017, 8, 1707. [Google Scholar] [CrossRef]
- da Rocha, J.T.; Trapani, L.; Segatto, M.; La Rosa, P.; Nogueira, C.W.; Zeni, G.; Pallottini, V. Molecular effects of diphenyl diselenide on cholesterol and glucose cell metabolism. Curr. Med. Chem. 2013, 20, 4426–4434. [Google Scholar] [CrossRef]
- Segatto, M.; Manduca, A.; Lecis, C.; Rosso, P.; Jozwiak, A.; Swiezewska, E.; Moreno, S.; Trezza, V.; Pallottini, V. Simvastatin treatment highlights a new role for the isoprenoid/cholesterol biosynthetic pathway in the modulation of emotional reactivity and cognitive performance in rats. Neuropsychopharmacology 2014, 39, 841–854. [Google Scholar] [CrossRef] [Green Version]
- Skeldal, S.; Matusica, D.; Nykjaer, A.; Coulson, E.J. Proteolytic processing of the p75 neurotrophin receptor: A prerequisite for signalling?: Neuronal life, growth and death signalling are crucially regulated by intra-membrane proteolysis and trafficking of p75(NTR). Bioessays 2011, 33, 614–625. [Google Scholar] [CrossRef]
- Schachtrup, C.; Ryu, J.K.; Mammadzada, K.; Khan, A.S.; Carlton, P.M.; Perez, A.; Christian, F.; Le Moan, N.; Vagena, E.; Baeza-Raja, B.; et al. Nuclear pore complex remodeling by p75(NTR) cleavage controls TGF-β signaling and astrocyte functions. Nat. Neurosci. 2015, 18, 1077–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, J.M.; Player, D.J.; Martin, N.R.W.; Capel, A.J.; Lewis, M.P.; Mudera, V. An Assessment of Myotube Morphology, Matrix Deformation, and Myogenic mRNA Expression in Custom-Built and Commercially Available Engineered Muscle Chamber Configurations. Front. Physiol. 2018, 9, 483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Guo, Y.; Jia, G.; Liu, G.; Zhao, H.; Huang, Z. Arginine promotes skeletal muscle fiber type transformation from fast-twitch to slow-twitch via Sirt1/AMPK pathway. J. Nutr. Biochem. 2018, 61, 155–162. [Google Scholar] [CrossRef]
- Dou, M.; Yao, Y.; Ma, L.; Wang, X.; Shi, X.; Yang, G.; Li, X. The long noncoding RNA MyHC IIA/X-AS contributes to skeletal muscle myogenesis and maintains the fast fiber phenotype. J. Biol. Chem. 2020, 295, 4937–4949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, N.; Narita, T.; Okita, N.; Kobayashi, M.; Furuta, Y.; Chujo, Y.; Sakai, M.; Yamada, A.; Takeda, K.; Konishi, T.; et al. Sterol regulatory element-binding protein-1c orchestrates metabolic remodeling of white adipose tissue by caloric restriction. Aging Cell 2017, 16, 508–517. [Google Scholar] [CrossRef]
- Yang, Y.P.; Lotta, L.; Beutner, G.; Li, X.; Schor, N.F. Induction of Expression of p75 Neurotrophin Receptor Intracellular Domain Does Not Induce Expression or Enhance Activity of Mitochondrial Complex II. Oxid. Med. Cell. Longev. 2016, 2016, 8752821. [Google Scholar] [CrossRef] [Green Version]
- Podlesniy, P.; Kichev, A.; Pedraza, C.; Saurat, J.; Encinas, M.; Perez, B.; Ferrer, I.; Espinet, C. Pro-NGF from Alzheimer’s disease and normal human brain displays distinctive abilities to induce processing and nuclear translocation of intracellular domain of p75NTR and apoptosis. Am. J. Pathol. 2006, 169, 119–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes, N.L.; Banks, G.B.; Tsang, M.; Margineantu, D.; Gu, H.; Djukovic, D.; Chan, J.; Torres, M.; Liggitt, H.D.; Hirenallur, S.D.K.; et al. Fnip1 regulates skeletal muscle fiber type specification, fatigue resistance, and susceptibility to muscular dystrophy. Proc. Natl. Acad. Sci. USA 2015, 112, 424–429. [Google Scholar] [CrossRef] [Green Version]
- Berdeaux, R.; Stewart, R. cAMP signaling in skeletal muscle adaptation: Hypertrophy, metabolism, and regeneration. Am. J. Physiol. Endocrinol. Metab. 2012, 303, E1–E17. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Meeker, R.B.; Massa, S.M.; Longo, F.M. Modulation of the p75 neurotrophin receptor suppresses age-related basal forebrain cholinergic neuron degeneration. Sci. Rep. 2019, 9, 5273. [Google Scholar] [CrossRef] [Green Version]
- Schneider, S.M.; Sridhar, V.; Bettis, A.K.; Heath-Barnett, H.; Balog-Alvarez, C.J.; Guo, L.J.; Johnson, R.; Jaques, S.; Vitha, S.; Glowcwski, A.C.; et al. Glucose Metabolism as a Pre-clinical Biomarker for the Golden Retriever Model of Duchenne Muscular Dystrophy. Mol. Imaging. Biol. 2018, 20, 780–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gayraud-Morel, B.; Chrétien, F.; Tajbakhsh, S. Skeletal muscle as a paradigm for regenerative biology and medicine. Regen Med. 2009, 4, 293–319. [Google Scholar] [CrossRef] [PubMed]
- Capitanio, D.; Moriggi, M.; Torretta, E.; Barbacini, P.; De Palma, S.; Viganò, A.; Lochmüller, H.; Muntoni, F.; Ferlini, A.; Mora, M.; et al. Comparative proteomic analyses of Duchenne muscular dystrophy and Becker muscular dystrophy muscles: Changes contributing to preserve muscle function in Becker muscular dystrophy patients. J. Cachexia Sarcopenia Muscle 2020, 11, 547–563. [Google Scholar] [CrossRef] [Green Version]
- Ljubicic, V.; Burt, M.; Lunde, J.A.; Jasmin, B.J. Resveratrol induces expression of the slow, oxidative phenotype in mdx mouse muscle together with enhanced activity of the SIRT1-PGC-1α axis. Am. J. Physiol. Cell Physiol. 2014, 307, C66–C82. [Google Scholar] [CrossRef] [Green Version]
- Blanchet, E.; Annicotte, J.S.; Pradelli, L.A.; Hugo, G.; Matecki, S.; Mornet, D.; Rivier, F.; Fajas, L. E2F transcription factor-1 deficiency reduces pathophysiology in the mouse model of Duchenne muscular dystrophy through increased muscle oxidative metabolism. Hum. Mol. Genet 2012, 21, 3910–3917. [Google Scholar] [CrossRef]
- Nitahara-Kasahara, Y.; Takeda, S.; Okada, T. Inflammatory predisposition predicts disease phenotypes in muscular dystrophy. Inflamm. Regen 2016, 36, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deponti, D.; François, S.; Baesso, S.; Sciorati, C.; Innocenzi, A.; Broccoli, V.; Muscatelli, F.; Meneveri, R.; Clementi, E.; Cossu, G.; et al. Necdin mediates skeletal muscle regeneration by promoting myoblast survival and differentiation. J. Cell Biol. 2007, 179, 305–319. [Google Scholar] [CrossRef] [Green Version]
- Khairallah, R.J.; Shi, G.; Sbrana, F.; Prosser, B.L.; Borroto, C.; Mazaitis, M.J.; Hoffman, E.P.; Mahurkar, A.; Sachs, F.; Sun, Y.; et al. Microtubules underlie dysfunction in duchenne muscular dystrophy. Sci. Signal 2012, 5, 56. [Google Scholar] [CrossRef] [Green Version]
- Lomen-Hoerth, C.; Shooter, E.M. Widespread neurotrophin receptor expression in the immune system and other nonneuronal rat tissues. J. Neurochem. 1995, 64, 1780–1789. [Google Scholar] [CrossRef]
- Halievski, K.; Xu, Y.; Haddad, Y.W.; Tang, Y.P.; Yamad, S.; Katsuno, M.; Adachi, H.; Sobue, G.; Breedlove, S.M.; Jordan, C.L. Muscle BDNF improves synaptic and contractile muscle strength in Kennedy’s disease mice in a muscle-type specific manner. J. Physiol. 2020, 598, 2719–2739. [Google Scholar] [CrossRef]
- Yang, X.; Brobst, D.; Chan, W.S.; Tse, M.C.L.; Herlea-Pana, O.; Ahuja, P.; Bi, X.; Zaw, A.M.; Kwong, Z.S.W.; Jia, W.H.; et al. Muscle-generated BDNF is a sexually dimorphic myokine that controls metabolic flexibility. Sci. Signal 2019, 12, 1468. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G. Energy sensing by the AMP-activated protein kinase and its effects on muscle metabolism. Proc. Nutr. Soc. 2011, 70, 92–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baeza-Raja, B.; Sachs, B.D.; Li, P.; Christian, F.; Vagena, E.; Davalos, D.; Le Moan, N.; Ryu, J.K.; Sikorski, S.L.; Chan, J.P.; et al. p75 Neurotrophin Receptor Regulates Energy Balance in Obesity. Cell Rep. 2016, 14, 255–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shanab, A.Y.; Mysona, B.A.; Matragoon, S.; El-Remessy, A.B. Silencing p75(NTR) prevents proNGF-induced endothelial cell death and development of acellular capillaries in rat retina. Mol. Ther. Methods Clin. Dev. 2015, 2, 15013. [Google Scholar] [CrossRef]
- Parkhurst, C.N.; Zampieri, N.; Chao, M.V. Nuclear localization of the p75 neurotrophin receptor intracellular domain. J. Biol. Chem. 2010, 285, 5361–5368. [Google Scholar] [CrossRef] [Green Version]
- Sciorati, C.; Touvier, T.; Buono, R.; Pessina, P.; François, S.; Perrotta, C.; Meneveri, R.; Clementi, E.; Brunelli, S. Necdin is expressed in cachectic skeletal muscle to protect fibers from tumor-induced wasting. J. Cell Sci. 2009, 122, 1119–1125. [Google Scholar] [CrossRef] [Green Version]
- Pessina, P.; Conti, V.; Pacelli, F.; Rosa, F.; Doglietto, G.B.; Brunelli, S.; Bossola, M. Skeletal muscle of gastric cancer patients expresses genes involved in muscle regeneration. Oncol. Rep. 2010, 24, 741–745. [Google Scholar]
- Pessina, P.; Conti, V.; Tonlorenzi, R.; Touvier, T.; Meneveri, R.; Cossu, G.; Brunelli, S. Necdin enhances muscle reconstitution of dystrophic muscle by vessel-associated progenitors, by promoting cell survival and myogenic differentiation. Cell Death Differ. 2012, 19, 827–838. [Google Scholar] [CrossRef] [Green Version]
- Webster, C.; Silberstein, L.; Hays, A.P.; Blau, H.M. Fast muscle fibers are preferentially affected in Duchenne muscular dystrophy. Cell 1988, 52, 503–513. [Google Scholar] [CrossRef]









© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pallottini, V.; Colardo, M.; Tonini, C.; Martella, N.; Strimpakos, G.; Colella, B.; Tirassa, P.; Bartolomeo, S.D.; Segatto, M. ProNGF/p75NTR Axis Drives Fiber Type Specification by Inducing the Fast-Glycolytic Phenotype in Mouse Skeletal Muscle Cells. Cells 2020, 9, 2232. https://doi.org/10.3390/cells9102232
Pallottini V, Colardo M, Tonini C, Martella N, Strimpakos G, Colella B, Tirassa P, Bartolomeo SD, Segatto M. ProNGF/p75NTR Axis Drives Fiber Type Specification by Inducing the Fast-Glycolytic Phenotype in Mouse Skeletal Muscle Cells. Cells. 2020; 9(10):2232. https://doi.org/10.3390/cells9102232
Chicago/Turabian StylePallottini, Valentina, Mayra Colardo, Claudia Tonini, Noemi Martella, Georgios Strimpakos, Barbara Colella, Paola Tirassa, Sabrina Di Bartolomeo, and Marco Segatto. 2020. "ProNGF/p75NTR Axis Drives Fiber Type Specification by Inducing the Fast-Glycolytic Phenotype in Mouse Skeletal Muscle Cells" Cells 9, no. 10: 2232. https://doi.org/10.3390/cells9102232