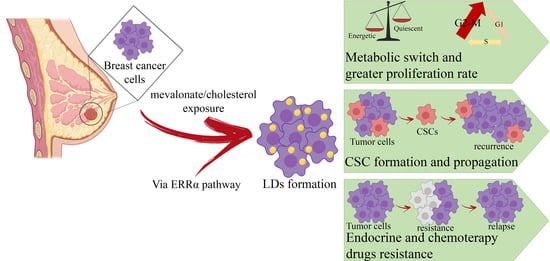
Cholesterol and Mevalonate: Two Metabolites Involved in Breast Cancer Progression and Drug Resistance through the ERRα Pathway
Abstract
:1. Introduction
2. Material and Methods
2.1. Cell Cultures
2.2. Immunoblot Analysis
2.3. Cell Cycle Analysis
2.4. Wound-Healing Assay
2.5. Seahorse XFe96 Metabolic Profile Analysis
2.6. Mitochondrial Membrane Potential and Mass Analysis
2.7. Mammosphere Formation Assay
2.8. Immunofluorescence Assay
2.9. Sulforhodamine B (SRB) Assay
2.10. Kaplan–Mayer Analysis
2.11. Statistical Analysis
3. Results
3.1. Mevalonate and Cholesterol Activate the ERRα Pathway, Increasing the Levels of Related Proteins
3.2. Cholesterol and Its Biosynthetic Precursor, Mevalonate, Lead Cell Cycle Progression
3.3. Mevalonate and Cholesterol Improve the Motility of Breast Cancer Cell Lines
3.4. Increase of Mitochondrial Activity and Energetic Metabolism in Mevalonate- and Cholesterol-Treated Breast Cancer Cells
3.5. Mevalonate and Cholesterol Increase Breast Cancer Stem Cell Propagation
3.6. Increase of Lipid Droplet Formation in Mevalonate- and Cholesterol-Treated Breast Cancer Cells
3.7. Mevalonate and Cholesterol Induce Chemotherapy Drug Resistance
3.8. High Levels of ERRα are Associated with a Poor Prognosis in Breast Cancer Patients
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musgrove, E.A.; Sutherland, R.L. Biological determinants of endocrine resistance in breast cancer. Nat. Rev. Cancer 2009, 9, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Goldhirsch, A.; Gelber, R.D.; Simes, R.J.; Glasziou, P.; Coates, A.S. Costs and benefits of adjuvant therapy in breast cancer: A quality-adjusted survival analysis. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 1989, 7, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y. Adipocyte and lipid metabolism in cancer drug resistance. J. Clin. Investig. 2019, 129, 3006–3017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz, A.L.S.; Barreto, E.A.; Fazolini, N.P.B.; Viola, J.P.B.; Bozza, P.T. Lipid droplets: Platforms with multiple functions in cancer hallmarks. Cell Death Dis. 2020, 11, 105. [Google Scholar] [CrossRef] [Green Version]
- Buhaescu, I.; Izzedine, H. Mevalonate pathway: A review of clinical and therapeutical implications. Clin. Biochem. 2007, 40, 575–584. [Google Scholar] [CrossRef]
- Clendening, J.W.; Pandyra, A.; Boutros, P.C.; El Ghamrasni, S.; Khosravi, F.; Trentin, G.A.; Martirosyan, A.; Hakem, A.; Hakem, R.; Jurisica, I.; et al. Dysregulation of the mevalonate pathway promotes transformation. Proc. Natl. Acad. Sci. USA 2010, 107, 15051–15056. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, J.L.; Brown, M.S. Regulation of the mevalonate pathway. Nature 1990, 343, 425–430. [Google Scholar] [CrossRef]
- Bjarnadottir, O.; Romero, Q.; Bendahl, P.O.; Jirstrom, K.; Ryden, L.; Loman, N.; Uhlen, M.; Johannesson, H.; Rose, C.; Grabau, D.; et al. Targeting HMG-CoA reductase with statins in a window-of-opportunity breast cancer trial. Breast Cancer Res. Treat. 2013, 138, 499–508. [Google Scholar] [CrossRef]
- Schointuch, M.N.; Gilliam, T.P.; Stine, J.E.; Han, X.; Zhou, C.; Gehrig, P.A.; Kim, K.; Bae-Jump, V.L. Simvastatin, an HMG-CoA reductase inhibitor, exhibits anti-metastatic and anti-tumorigenic effects in endometrial cancer. Gynecol. Oncol. 2014, 134, 346–355. [Google Scholar] [CrossRef] [Green Version]
- Hubner, C.; Hoffmann, G.F.; Charpentier, C.; Gibson, K.M.; Finckh, B.; Puhl, H.; Lehr, H.A.; Kohlschutter, A. Decreased plasma ubiquinone-10 concentration in patients with mevalonate kinase deficiency. Pediatr. Res. 1993, 34, 129–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maltese, W.A.; Aprille, J.R. Relation of mevalonate synthesis to mitochondrial ubiquinone content and respiratory function in cultured neuroblastoma cells. J. Biol. Chem. 1985, 260, 11524–11529. [Google Scholar] [PubMed]
- Mullen, P.J.; Yu, R.; Longo, J.; Archer, M.C.; Penn, L.Z. The interplay between cell signalling and the mevalonate pathway in cancer. Nat. Rev. Cancer 2016, 16, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Swanson, K.M.; Hohl, R.J. Anti-cancer therapy: Targeting the mevalonate pathway. Curr. Cancer Drug Targets 2006, 6, 15–37. [Google Scholar] [CrossRef]
- Gobel, A.; Breining, D.; Rauner, M.; Hofbauer, L.C.; Rachner, T.D. Induction of 3-hydroxy-3-methylglutaryl-CoA reductase mediates statin resistance in breast cancer cells. Cell Death Dis. 2019, 10, 91. [Google Scholar] [CrossRef]
- Gazzerro, P.; Proto, M.C.; Gangemi, G.; Malfitano, A.M.; Ciaglia, E.; Pisanti, S.; Santoro, A.; Laezza, C.; Bifulco, M. Pharmacological actions of statins: A critical appraisal in the management of cancer. Pharm. Rev. 2012, 64, 102–146. [Google Scholar] [CrossRef]
- Ahern, T.P.; Lash, T.L.; Damkier, P.; Christiansen, P.M.; Cronin-Fenton, D.P. Statins and breast cancer prognosis: Evidence and opportunities. Lancet Oncol. 2014, 15, e461–e468. [Google Scholar] [CrossRef] [Green Version]
- Osmak, M. Statins and cancer: Current and future prospects. Cancer Lett. 2012, 324, 1–12. [Google Scholar] [CrossRef]
- Garwood, E.R.; Kumar, A.S.; Baehner, F.L.; Moore, D.H.; Au, A.; Hylton, N.; Flowers, C.I.; Garber, J.; Lesnikoski, B.A.; Hwang, E.S.; et al. Fluvastatin reduces proliferation and increases apoptosis in women with high grade breast cancer. Breast Cancer Res. Treat. 2010, 119, 137–144. [Google Scholar] [CrossRef] [Green Version]
- Llaverias, G.; Danilo, C.; Mercier, I.; Daumer, K.; Capozza, F.; Williams, T.M.; Sotgia, F.; Lisanti, M.P.; Frank, P.G. Role of cholesterol in the development and progression of breast cancer. Am. J. Pathol. 2011, 178, 402–412. [Google Scholar] [CrossRef]
- Poirot, M.; Silvente-Poirot, S.; Weichselbaum, R.R. Cholesterol metabolism and resistance to tamoxifen. Curr. Opin. Pharm. 2012, 12, 683–689. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Schwaid, A.G.; Wang, X.; Wang, X.; Chen, S.; Chu, Q.; Saghatelian, A.; Wan, Y. Ligand Activation of ERRalpha by Cholesterol Mediates Statin and Bisphosphonate Effects. Cell Metab. 2016, 23, 479–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casaburi, I.; Avena, P.; De Luca, A.; Chimento, A.; Sirianni, R.; Malivindi, R.; Rago, V.; Fiorillo, M.; Domanico, F.; Campana, C.; et al. Estrogen related receptor alpha (ERRalpha) a promising target for the therapy of adrenocortical carcinoma (ACC). Oncotarget 2015, 6, 25135–25148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Marcos, P.J.; Auwerx, J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr. 2011, 93, 884S–890S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schreiber, S.N.; Emter, R.; Hock, M.B.; Knutti, D.; Cardenas, J.; Podvinec, M.; Oakeley, E.J.; Kralli, A. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. USA 2004, 101, 6472–6477. [Google Scholar] [CrossRef] [Green Version]
- Yun, C.W.; Han, Y.S.; Lee, S.H. PGC-1alpha Controls Mitochondrial Biogenesis in Drug-Resistant Colorectal Cancer Cells by Regulating Endoplasmic Reticulum Stress. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Fiorillo, M.; Peiris-Pages, M.; Sanchez-Alvarez, R.; Bartella, L.; Di Donna, L.; Dolce, V.; Sindona, G.; Sotgia, F.; Cappello, A.R.; Lisanti, M.P. Bergamot natural products eradicate cancer stem cells (CSCs) by targeting mevalonate, Rho-GDI-signalling and mitochondrial metabolism. Biochim. Et Biophys. Acta Bioenerg. 2018, 1859, 984–996. [Google Scholar] [CrossRef]
- Shin, M.K.; Cheong, J.H. Mitochondria-centric bioenergetic characteristics in cancer stem-like cells. Arch. Pharmacal Res. 2019, 42, 113–127. [Google Scholar] [CrossRef] [Green Version]
- Ginestier, C.; Monville, F.; Wicinski, J.; Cabaud, O.; Cervera, N.; Josselin, E.; Finetti, P.; Guille, A.; Larderet, G.; Viens, P.; et al. Mevalonate metabolism regulates Basal breast cancer stem cells and is a potential therapeutic target. Stem Cells 2012, 30, 1327–1337. [Google Scholar] [CrossRef]
- Frattaruolo, L.; Fiorillo, M.; Brindisi, M.; Curcio, R.; Dolce, V.; Lacret, R.; Truman, A.W.; Sotgia, F.; Lisanti, M.P.; Cappello, A.R. Thioalbamide, A Thioamidated Peptide from Amycolatopsis alba, Affects Tumor Growth and Stemness by Inducing Metabolic Dysfunction and Oxidative Stress. Cells 2019, 8, 1408. [Google Scholar] [CrossRef] [Green Version]
- Frattaruolo, L.; Lacret, R.; Cappello, A.R.; Truman, A.W. A Genomics-Based Approach Identifies a Thioviridamide-Like Compound with Selective Anticancer Activity. ACS Chem. Biol. 2017, 12, 2815–2822. [Google Scholar] [CrossRef] [PubMed]
- Bonesi, M.; Brindisi, M.; Armentano, B.; Curcio, R.; Sicari, V.; Loizzo, M.R.; Cappello, M.S.; Bedini, G.; Peruzzi, L.; Tundis, R. Exploring the anti-proliferative, pro-apoptotic, and antioxidant properties of Santolina corsica Jord. & Fourr. (Asteraceae). Biomed. Pharmacother. 2018, 107, 967–978. [Google Scholar] [CrossRef]
- Mazzotta, S.; Frattaruolo, L.; Brindisi, M.; Ulivieri, C.; Vanni, F.; Brizzi, A.; Carullo, G.; Cappello, A.R.; Aiello, F. 3-Amino-alkylated indoles: Unexplored green products acting as anti-inflammatory agents. Future Med. Chem. 2019. [Google Scholar] [CrossRef] [PubMed]
- Frattaruolo, L.; Carullo, G.; Brindisi, M.; Mazzotta, S.; Bellissimo, L.; Rago, V.; Curcio, R.; Dolce, V.; Aiello, F.; Cappello, A.R. Antioxidant and Anti-Inflammatory Activities of Flavanones from Glycyrrhiza glabra L. (licorice) Leaf Phytocomplexes: Identification of Licoflavanone as a Modulator of NF-kB/MAPK Pathway. Antioxidants 2019, 8, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiorillo, M.; Sotgia, F.; Lisanti, M.P. “Energetic” Cancer Stem Cells (e-CSCs): A New Hyper-Metabolic and Proliferative Tumor Cell Phenotype, Driven by Mitochondrial Energy. Front. Oncol. 2018, 8, 677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armentano, B.C.; Curcio, R.; Brindisi, M.; Mancuso, R.; Rago, V.; Ziccarelli, I.; Frattaruolo, L.; Fiorillo, M.; Dolce, V.; Gabriele, B.; et al. 5-(Carbamoylmethylene)-oxazolidin-2-ones as a Promising Class of Heterocycles Inducing Apoptosis Triggered by Increased ROS Levels and Mitochondrial Dysfunction in Breast and Cervical Cancer. Biomedicines 2020, 8, 35. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.C.; Park, A.Y.; Guan, J.L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef] [Green Version]
- Fiorillo, M.; Lamb, R.; Tanowitz, H.B.; Mutti, L.; Krstic-Demonacos, M.; Cappello, A.R.; Martinez-Outschoorn, U.E.; Sotgia, F.; Lisanti, M.P. Repurposing atovaquone: Targeting mitochondrial complex III and OXPHOS to eradicate cancer stem cells. Oncotarget 2016, 7, 34084–34099. [Google Scholar] [CrossRef] [Green Version]
- Lamb, R.; Bonuccelli, G.; Ozsvari, B.; Peiris-Pages, M.; Fiorillo, M.; Smith, D.L.; Bevilacqua, G.; Mazzanti, C.M.; McDonnell, L.A.; Naccarato, A.G.; et al. Mitochondrial mass, a new metabolic biomarker for stem-like cancer cells: Understanding WNT/FGF-driven anabolic signaling. Oncotarget 2015, 6, 30453–30471. [Google Scholar] [CrossRef] [Green Version]
- Fiorillo, M.; Lamb, R.; Tanowitz, H.B.; Cappello, A.R.; Martinez-Outschoorn, U.E.; Sotgia, F.; Lisanti, M.P. Bedaquiline, an FDA-approved antibiotic, inhibits mitochondrial function and potently blocks the proliferative expansion of stem-like cancer cells (CSCs). Aging 2016, 8, 1593–1607. [Google Scholar] [CrossRef] [Green Version]
- Ozsvari, B.; Fiorillo, M.; Bonuccelli, G.; Cappello, A.R.; Frattaruolo, L.; Sotgia, F.; Trowbridge, R.; Foster, R.; Lisanti, M.P. Mitoriboscins: Mitochondrial-based therapeutics targeting cancer stem cells (CSCs), bacteria and pathogenic yeast. Oncotarget 2017, 8, 67457–67472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2009, 123, 725–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perri, F.; Frattaruolo, L.; Haworth, I.; Brindisi, M.; El-magboub, A.; Ferrario, A.; Gomer, C.; Aiello, F.; Adams, J.D. Naturally occurring sesquiterpene lactones and their semi-synthetic derivatives modulate PGE2 levels by decreasing COX2 activity and expression. Heliyon 2019, 5, e01366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deblois, G.; St-Pierre, J.; Giguere, V. The PGC-1/ERR signaling axis in cancer. Oncogene 2013, 32, 3483–3490. [Google Scholar] [CrossRef] [Green Version]
- Harari, D.; Yarden, Y. Molecular mechanisms underlying ErbB2/HER2 action in breast cancer. Oncogene 2000, 19, 6102–6114. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.Y.; Kazmin, D.; Jasper, J.S.; Kunder, R.; Zuercher, W.J.; McDonnell, D.P. The metabolic regulator ERRalpha, a downstream target of HER2/IGF-1R, as a therapeutic target in breast cancer. Cancer Cell 2011, 20, 500–510. [Google Scholar] [CrossRef] [Green Version]
- Kar, R.; Jha, N.K.; Jha, S.K.; Sharma, A.; Dholpuria, S.; Asthana, N.; Chaurasiya, K.; Singh, V.K.; Burgee, S.; Nand, P. A “NOTCH” Deeper into the Epithelial-To-Mesenchymal Transition (EMT) Program in Breast Cancer. Genes (Basel) 2019, 10, 961. [Google Scholar] [CrossRef] [Green Version]
- Robinson, B.D.; Jones, J.G. Tumor microenvironment of metastasis (TMEM): A novel tissue-based assay for metastatic risk in breast cancer. Future Oncol. 2009, 5, 919–921. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, C.A.; Ross, A.H. Cancer stem cells: Cell culture, markers, and targets for new therapies. J. Cell. Biochem. 2009, 108, 1031–1038. [Google Scholar] [CrossRef] [Green Version]
- Martin, S.; Parton, R.G. Lipid droplets: A unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 2006, 7, 373–378. [Google Scholar] [CrossRef]
- Kamili, A.; Roslan, N.; Frost, S.; Cantrill, L.C.; Wang, D.; Della-Franca, A.; Bright, R.K.; Groblewski, G.E.; Straub, B.K.; Hoy, A.J.; et al. TPD52 expression increases neutral lipid storage within cultured cells. J. Cell Sci. 2015, 128, 3223–3238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, U.J.; Lee, J.H.; Shim, J.; Yoon, K.; Goh, S.H.; Yi, E.H.; Ye, S.K.; Lee, J.S.; Lee, H.; Park, J.; et al. Anti-cancer effect of doxorubicin is mediated by downregulation of HMG-Co A reductase via inhibition of EGFR/Src pathway. Lab. Investig. 2019, 99, 1157–1172. [Google Scholar] [CrossRef] [PubMed]
- Soerjomataram, I.; Louwman, M.W.; Ribot, J.G.; Roukema, J.A.; Coebergh, J.W. An overview of prognostic factors for long-term survivors of breast cancer. Breast Cancer Res. Treat. 2008, 107, 309–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, E.R.; Chang, C.Y.; McDonnell, D.P. Cholesterol and breast cancer pathophysiology. Trends Endocrinol. Metab. TEM 2014, 25, 649–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yambire, K.F.; Fernandez-Mosquera, L.; Steinfeld, R.; Muhle, C.; Ikonen, E.; Milosevic, I.; Raimundo, N. Mitochondrial biogenesis is transcriptionally repressed in lysosomal lipid storage diseases. eLife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Roslan, N.; Bieche, I.; Bright, R.K.; Lidereau, R.; Chen, Y.; Byrne, J.A. TPD52 represents a survival factor in ERBB2-amplified breast cancer cells. Mol. Carcinog. 2014, 53, 807–819. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Phan, L.M.; Yeung, S.C.; Lee, M.H. Cancer metabolic reprogramming: Importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol. Med. 2014, 11, 1–19. [Google Scholar] [CrossRef]
- Seyfried, T.N.; Shelton, L.M. Cancer as a metabolic disease. Nutr. Metab. 2010, 7, 7. [Google Scholar] [CrossRef] [Green Version]
- Pelicano, H.; Martin, D.S.; Xu, R.H.; Huang, P. Glycolysis inhibition for anticancer treatment. Oncogene 2006, 25, 4633–4646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chae, Y.C.; Kim, J.H. Cancer stem cell metabolism: Target for cancer therapy. BMB Rep. 2018, 51, 319–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snyder, V.; Reed-Newman, T.C.; Arnold, L.; Thomas, S.M.; Anant, S. Cancer Stem Cell Metabolism and Potential Therapeutic Targets. Front. Oncol. 2018, 8, 203. [Google Scholar] [CrossRef] [PubMed]
- Vlashi, E.; Pajonk, F. Cancer stem cells, cancer cell plasticity and radiation therapy. Semin. Cancer Biol. 2015, 31, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Bozza, P.T.; Viola, J.P. Lipid droplets in inflammation and cancer. Prostaglandins Leukot. Essent. Fat. Acids 2010, 82, 243–250. [Google Scholar] [CrossRef]
- Santos, C.R.; Schulze, A. Lipid metabolism in cancer. FEBS J. 2012, 279, 2610–2623. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, Q.; Halim, A.; Song, G. Targeting lipid metabolism of cancer cells: A promising therapeutic strategy for cancer. Cancer Lett. 2017, 401, 39–45. [Google Scholar] [CrossRef]
- Chen, Y.; Frost, S.; Khushi, M.; Cantrill, L.C.; Yu, H.; Arthur, J.W.; Bright, R.K.; Groblewski, G.E.; Byrne, J.A. Delayed recruiting of TPD52 to lipid droplets—Evidence for a “second wave” of lipid droplet-associated proteins that respond to altered lipid storage induced by Brefeldin A treatment. Sci. Rep. 2019, 9, 9790. [Google Scholar] [CrossRef]
- Jarzabek, K.; Koda, M.; Kozlowski, L.; Sulkowski, S.; Kottler, M.L.; Wolczynski, S. The significance of the expression of ERRalpha as a potential biomarker in breast cancer. J. Steroid Biochem. Mol. Biol. 2009, 113, 127–133. [Google Scholar] [CrossRef]
- Suzuki, T.; Miki, Y.; Moriya, T.; Shimada, N.; Ishida, T.; Hirakawa, H.; Ohuchi, N.; Sasano, H. Estrogen-related receptor alpha in human breast carcinoma as a potent prognostic factor. Cancer Res. 2004, 64, 4670–4676. [Google Scholar] [CrossRef] [Green Version]















| Symbol | Gene Probe | HR (Hazard Ratio) | p-Value (Log Rank Test) |
|---|---|---|---|
| All Breast Cancer Subtypes n = 3571 | |||
| ERRα/ESRRA | 203193_at | 0.87 | 0.023 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brindisi, M.; Fiorillo, M.; Frattaruolo, L.; Sotgia, F.; Lisanti, M.P.; Cappello, A.R. Cholesterol and Mevalonate: Two Metabolites Involved in Breast Cancer Progression and Drug Resistance through the ERRα Pathway. Cells 2020, 9, 1819. https://doi.org/10.3390/cells9081819
Brindisi M, Fiorillo M, Frattaruolo L, Sotgia F, Lisanti MP, Cappello AR. Cholesterol and Mevalonate: Two Metabolites Involved in Breast Cancer Progression and Drug Resistance through the ERRα Pathway. Cells. 2020; 9(8):1819. https://doi.org/10.3390/cells9081819
Chicago/Turabian StyleBrindisi, Matteo, Marco Fiorillo, Luca Frattaruolo, Federica Sotgia, Michael P. Lisanti, and Anna Rita Cappello. 2020. "Cholesterol and Mevalonate: Two Metabolites Involved in Breast Cancer Progression and Drug Resistance through the ERRα Pathway" Cells 9, no. 8: 1819. https://doi.org/10.3390/cells9081819