Mixing and Matching Chromosomes during Female Meiosis
Abstract
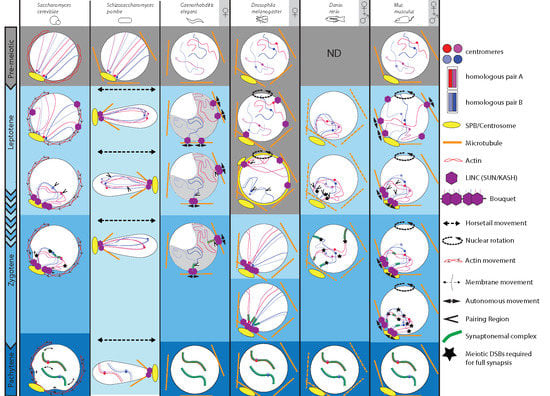
:1. Introduction
1.1. Caenorhabditis Elegans
1.2. Drosophila Melanogaster
1.3. Danio Rerio
1.4. Mus Musculus
2. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Richardson, B.E.; Lehmann, R. Mechanisms guiding primordial germ cell migration: Strategies from different organisms. Nat. Rev. Mol. Cell Boil. 2010, 11, 37–49. [Google Scholar] [CrossRef] [Green Version]
- Zickler, D.; Kleckner, N. Recombination, Pairing, and Synapsis of Homologs during Meiosis. Cold Spring Harb. Perspect. Boil. 2015, 7, a016626. [Google Scholar] [CrossRef] [Green Version]
- Zickler, D.; Kleckner, N. A few of our favorite things: Pairing, the bouquet, crossover interference and evolution of meiosis. Semin. Cell Dev. Boil. 2016, 54, 135–148. [Google Scholar] [CrossRef] [Green Version]
- Tsubouchi, T.; Roeder, G.S. A Synaptonemal Complex Protein Promotes Homology-Independent Centromere Coupling. Science 2005, 308, 870–873. [Google Scholar] [CrossRef] [PubMed]
- Obeso, D.; Dawson, D. Temporal Characterization of Homology-Independent Centromere Coupling in Meiotic Prophase. PLoS ONE 2010, 5, e10336. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.-Q.; Yamamoto, A.; Haraguchi, T.; Hiraoka, Y. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell 2004, 6, 329–341. [Google Scholar] [CrossRef] [Green Version]
- Scherthan, H.; Bahler, J.; Kohli, J. Dynamics of chromosome organization and pairing during meiotic prophase in fission yeast. J. Cell Boil. 1994, 127, 273–285. [Google Scholar] [CrossRef]
- Sharif, W.D.; Glick, G.G.; Davidson, M.K.; Wahls, W.P. Distinct functions of S. pombe Rec12 (Spo11) protein and Rec12-dependent crossover recombination (chiasmata) in meiosis I; and a requirement for Rec12 in meiosis II. Cell Chromosom. 2002, 1, 1. [Google Scholar] [CrossRef]
- Davis, L.; Smith, G.R. Nonrandom homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics 2003, 163, 857–874. [Google Scholar]
- Wells, J.L.; Pryce, D.; McFarlane, R. Homologous chromosome pairing inSchizosaccharomyces pombe. Yeast 2006, 23, 977–989. [Google Scholar] [CrossRef] [Green Version]
- Phillips, C.; Meng, X.; Zhang, L.; Chretien, J.H.; Urnov, F.D.; Dernburg, A.F. Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nat. Cell Biol. 2009, 11, 934–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacQueen, A.J.; Phillips, C.; Bhalla, N.; Weiser, P.; Villeneuve, A.M.; Dernburg, A.F. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 2005, 123, 1037–1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgley, M.L.; Baillie, D.L.; Riddle, D.L.; Rose, A.M. Genetic balancers. Methods Cell Biol. 1995, 48, 147–184. [Google Scholar] [PubMed]
- Rog, O.; Dernburg, A.F. Chromosome pairing and synapsis during Caenorhabditis elegans meiosis. Curr. Opin. Cell Boil. 2013, 25, 349–356. [Google Scholar] [CrossRef] [Green Version]
- Christophorou, N.; Rubin, T.; Huynh, J.-R. Synaptonemal Complex Components Promote Centromere Pairing in Pre-meiotic Germ Cells. PLoS Genet. 2013, 9, e1004012. [Google Scholar] [CrossRef] [Green Version]
- Joyce, E.F.; Apostolopoulos, N.; Beliveau, B.J.; Wu, C.-T. Germline Progenitors Escape the Widespread Phenomenon of Homolog Pairing during Drosophila Development. PLoS Genet. 2013, 9, e1004013. [Google Scholar] [CrossRef] [Green Version]
- Blokhina, Y.P.; Nguyen, A.; Draper, B.W.; Burgess, S. The telomere bouquet is a hub where meiotic double-strand breaks, synapsis, and stable homolog juxtaposition are coordinated in the zebrafish, Danio rerio. PLoS Genet. 2019, 15, e1007730. [Google Scholar] [CrossRef] [Green Version]
- Scherthan, H.; Weich, S.; Schwegler, H.; Heyting, C.; Härle, M.; Cremer, T. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J. Cell Boil. 1996, 134, 1109–1125. [Google Scholar] [CrossRef] [Green Version]
- Tankimanova, M.; Tease, C.; Hultén, M. The initiation of homologous chromosome synapsis in mouse fetal oocytes is not directly driven by centromere and telomere clustering in the bouquet. Cytogenet. Genome Res. 2004, 105, 172–181. [Google Scholar] [CrossRef]
- Chikashige, Y.; Ding, D.-Q.; Funabiki, H.; Haraguchi, T.; Mashiko, S.; Yanagida, M.; Hiraoka, Y. Telomere-led premeiotic chromosome movement in fission yeast. Science 1994, 264, 270–273. [Google Scholar] [CrossRef]
- Crittenden, S.L.; Leonhard, K.A.; Byrd, D.T.; Kimble, J. Cellular Analyses of the Mitotic Region in the Caenorhabditis elegans Adult Germ Line. Mol. Boil. Cell 2006, 17, 3051–3061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boateng, K.A.; Bellani, M.A.; Gregoretti, I.V.; Pratto, F.; Camerini-Otero, R.D. Homologous pairing preceding SPO11-mediated double-strand breaks in mice. Dev. Cell 2013, 24, 196–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishiguro, K.-I.; Kim, J.; Shibuya, H.; Hernández-Hernández, A.; Suzuki, A.; Fukagawa, T.; Shioi, G.; Kiyonari, H.; Li, X.C.; Schimenti, J.; et al. Meiosis-specific cohesin mediates homolog recognition in mouse spermatocytes. Genes Dev. 2014, 28, 594–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherthan, H.; Schöfisch, K.; Dell, T.; Illner, D. Contrasting behavior of heterochromatic and euchromatic chromosome portions and pericentric genome separation in pre-bouquet spermatocytes of hybrid mice. Chromosoma 2014, 123, 609–624. [Google Scholar] [CrossRef] [Green Version]
- Giroux, C.N.; Dresser, M.E.; Tiano, H.F. Genetic control of chromosome synapsis in yeast meiosis. Genome 1989, 31, 88–94. [Google Scholar] [CrossRef]
- Dernburg, A.F.; McDonald, K.; Moulder, G.; Barstead, R.; Dresser, M.; Villeneuve, A.M. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 1998, 94, 387–398. [Google Scholar] [CrossRef] [Green Version]
- McKim, K.S.; Buck, E.; Li, J.; Chen, Y.; Weng, G.; Scarlata, S.; Iyengar, R. Meiotic Synapsis in the Absence of Recombination. Science 1998, 279, 876–878. [Google Scholar] [CrossRef] [Green Version]
- Trelles-Sticken, E.; Loidl, J.; Scherthan, H. Bouquet formation in budding yeast: Initiation of recombination is not required for meiotic telomere clustering. J. Cell Sci. 1999, 112, 651–658. [Google Scholar]
- Woglar, A.; Jantsch, V. Chromosome movement in meiosis I prophase of Caenorhabditis elegans. Chromosoma 2013, 123, 15–24. [Google Scholar] [CrossRef] [Green Version]
- Mlynarczyk-Evans, S.; Villeneuve, A.M. Time-Course Analysis of Early Meiotic Prophase Events Informs Mechanisms of Homolog Pairing and Synapsis inCaenorhabditis elegans. Genetics 2017, 207, 103–114. [Google Scholar] [CrossRef] [Green Version]
- Takeo, S.; Lake, C.M.; De Sá, E.M.; Sunkel, C.; Hawley, R.S. Synaptonemal Complex-Dependent Centromeric Clustering and the Initiation of Synapsis in Drosophila Oocytes. Curr. Boil. 2011, 21, 1845–1851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanneti, N.S.; Landy, K.; Joyce, E.F.; McKim, K.S. A Pathway for Synapsis Initiation during Zygotene in Drosophila Oocytes. Curr. Boil. 2011, 21, 1852–1857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liebe, B.; Petukhova, G.; Barchi, M.; Bellani, M.; Braselmann, H.; Nakano, T.; Pandita, T.K.; Jasin, M.; Fornace, A.J.; Meistrich, M.; et al. Mutations that affect meiosis in male mice influence the dynamics of the mid-preleptotene and bouquet stages. Exp. Cell Res. 2006, 312, 3768–3781. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, H.; Morimoto, A.; Watanabe, Y. The Dissection of Meiotic Chromosome Movement in Mice Using an In Vivo Electroporation Technique. PLoS Genet. 2014, 10, e1004821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.-Y.; Horn, H.; Stewart, C.L.; Burke, B.; Bolcun-Filas, E.; Schimenti, J.C.; Dresser, M.E.; Pezza, R.J. Mechanism and regulation of rapid telomere prophase movements in mouse meiotic chromosomes. Cell Rep. 2015, 11, 551–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherthan, H.; Wang, H.; Adelfalk, C.; White, E.J.; Cowan, C.; Cande, W.Z.; Kaback, D. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2007, 104, 16934–16939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koszul, R.; Kim, K.P.; Prentiss, M.; Kleckner, N.; Kameoka, S. Meiotic Chromosomes Move by Linkage to Dynamic Actin Cables with Transduction of Force through the Nuclear Envelope. Cell 2008, 133, 1188–1201. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.S.; Zanders, S.E.; Alani, E. Sustained and Rapid Chromosome Movements Are Critical for Chromosome Pairing and Meiotic Progression in Budding Yeast. Genetics 2011, 188, 21–32. [Google Scholar] [CrossRef] [Green Version]
- Christophorou, N.; Rubin, T.; Bonnet, I.; Piolot, T.; Arnaud, M.; Huynh, J.-R. Microtubule-driven nuclear rotations promote meiotic chromosome dynamics. Nat. Cell Biol. 2015, 17, 1388–1400. [Google Scholar] [CrossRef] [Green Version]
- Trelles-Sticken, E.; Adelfalk, C.; Loidl, J.; Scherthan, H. Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J. Cell Boil. 2005, 170, 213–223. [Google Scholar] [CrossRef]
- Baudrimont, A.; Penkner, A.; Woglar, A.; Machacek, T.; Wegrostek, C.; Gloggnitzer, J.; Fridkin, A.; Klein, F.; Gruenbaum, Y.; Pasierbek, P.; et al. Leptotene/Zygotene Chromosome Movement Via the SUN/KASH Protein Bridge in Caenorhabditis elegans. PLoS Genet. 2010, 6, e1001219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wynne, D.; Rog, O.; Carlton, P.M.; Dernburg, A.F. Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J. Cell Boil. 2012, 196, 47–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conrad, M.N.; Lee, C.-Y.; Wilkerson, J.L.; Dresser, M.E. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2007, 104, 8863–8868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conrad, M.N.; Lee, C.-Y.; Chao, G.; Shinohara, M.; Kosaka, H.; Shinohara, A.; Conchello, J.-A.; Dresser, M.E. Rapid Telomere Movement in Meiotic Prophase Is Promoted By NDJ1, MPS3, and CSM4 and Is Modulated by Recombination. Cell 2008, 133, 1175–1187. [Google Scholar] [CrossRef] [Green Version]
- Ding, D.Q.; Chikashige, Y.; Haraguchi, T.; Hiraoka, Y. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci. 1998, 111, 111. [Google Scholar]
- Sato, A.; Isaac, B.; Phillips, C.; Rillo, R.; Carlton, P.M.; Wynne, D.; Kasad, R.A.; Dernburg, A.F. Cytoskeletal Forces Span the Nuclear Envelope to Coordinate Meiotic Chromosome Pairing and Synapsis. Cell 2009, 139, 907–919. [Google Scholar] [CrossRef] [Green Version]
- Elkouby, Y.; Jamieson-Lucy, A.; Mullins, M.C. Oocyte Polarization Is Coupled to the Chromosomal Bouquet, a Conserved Polarized Nuclear Configuration in Meiosis. PLoS Boil. 2016, 14, e1002335. [Google Scholar] [CrossRef] [Green Version]
- Chikashige, Y.; Tsutsumi, C.; Yamane, M.; Okamasa, K.; Haraguchi, T.; Hiraoka, Y. Meiotic Proteins Bqt1 and Bqt2 Tether Telomeres to Form the Bouquet Arrangement of Chromosomes. Cell 2006, 125, 59–69. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, M.; Katsuyama, S.; Tateho, K.; Nakamura, H.; Miyoshi, J.; Ohba, T.; Matsuhara, H.; Miki, F.; Okazaki, K.; Haraguchi, T.; et al. Microtubule-organizing center formation at telomeres induces meiotic telomere clustering. J. Cell Boil. 2013, 200, 385–395. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, T.G.; Chikashige, Y.; Ozoe, F.; Kawamukai, M.; Hiraoka, Y. Activation of the pheromone-responsive MAP kinase drives haploid cells to undergo ectopic meiosis with normal telomere clustering and sister chromatid segregation in fission yeast. J. Cell Sci. 2004, 117, 3875–3886. [Google Scholar] [CrossRef] [Green Version]
- Phillips, C.; Wong, C.; Bhalla, N.; Carlton, P.M.; Weiser, P.; Meneely, P.M.; Dernburg, A.F. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 2005, 123, 1051–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, C.; Dernburg, A.F. A Family of Zinc-Finger Proteins Is Required for Chromosome-Specific Pairing and Synapsis during Meiosis in C. elegans. Dev. Cell 2006, 11, 817–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibuya, H.; Ishiguro, K.-I.; Watanabe, Y. The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nat. Cell Biol. 2014, 16, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Daniel, K.; Tränkner, D.; Wojtasz, L.; Shibuya, H.; Watanabe, Y.; Alsheimer, M.; Toth, A. Mouse CCDC79 (TERB1) is a meiosis-specific telomere associated protein. BMC Cell Boil. 2014, 15, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibuya, H.; Hernández-Hernández, A.; Morimoto, A.; Negishi, L.; Höög, C.; Watanabe, Y. MAJIN Links Telomeric DNA to the Nuclear Membrane by Exchanging Telomere Cap. Cell 2015, 163, 1252–1266. [Google Scholar] [CrossRef] [Green Version]
- Jaspersen, S.L.; Martin, A.E.; Glazko, G.; Giddings, T.H.; Morgan, G.; Mushegian, A.R.; Winey, M. The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J. Cell Boil. 2006, 174, 665–675. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.-Y.; Bisig, C.G.; Conrad, M.M.; Ditamo, Y.; De Almeida, L.P.; Dresser, M.E.; Pezza, R.J. Extranuclear Structural Components that Mediate Dynamic Chromosome Movements in Yeast Meiosis. Curr. Boil. 2020. [Google Scholar] [CrossRef]
- Hagan, I.; Yanagida, M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Boil. 1995, 129, 1033–1047. [Google Scholar] [CrossRef] [Green Version]
- Miki, F.; Kurabayashi, A.; Tange, Y.; Okazaki, K.; Shimanuki, M.; Niwa, O. Two-hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol. Genet. Genom. 2003, 270, 449–461. [Google Scholar] [CrossRef]
- Wälde, S.; King, M.C. The KASH protein Kms2 coordinates mitotic remodeling of the spindle pole body. J. Cell Sci. 2014, 127, 3625–3640. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.; Xu, R.; Yu, J.; Xu, T.; Zhuang, Y.; Han, M. SUN1 Is Required for Telomere Attachment to Nuclear Envelope and Gametogenesis in Mice. Dev. Cell 2007, 12, 863–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Link, J.; Leubner, M.; Schmitt, J.; Göb, E.; Benavente, R.; Jeang, K.-T.; Xu, R.; Alsheimer, M. Analysis of Meiosis in SUN1 Deficient Mice Reveals a Distinct Role of SUN2 in Mammalian Meiotic LINC Complex Formation and Function. PLoS Genet. 2014, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morimoto, A.; Shibuya, H.; Zhu, X.; Kim, J.; Ishiguro, K.-I.; Han, M.; Watanabe, Y. A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J. Cell Boil. 2012, 198, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Harigaya, Y.; Yamamoto, M. Molecular mechanisms underlying the mitosis–meiosis decision. Chromosom. Res. 2007, 15, 523–537. [Google Scholar] [CrossRef]
- Labella, S.; Woglar, A.; Jantsch, V.; Zetka, M. Polo Kinases Establish Links between Meiotic Chromosomes and Cytoskeletal Forces Essential for Homolog Pairing. Dev. Cell 2011, 21, 948–958. [Google Scholar] [CrossRef] [Green Version]
- Harper, N.C.; Rillo, R.; Jover-Gil, S.; Assaf, Z.J.; Bhalla, N.; Dernburg, A.F. Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Dev. Cell 2011, 21, 934–947. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Miley, N.; Zastrow, M.S.; MacQueen, A.J.; Sato, A.; Nabeshima, K.; Martinez-Perez, E.; Mlynarczyk-Evans, S.; Carlton, P.M.; Villeneuve, A.M. HAL-2 Promotes Homologous Pairing during Caenorhabditis elegans Meiosis by Antagonizing Inhibitory Effects of Synaptonemal Complex Precursors. PLoS Genet. 2012, 8, e1002880. [Google Scholar] [CrossRef] [Green Version]
- Roelens, B.; Barroso, C.; Montoya, A.; Cutillas, P.; Zhang, W.; Woglar, A.; Girard, C.; Martinez-Perez, E.; Villeneuve, A.M. Spatial Regulation of Polo-Like Kinase Activity During Caenorhabditis elegans Meiosis by the Nucleoplasmic HAL-2/HAL-3 Complex. Genetics 2019, 213, 79–96. [Google Scholar] [CrossRef]
- Alleva, B.; Balukoff, N.; Peiper, A.; Smolikove, S. Regulating chromosomal movement by the cochaperone FKB-6 ensures timely pairing and synapsis. J. Cell Boil. 2017, 216, 393–408. [Google Scholar] [CrossRef]
- Olgun, A.; Aleksenko, T.; Pereira-Smith, O.M.; Vassilatis, D.K. Functional analysis of MRG-1: The ortholog of human MRG15 in Caenorhabditis elegans. Journals Gerontol. Ser. A: Boil. Sci. Med Sci. 2005, 60, 543–548. [Google Scholar] [CrossRef] [Green Version]
- Dombecki, C.R.; Chiang, A.C.; Kang, H.-J.; Bilgir, C.; Stefanski, N.A.; Neva, B.J.; Klerkx, E.P.; Nabeshima, K. The Chromodomain Protein MRG-1 Facilitates SC-Independent Homologous Pairing during Meiosis in Caenorhabditis elegans. Dev. Cell 2011, 21, 1092–1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Sun, X.; Jing, Y.; Wang, M.; Liu, K.; Jian, Y.; Yang, M.; Cheng, Z.; Yang, C. MRG-1 is required for genomic integrity in Caenorhabditis elegans germ cells. Cell Res. 2012, 22, 886–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato-Carlton, A.; Li, X.; Crawley, O.; Testori, S.; Martinez-Perez, E.; Sugimoto, A.; Carlton, P.M. Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age. PLoS Genet. 2014, 10, e1004638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikolcevic, P.; Isoda, M.; Shibuya, H.; Barrantes, I.D.B.; Igea, A.; Suja, J.A.; Shackleton, S.; Watanabe, Y.; Nebreda, A.R. Essential role of the Cdk2 activator RingoA in meiotic telomere tethering to the nuclear envelope. Nat. Commun. 2016, 7, 11084. [Google Scholar] [CrossRef] [Green Version]
- Tu, Z.; Bayazit, M.B.; Liu, H.; Zhang, J.; Busayavalasa, K.; Risal, S.; Shao, J.; Satyanarayana, A.; Coppola, V.; Tessarollo, L.; et al. Speedy A–Cdk2 binding mediates initial telomere–nuclear envelope attachment during meiotic prophase I independent of Cdk2 activation. Proc. Natl. Acad. Sci. USA 2016, 114, 592–597. [Google Scholar] [CrossRef] [Green Version]
- Alleva, B.; Smolikove, S. Moving and stopping: Regulation of chromosome movement to promote meiotic chromosome pairing and synapsis. Nucleus 2017, 8, 613–624. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Shao, Y.; Jin, H.; Yu, H.-G. Ndj1, a telomere-associated protein, regulates centrosome separation in budding yeast meiosis. J. Cell Boil. 2015, 209, 247–259. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, A.; West, R.R.; McIntosh, J.R.; Hiraoka, Y. A Cytoplasmic Dynein Heavy Chain Is Required for Oscillatory Nuclear Movement of Meiotic Prophase and Efficient Meiotic Recombination in Fission Yeast. J. Cell Boil. 1999, 145, 1233–1250. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, A.; Tsutsumi, C.; Kojima, H.; Oiwa, K.; Hiraoka, Y. Dynamic Behavior of Microtubules during Dynein-dependent Nuclear Migrations of Meiotic Prophase in Fission Yeast. Mol. Boil. Cell 2001, 12, 3933–3946. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Álvarez, A.; Bez, C.; O’Toole, E.T.; Morphew, M.; Cooper, J.P. Mitotic Nuclear Envelope Breakdown and Spindle Nucleation Are Controlled by Interphase Contacts between Centromeres and the Nuclear Envelope. Dev. Cell 2016, 39, 544–559. [Google Scholar] [CrossRef] [Green Version]
- Katsumata, K.; Nishi, E.; Afrin, S.; Narusawa, K.; Yamamoto, A. Position matters: Multiple functions of LINC-dependent chromosome positioning during meiosis. Curr. Genet. 2017, 27, 117–1052. [Google Scholar] [CrossRef] [PubMed]
- Penkner, A.; Tang, L.; Novatchkova, M.; Ladurner, M.; Fridkin, A.; Gruenbaum, Y.; Schweizer, D.; Loidl, J.; Jantsch, V. The Nuclear Envelope Protein Matefin/SUN-1 Is Required for Homologous Pairing in C. elegans Meiosis. Dev. Cell 2007, 12, 873–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labrador, L.; Barroso, C.; Lightfoot, J.W.; Müller-Reichert, T.; Flibotte, S.; Taylor, J.; Moerman, N.G.; Villeneuve, A.M.; Martinez-Perez, E. Chromosome Movements Promoted by the Mitochondrial Protein SPD-3 Are Required for Homology Search during Caenorhabditis elegans Meiosis. PLoS Genet. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, T.; Liu, Z.; Habara, Y.; Nishiwaki, K.; Nakayama, J.-I.; Inoue, K.; Sakamoto, H.; Strome, S. MRG-1, an autosome-associated protein, silences X-linked genes and protects germline immortality in Caenorhabditis elegans. Development 2007, 134, 757–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, M.; Mlynarczyk-Evans, S.; Villeneuve, A.M. The synaptonemal complex shapes the crossover landscape through cooperative assembly, crossover promotion and crossover inhibition during Caenorhabditis elegans meiosis. Genetics 2010, 186, 45–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rog, O.; Köhler, S.; Dernburg, A.F. The synaptonemal complex has liquid crystalline properties and spatially regulates meiotic recombination factors. Elife 2017, 6, 4482. [Google Scholar] [CrossRef]
- Kim, Y.; Kostow, N.; Dernburg, A.F. The Chromosome Axis Mediates Feedback Control of CHK-2 to Ensure Crossover Formation in C. elegans. Dev. Cell 2015, 35, 247–261. [Google Scholar] [CrossRef] [Green Version]
- Grell, R.F.; Day, J.W. Chromosome pairing in the oogonial cells of Drosophila melanogaster. Chromosoma 1970, 31, 434–445. [Google Scholar] [CrossRef]
- Blumenstiel, J.; Fu, R.; Theurkauf, W.E.; Hawley, R.S. Components of the RNAi Machinery That Mediate Long-Distance Chromosomal Associations Are Dispensable for Meiotic and Early Somatic Homolog Pairing in Drosophila melanogaster. Genetics 2008, 180, 1355–1365. [Google Scholar] [CrossRef] [Green Version]
- Gong, W.J.; McKim, K.S.; Hawley, R.S. All Paired Up with No Place to Go: Pairing, Synapsis, and DSB Formation in a Balancer Heterozygote. PLoS Genet. 2005, 1. [Google Scholar] [CrossRef] [Green Version]
- Sherizen, D.; Jang, J.K.; Bhagat, R.; Kato, N.; McKim, K.S. Meiotic Recombination in Drosophila Females Depends on Chromosome Continuity Between Genetically Defined Boundaries. Genetics 2004, 169, 767–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vazquez, J.; Belmont, A.S.; Sedat, J.W. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr. Boil. 2002, 12, 1473–1483. [Google Scholar] [CrossRef] [Green Version]
- Spradling, A. Developmental genetics of oogenesis. In The development of Drosophila melanogaster; Bate, M., Martinez-Arias, A., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1993; pp. 1–70. [Google Scholar]
- Carpenter, A.T.C. Electron microscopy of meiosis in Drosophila melanogaster females. Chromosoma 1975, 51, 157–182. [Google Scholar] [CrossRef] [PubMed]
- Huynh, J.-R.; Johnston, D.S. The Origin of Asymmetry: Early Polarisation of the Drosophila Germline Cyst and Oocyte. Curr. Boil. 2004, 14, R438–R449. [Google Scholar] [CrossRef] [Green Version]
- McKee, B.D.; Karpen, G.H. Drosophila ribosomal RNA genes function as an X-Y pairing site during male meiosis. Cell 1990, 61, 61–72. [Google Scholar] [CrossRef]
- Jagannathan, M.; Cummings, R.; Yamashita, Y.M. The modular mechanism of chromocenter formation in Drosophila. eLife 2019, 8, 8. [Google Scholar] [CrossRef]
- Viets, K.; Sauria, M.E.; Chernoff, C.; Viales, R.R.; Echterling, M.; Anderson, C.; Tran, S.; Dove, A.; Goyal, R.; Voortman, L.; et al. Characterization of Button Loci that Promote Homologous Chromosome Pairing and Cell-Type-Specific Interchromosomal Gene Regulation. Dev. Cell 2019, 51, 341–356.e7. [Google Scholar] [CrossRef]
- Sola, L.; Gornung, E. Classical and molecular cytogenetics of the zebrafish, Danio rerio (Cyprinidae, Cypriniformes): An overview. Genetics 2001, 111, 397–412. [Google Scholar]
- Saito, K.; Sakai, C.; Kawasaki, T.; Sakai, N. Telomere distribution pattern and synapsis initiation during spermatogenesis in zebrafish. Dev. Dyn. 2014, 243, 1448–1456. [Google Scholar] [CrossRef]
- Kochakpour, N.; Moens, P.B. Sex-specific crossover patterns in Zebrafish (Danio rerio). Heredity 2008, 100, 489–495. [Google Scholar] [CrossRef] [Green Version]
- Singer, A.; Perlman, H.; Yan, Y.; Walker, C.; Corley-Smith, G.; Brandhorst, B.; Postlethwait, J. Sex-specific recombination rates in zebrafish (Danio rerio). Genetics 2002, 160, 649–657. [Google Scholar] [PubMed]
- Anderson, J.L.; Marí, A.R.; Braasch, I.; Amores, Á.; Hohenlohe, P.; Batzel, P.; Postlethwait, J.H. Multiple Sex-Associated Regions and a Putative Sex Chromosome in Zebrafish Revealed by RAD Mapping and Population Genomics. PLoS ONE 2012, 7, e40701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molyneaux, K.A.; Stallock, J.; Schaible, K.; Wylie, C. Time-Lapse Analysis of Living Mouse Germ Cell Migration. Dev. Boil. 2001, 240, 488–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donovan, P. Migratory and postmigratory mouse primordial germ cells behave differently in culture. Cell 1986, 44, 831–838. [Google Scholar] [CrossRef]
- Enders, G.C.; May, J.J. Developmentally Regulated Expression of a Mouse Germ Cell Nuclear Antigen Examined from Embryonic Day 11 to Adult in Male and Female Mice. Dev. Boil. 1994, 163, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Hilscher, B.; Hilscher, W.; Birke, A.; Pelzer, H.; Gauss, G. Kinetics of gametogenesis. Cell Tissue Res. 1974, 154, 443–470. [Google Scholar] [CrossRef]
- Speed, R.M. Meiosis in the foetal mouse ovary. Chromosoma 1982, 85, 427–437. [Google Scholar] [CrossRef]
- Western, P.; Miles, D.C.; Bergen, J.A.V.D.; Burton, M.; Sinclair, A. Dynamic Regulation of Mitotic Arrest in Fetal Male Germ Cells. STEM CELLS 2008, 26, 339–347. [Google Scholar] [CrossRef]
- Yoshida, S. Stem cells in mammalian spermatogenesis. Dev. Growth Differ. 2010, 52, 311–317. [Google Scholar] [CrossRef]
- Spradling, A.; Fuller, M.T.; Braun, R.E.; Yoshida, S. Germline Stem Cells. Cold Spring Harb. Perspect. Boil. 2011, 3, a002642. [Google Scholar] [CrossRef] [Green Version]
- Evans, E.; Hogarth, C.; Mitchell, D.; Griswold, M.D. Riding the spermatogenic wave: Profiling gene expression within neonatal germ and sertoli cells during a synchronized initial wave of spermatogenesis in mice. Boil. Reprod. 2014, 90, 108. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.J.; McCarrey, J.R.; Yang, F.; Page, D.C. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 2001, 27, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Baudat, F.; Manova, K.; Yuen, J.P.; Jasin, M.; Keeney, S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 2000, 6, 989–998. [Google Scholar] [CrossRef]
- Enguita-Marruedo, A.; Van Cappellen, W.A.; Hoogerbrugge, J.W.; Carofiglio, F.; Wassenaar, E.; Slotman, J.A.; Houtsmuller, A.; Baarends, W.M. Live cell analyses of synaptonemal complex dynamics and chromosome movements in cultured mouse testis tubules and embryonic ovaries. Chromosoma 2018, 127, 341–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bisig, C.G.; Guiraldelli, M.F.; Kouznetsova, A.; Scherthan, H.; Höög, C.; Dawson, D.; Pezza, R.J. Synaptonemal Complex Components Persist at Centromeres and Are Required for Homologous Centromere Pairing in Mouse Spermatocytes. PLoS Genet. 2012, 8, e1002701. [Google Scholar] [CrossRef] [Green Version]
- Qiao, H.; Chen, J.K.; Reynolds, A.; Höög, C.; Paddy, M.; Hunter, N. Interplay between Synaptonemal Complex, Homologous Recombination, and Centromeres during Mammalian Meiosis. PLoS Genet. 2012, 8, e1002790. [Google Scholar] [CrossRef] [Green Version]
- Lei, L.; Spradling, A.C. Mouse primordial germ cells produce cysts that partially fragment prior to meiosis. Development 2013, 140, 2075–2081. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, J.; Benavente, R.; Hodzic, D.; Höög, C.; Stewart, C.L.; Alsheimer, M. Transmembrane protein Sun2 is involved in tethering mammalian meiotic telomeres to the nuclear envelope. Proc. Natl. Acad. Sci. USA 2007, 104, 7426–7431. [Google Scholar] [CrossRef] [Green Version]
- Horn, H.; Kim, D.I.; Wright, G.; Wong, E.S.M.; Stewart, C.L.; Burke, B.; Roux, K.J. A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J. Cell Boil. 2013, 202, 1023–1039. [Google Scholar] [CrossRef] [Green Version]
- Sheehan, M.; Pawlowski, W.P. Live imaging of rapid chromosome movements in meiotic prophase I in maize. Proc. Natl. Acad. Sci. USA 2009, 106, 20989–20994. [Google Scholar] [CrossRef] [Green Version]
- Varas, J.; Graumann, K.; Osman, K.; Pradillo, M.; Evans, D.; Santos, J.L.; Armstrong, S.J. Absence of SUN1 and SUN2 proteins inArabidopsis thalianaleads to a delay in meiotic progression and defects in synapsis and recombination. Plant J. 2015, 81, 329–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| S. Cerevisiae | S. Pombe | C. Elegans | D. Melanogaster | D. Rerio | M. Musculus | |
|---|---|---|---|---|---|---|
| Chomosome number (2n) | 32 | 6 | 12 | 8 | 50 | 40 |
| Localisation of initial homologous pairing | Centromere [4,5] | Arms [6,7] Centromeres [8,9,10] | Pairing Center [11,12,13,14] | X-Y rDNA Centromeres [15] Euchromatic Loci [15,16] | Sub Telomere [17] | ♂ Telomeres [18] ♀ Distal/Interstitial Regions [19] |
| Timing of initial homologous pairing | Zygotene [4] | Prophase [20] | Transition Zone [21] | Mitotic Region [15,16] | Leptotene/Early Zygotene [17] | Premeiotic S-phase [22]/Early Leptotene [23,24] |
| Meiotic DSB dependent synapsis | Yes [25] | NA | No [26] | No [27] | Yes [17] | Yes [22,23,24] |
| Type of bouquet | Telomeres [28] | Telomere [20] | Half-moon Shape [21,29,30] | Centromere Cluster [31,32] | Centromere Telomere [17] | Centromere [18,33] Telomere [34] |
| Bouquet stage | Lept/Zyg Transition [28] | Prophase [20] | NA | NA | Leptotene [17] | ♂ Lept/Zyg Transition [34,35] ♀ Zyg/Pachytene Transition [19] |
| Chromosome movement type | Coordinated-[36,37] autonomous [37,38] | Horsetail Movements [20] | Autonomous [30] | Nuclear Rotation [39] | ND | Nuclear Rotation Autonomous [35] |
| Chromosome movement stages | Prophase [36,40] | Horsetail Stage [20] | Leptotene-zygotene [41,42] | 8-cell cyst [39] | ND | Leptotene-zygotene [35] |
| Force-inducing cytoskeleton | Actin [36,43,44] | Microtubules [45] | Microtubules [46] | Microtubules [39] | Microtubules? [47] | Microtubules [34,35] |
| Speed (nm/sec) | 300–500 [37,44] | 83 [20] | 125–400 [41,42] | 300 [39] | ND | 109–120 [35] |
| Adaptors to nuclear envelope | Ndj1p/Csm4p [36,43,44] | Taz1/Rap1 Bqt1/2 [48,49,50] | HIM-8/ZIM-2/ZIM-1/ZIM-3 [51,52] | ND | ND | TERB1-TRF1 [53,54] TERB2-MAJIN [55]. |
| LINC | Mps3p/Mps2p [44,56,57] | Sad1/Kms1/Kms2 [58,59,60] | SUN-1/ZYG-12 [46] | Klarsicht, Klaroid [39] | ND | SUN1/SUN2/ [61,62] KASH5 [63] |
| Motors | ND | Dynein [48,49,50] | Dynein [46] | Dynein [39] | ND | Dynein [34,35] |
| Other regulators | ND | MAP/Pat1 [20,50,64] | CHK-2/PLK-2 [65,66]/HAL-2/HAL-3 [67,68] FKB-6 [69] MRG-1 [70,71,72] PPH-4.1 [73] | ND | ND | Rad21 [23] CDK/Cdk2 [74]/ SpeedyA [74,75]. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubin, T.; Macaisne, N.; Huynh, J.-R. Mixing and Matching Chromosomes during Female Meiosis. Cells 2020, 9, 696. https://doi.org/10.3390/cells9030696
Rubin T, Macaisne N, Huynh J-R. Mixing and Matching Chromosomes during Female Meiosis. Cells. 2020; 9(3):696. https://doi.org/10.3390/cells9030696
Chicago/Turabian StyleRubin, Thomas, Nicolas Macaisne, and Jean-René Huynh. 2020. "Mixing and Matching Chromosomes during Female Meiosis" Cells 9, no. 3: 696. https://doi.org/10.3390/cells9030696