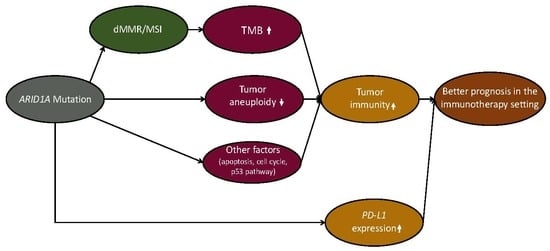
ARID1A Mutations Are Associated with Increased Immune Activity in Gastrointestinal Cancer
Abstract
:1. Introduction
2. Results
2.1. ARID1A Mutations Are Associated with Elevated Immune Activity in GI Cancers
2.2. ARID1A Mutations Are Associated with Increased TMB and Reduced Tumor Aneuploidy Levels in GI Cancers
2.3. ARID1A Mutations Are Associated with the Microsatellite Instability (MSI) Genomic Feature of GI Cancers
2.4. ARID1A Mutations Are Associated with Elevated PD-L1 Expression in GI Cancers and Favorable Immune Checkpoint Blockade Therapy Response in Cancer
3. Discussion
4. Conclusions
5. Methods
5.1. Materials
5.2. Comparisons of Immune Signature Enrichment Levels between Two Groups of Samples
5.3. Gene-Set Enrichment Analysis
5.4. Evaluation of Tumor Aneuploidy Levels
5.5. Prediction of Tumor Immune Activity Using ARID1A Mutation, TMB, and Tumor Aneuploidy Level
5.6. Survival Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACRG | Asian Cancer Research Group |
| ARID1A | AT-rich interaction domain 1A |
| ATP | Adenosine triphosphate |
| COAD | Colon adenocarcinoma |
| CTLA4 | Cytotoxic T-lymphocyte-associated protein 4 |
| FDR | False discovery rate |
| GDC | Genomic Data Commons |
| GEO | Gene Expression Omnibus |
| GSEA | Gene-Set Enrichment Analysis |
| GI | Gastrointestinal |
| HLA | Human leukocyte antigen |
| IgA | Immunoglobulin A |
| MHC | Major histocompatibility complex |
| MMR | Mismatch repair |
| MSI | Microsatellite instability |
| MSS | Microsatellite stable |
| OR | Odds ratio |
| OS | Overall survival |
| PD-1 | Programmed cell death protein 1 |
| PD-L1 | Programmed cell death 1 ligand |
| ssGSEA | Single-sample gene-set enrichment analysis |
| STAD | Stomach adenocarcinoma |
| TCGA | The Cancer Genome Atlas |
| TMB | Tumor mutation burden |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirk, R. Gastrointestinal cancer: New drug shows promise in refractory colorectal cancer. Nat. Rev. Clin. Oncol. 2012, 9, 610. [Google Scholar] [CrossRef] [PubMed]
- Van Allen, E.M.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Geukes Foppen, M.H.; Goldinger, S.M.; et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015, 350, 207–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herbst, R.S.; Baas, P.; Kim, D.W.; Felip, E.; Pérez-Gracia, J.L.; Han, J.Y.; Molina, J.; Kim, J.-H.; Dubos Arvis, K.; Ahn, M.-J. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet 2016, 387, 1540–1550. [Google Scholar] [CrossRef]
- Bauml, J.; Seiwert, T.Y.; Pfister, D.G.; Worden, F.; Liu, S.V.; Gilbert, J.; Saba, N.F.; Weiss, J.; Wirth, L.; Sukari, A. Pembrolizumab for platinum-and cetuximab-refractory head and neck cancer: Results from a single-arm, phase II study. J. Clin. Oncol. 2017, 35, 1542–1549. [Google Scholar] [CrossRef] [PubMed]
- Tomita, Y.; Fukasawa, S.; Shinohara, N.; Kitamura, H.; Oya, M.; Eto, M.; Tanabe, K.; Kimura, G.; Yonese, J.; Yao, M.; et al. Nivolumab versus everolimus in advanced renal cell carcinoma: Japanese subgroup analysis from the CheckMate 025 study. Jpn. J. Clin. Oncol. 2017, 47, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Banobre, J.; Goel, A. DNA Mismatch repair deficiency and immune checkpoint inhibitors in gastrointestinal cancers. Gastroenterology 2019, 156, 890–903. [Google Scholar] [CrossRef]
- Taube, J.M.; Klein, A.; Brahmer, J.R.; Xu, H.; Pan, X.; Kim, H.H.; Chen, L.; Pardoll, D.M.; Topalian, S.L.; Anders, R.A.; et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin. Cancer Res. 2014, 20, 5064–5074. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Pharm, M.; Skora, A.D.; Luber, B.S.; Azad, N.S.; et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef]
- Goodman, A.M.; Kato, S.; Bazhenova, L.; Patel, S.P.; Frampton, G.M.; Miller, V.; Stephens, P.J.; Daniels, G.A.; Kurzrock, R. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol. Cancer Ther. 2017, 16, 2598–2608. [Google Scholar] [CrossRef]
- Davoli, T.; Uno, H.; Wooten, E.C.; Elledge, S.J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017, 355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Z.-Y.; Zhong, W.Z.; Zhang, X.C.; Su, J.; Xie, Z.; Liu, S.-Y.; Tu, H.-T.; Chen, H.-J.; Sun, Y.-L.; Zhou, Q.; et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin. Cancer Res. 2017, 23, 3012–3024. [Google Scholar] [CrossRef] [PubMed]
- Bitler, B.G.; Fatkhutdinov, N.; Zhang, R. Potential therapeutic targets in ARID1A-mutated cancers. Expert Opin. Ther. Targets 2015, 19, 1419–1422. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Li, J.; Xu, J.; Zhang, S.; Xu, Y.; Zhao, W.X.; Yin, Z.Y.; Wang, X.M. Decreased expression of ARID1A associates with poor prognosis and promotes metastases of hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 47. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.D.; Lee, J.E.; Jung, H.Y.; Oh, M.-H.; Lee, J.-H.; Jang, S.-H.; Kim, K.-J.; Han, S.W.; Kim, S.Y.; Kim, H.J.; et al. Loss of tumor suppressor ARID1A protein expression correlates with poor prognosis in patients with primary breast cancer. J. Breast Cancer 2015, 18, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Ju, Z.; Zhao, W.; Wang, L.; Peng, Y.; Ge, Z.; Nagel, Z.D.; Zou, J.; Wang, C.; Kapoor, P.; et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat. Med. 2018, 24, 556–562. [Google Scholar] [CrossRef]
- Zhu, Y.P.; Sheng, L.L.; Wu, J.; Yang, M.; Cheng, X.F.; Wu, N.N.; Ye, X.B.; Cai, J.; Wang, L.; Shen, Q.; et al. Loss of ARID1A expression is associated with poor prognosis in patients with gastric cancer. Hum. Pathol. 2018, 78, 28–35. [Google Scholar] [CrossRef]
- Mathur, R.; Alver, B.H.; San Roman, A.K.; Wilson, B.G.; Wang, X.; Agoston, A.T.; Park, P.J.; Shivdasani, R.A.; Roberts, C.W. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat. Genet. 2017, 49, 296–302. [Google Scholar] [CrossRef]
- Kim, K.J.; Jung, H.Y.; Oh, M.H.; Cho, H.; Lee, J.H.; Jang, S.-H.; Lee, M.S. Loss of ARID1A expression in gastric cancer: Correlation with mismatch repair deficiency and clinicopathologic features. J. Gastric Cancer 2015, 15, 201–208. [Google Scholar] [CrossRef]
- Han, N.; Kim, M.A.; Lee, H.S.; Kim, W.H. Loss of ARID1A Expression is Related to Gastric Cancer Progression, Epstein-Barr Virus Infection, and Mismatch Repair Deficiency. Appl. Immunohistochem. Mol. Morphol. 2016, 24, 320–325. [Google Scholar] [CrossRef]
- Yang, L.; Wei, S.; Zhao, R.; Wu, Y.; Qiu, H.; Xiong, H. Loss of ARID1A expression predicts poor survival prognosis in gastric cancer: A systematic meta-analysis from 14 studies. Sci. Rep. 2016, 6, 28919. [Google Scholar] [CrossRef] [PubMed]
- Buglioni, S.; Melucci, E.; Sperati, F.; Pallocca, M.; Terrenato, I.; Nicola, F.D.; Goeman, F.; Casini, B.; Amoreo, C.A.; Gallo, E.; et al. The clinical significance of PD-L1 in advanced gastric cancer is dependent on ARID1A mutations and ATM expression. Oncoimmunology 2018, 7, e1457602. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Ahn, J.M.; Bae, W.J.; Sung, C.O.; Lee, D. Functional loss of ARID1A is tightly associated with high PD-L1 expression in gastric cancer. Int. J. Cancer 2019, 145, 916–926. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research, N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gilette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Wang, X.; Li, M. Correlate tumor mutation burden with immune signatures in human cancers. BMC Immunol. 2019, 20, 4. [Google Scholar] [CrossRef]
- Carter, S.L.; Cibulskis, K.; Helman, E.; McKenna, A.; Shen, H.; Zack, T.; Laird, P.W.; Onofrio, R.C.; Winckler, W.; Weir, B.A.; et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012, 30, 413–421. [Google Scholar] [CrossRef]
- Hugo, W.; Zaretsky, J.M.; Sun, L.; Song, C.; Moreno, B.H.; Lieskovan, S.H.; Berent-Maoz, B.; Pang, J.; Chimielowski, B.; Cherry, G.; et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016, 165, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Samstein, R.M.; Lee, C.H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A.; et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Kobayashi, A.; Jiang, P.; Ferrari de Andrade, L.; Tay, R.E.; Luoma, A.M.; Tsoucas, D.; Qiu, X.; Lim, K.; Rao, P.; et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 2018, 359, 770–775. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Lyu, J.; Yang, E.J.; Liu, Y.; Zhang, B.; Shim, J.S. Targeting AURKA-CDC25C axis to induce synthetic lethality in ARID1A-deficient colorectal cancer cells. Nat. Commun. 2018, 9, 3212. [Google Scholar] [CrossRef] [PubMed]
- Williamson, C.T.; Miller, R.; Pemberton, H.N.; Jones, S.E.; Campbell, J.; Konde, A.; Badham, N.; Rafiq, R.; Brough, R.; Gulati, A.; et al. ATR inhibitors as a synthetic lethal therapy for tumours deficient in ARID1A. Nat. Commun. 2016, 7, 13837. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Peng, Y.; Wei, L.; Zhang, W.; Yang, L.; Lan, L.; Kapoor, P.; Ju, Z.; Mo, Q.; Shih, L.-M.; et al. ARID1A deficiency impairs the DNA Damage checkpoint and sensitizes cells to PARP inhibitors. Cancer Discov. 2015, 5, 752–767. [Google Scholar] [CrossRef] [PubMed]
- Allo, G.; Bernardini, M.Q.; Wu, R.C.; Shih Ie, M.; Kalloger, S.; Pollet, A.; Gilks, C.B.; Clarke, B.A. ARID1A loss correlates with mismatch repair deficiency and intact p53 expression in high-grade endometrial carcinomas. Mod. Pathol. 2015, 27, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Liu, Z.; Li, M.; Chen, C.; Wang, X. Increased glycolysis correlates with elevated immune activity in tumor immune microenvironment. EBioMedicine 2019, 42, 431–442. [Google Scholar] [CrossRef] [Green Version]
- Goel, S.; DeCristo, M.J.; Watt, A.C.; BrinJones, H.; Sceneay, J.; Li, B.B.; Khan, N.; Ubellacker, J.M.; Xie, S.; Metzger-Filho, O.; et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 2017, 548, 471–475. [Google Scholar] [CrossRef]
- Gasser, S.; Raulet, D.H. The DNA damage response arouses the immune system. Cancer Res. 2006, 66, 3959–3962. [Google Scholar] [CrossRef]
- Jiang, Z.; Liu, Z.; Li, M.; Chen, C.; Wang, X. Immunogenomics analysis reveals that TP53 mutations inhibit tumor immunity in gastric cancer. Transl. Oncol. 2018, 11, 1171–1187. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Ru, B.; Wong, C.N.; Tong, Y.; Yi Zhong, J.; Wa Zhong, S.S.; Wu, W.C.; Chi, K.C.; Wong, C.Y.; Lau, C.Y.L.; Chen, I.; et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 2019. [Google Scholar] [CrossRef] [PubMed]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Li, M.; Jiang, Z.; Wang, X. ARID1A Mutations Are Associated with Increased Immune Activity in Gastrointestinal Cancer. Cells 2019, 8, 678. https://doi.org/10.3390/cells8070678
Li L, Li M, Jiang Z, Wang X. ARID1A Mutations Are Associated with Increased Immune Activity in Gastrointestinal Cancer. Cells. 2019; 8(7):678. https://doi.org/10.3390/cells8070678
Chicago/Turabian StyleLi, Lin, Mengyuan Li, Zehang Jiang, and Xiaosheng Wang. 2019. "ARID1A Mutations Are Associated with Increased Immune Activity in Gastrointestinal Cancer" Cells 8, no. 7: 678. https://doi.org/10.3390/cells8070678