Extracellular Vesicles from Uterine Aspirates Represent a Promising Source for Screening Markers of Gynecologic Cancers
Abstract
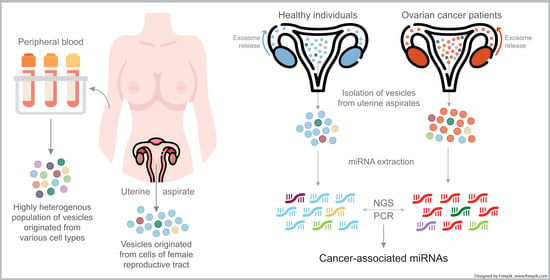
:1. Introduction
2. Materials and Methods
2.1. Clinical Specimens and Patient Consent
2.2. Sample Processing
2.3. Isolation of EVs
2.4. Particle Size Distribution and Quantification
2.5. Transmission Electron Microscopy
2.6. Immunoblotting and Antibodies
2.7. RNA Extraction and Measurement
2.8. Small RNA Deep Sequencing
2.9. Bioinformatics
2.10. Reverse Transcription and Quantitative Real-Time PCR
2.11. Statistical Analysis
3. Results
3.1. Characterization of EVs from Uterine Aspirates
3.2. Characterization of Small RNAs Obtained from EVs Isolated from Uterine Aspirates
3.3. Comparison of EV miRNAs Content in UA from EOC Patients and Healthy Individuals
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Amintas, S.; Vendrely, V.; Dupin, C.; Buscail, L.; Laurent, C.; Bournet, B.; Merlio, J.-P.; Bedel, A.; Moreau-Gaudry, F.; Boutin, J.; et al. Next-Generation Cancer Biomarkers: Extracellular Vesicle DNA as a Circulating Surrogate of Tumor DNA. Front. Cell Dev. Biol. 2021, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Abhange, K.; Makler, A.; Wen, Y.; Ramnauth, N.; Mao, W.; Asghar, W.; Wan, Y. Small extracellular vesicles in cancer. Bioact. Mater. 2021, 6, 3705–3743. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; He, X.; Li, Q.; Lai, H.; Zhang, H.; Hu, Z.; Li, Y.; Huang, S. EV-origin: Enumerating the tissue-cellular origin of circulating extracellular vesicles using exLR profile. Comput. Struct. Biotechnol. J. 2020, 18, 2851–2859. [Google Scholar] [CrossRef]
- Rikkert, L.G.; Beekman, P.; Caro, J.; Coumans, F.A.W.; Enciso-Martinez, A.; Jenster, G.; Le Gac, S.; Lee, W.; Van Leeuwen, T.G.; Loozen, G.B.; et al. Cancer-ID: Toward Identification of Cancer by Tumor-Derived Extracellular Vesicles in Blood. Front. Oncol. 2020, 10, 608. [Google Scholar] [CrossRef]
- Campoy, I.; Lanau, L.; Altadill, T.; Sequeiros, T.; Cabrera, S.; Cubo-Abert, M.; Pérez-Benavente, A.; Garcia, A.; Borrós, S.; Santamaria, A.; et al. Exosome-like vesicles in uterine aspirates: A comparison of ultracentrifugation-based isolation protocols. J. Transl. Med. 2016, 14, 180. [Google Scholar] [CrossRef] [Green Version]
- Luddi, A.; Zarovni, N.; Maltinti, E.; Governini, L.; De Leo, V.; Cappelli, V.; Quintero, L.; Paccagnini, E.; Loria, F.; Piomboni, P. Clues to Non-Invasive Implantation Window Monitoring: Isolation and Characterisation of Endometrial Exosomes. Cells 2019, 8, 811. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Greenblatt, E.M.; Shin, M.E.; Brown, T.J.; Chan, C. Cargo small non-coding RNAs of extracellular vesicles isolated from uterine fluid associate with endometrial receptivity and implantation success. Fertil. Steril. 2021, 115, 1327–1336. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Otsuka, I.; Kameda, S.; Hoshi, K. Early detection of ovarian and fallopian tube cancer by examination of cytological samples from the endometrial cavity. Br. J. Cancer 2013, 109, 603–609. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Douville, C.; Cohen, J.D.; Yen, T.-T.; Kinde, I.; Sundfelt, K.; Kjær, S.K.; Hruban, R.H.; Shih, I.-M.; et al. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Sci. Transl. Med. 2018, 10, eaap8793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maritschnegg, E.; Wang, Y.; Pecha, N.; Horvat, R.; Van Nieuwenhuysen, E.; Vergote, I.; Heitz, F.; Sehouli, J.; Kinde, I.; Diaz, L.A.; et al. Lavage of the Uterine Cavity for Molecular Detection of Müllerian Duct Carcinomas: A Proof-of-Concept Study. J. Clin. Oncol. 2015, 33, 4293–4300. [Google Scholar] [CrossRef] [PubMed]
- Kinde, I.; Bettegowda, C.; Wang, Y.; Wu, J.; Agrawal, N.; Shih, I.-M.; Kurman, R.; Dao, F.; Levine, D.A.; Giuntoli, R.; et al. Evaluation of DNA from the Papanicolaou Test to Detect Ovarian and Endometrial Cancers. Sci. Transl. Med. 2013, 5, 167ra4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thery, C.; Clayton, A.; Amigorena, S.; Raposo, G. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. In Current Protocols in Cell Biology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006. [Google Scholar]
- Skryabin, G.O.; Komelkov, A.V.; Galetsky, S.A.; Bagrov, D.V.; Evtushenko, E.G.; Nikishin, I.I.; Zhordaniia, K.I.; Savelyeva, E.E.; Akselrod, M.E.; PaIanidi, I.G.; et al. Stomatin is highly expressed in exosomes of different origin and is a promising candidate as an exosomal marker. J. Cell. Biochem. 2021, 122, 100–115. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Tamura, T.; Yoshioka, Y.; Sakamoto, S.; Ichikawa, T.; Ochiya, T. Extracellular vesicles as a promising biomarker resource in liquid biopsy for cancer. Extracell. Vesicles Circ. Nucleic Acids 2021, 2, 148–174. [Google Scholar] [CrossRef]
- Alberro, A.; Iparraguirre, L.; Fernandes, A.; Otaegui, D. Extracellular Vesicles in Blood: Sources, Effects, and Applications. Int. J. Mol. Sci. 2021, 22, 8163. [Google Scholar] [CrossRef]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 2014, 3, 23743. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef] [Green Version]
- Nolte’T Hoen, E.N.M.; Buermans, H.P.J.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.M.; ’t Hoen, P.A.C. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, H.; Chen, Q.; Lin, L.; Sha, C.; Li, T.; Liu, Y.; Yin, X.; Xu, Y.; Chen, L.; Gao, W.; et al. Regulation of exosome production and cargo sorting. Int. J. Biol. Sci. 2021, 17, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Guduric-Fuchs, J.; O’Connor, A.; Camp, B.; O’Neill, C.L.; Medina, R.J.; A Simpson, D. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genom. 2012, 13, 357. [Google Scholar] [CrossRef] [Green Version]
- Lunavat, T.R.; Cheng, L.; Kim, D.-K.; Bhadury, J.; Jang, S.C.; Lässer, C.; Sharples, R.A.; Lopez, M.D.; Nilsson, J.; Gho, Y.S.; et al. Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells–Evidence of unique microRNA cargos. RNA Biol. 2015, 12, 810–823. [Google Scholar] [CrossRef] [Green Version]
- Palma, J.; Yaddanapudi, S.C.; Pigati, L.; Havens, M.A.; Jeong, S.; Weiner, G.A.; Weimer, K.M.E.; Stern, B.; Hastings, M.L.; Duelli, D.M. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012, 40, 9125–9138. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Front. Genet. 2013, 4, 142. [Google Scholar] [CrossRef] [Green Version]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of MicroRNAs in living cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef] [Green Version]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral Sphingomyelinase 2 (nSMase2)-dependent Exosomal Transfer of Angiogenic MicroRNAs Regulate Cancer Cell Metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef] [Green Version]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.M.; Abdelmohsen, K.; Mustapic, M.; Kapogiannis, D.; Gorospe, M. RNA in Extracellular Vesicles. Wiley Interdiscip. Rev. RNA 2017, 8, e1413. [Google Scholar] [CrossRef] [PubMed]
- Koppers-Lalic, D.; Hackenberg, M.; Bijnsdorp, I.V.; van Eijndhoven, M.A.; Sadek, P.; Sie, D.; Zini, N.; Middeldorp, J.; Ylstra, B.; de Menezes, R.X.; et al. Nontemplated Nucleotide Additions Distinguish the Small RNA Composition in Cells from Exosomes. Cell Rep. 2014, 8, 1649–1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Gibbings, D.J.; Ciaudo, C.; Erhardt, M.; Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009, 11, 1143–1149. [Google Scholar] [CrossRef]
- Frank, F.; Sonenberg, N.; Nagar, B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 2010, 465, 818–822. [Google Scholar] [CrossRef]
- Lee, Y.S.; Pressman, S.; Andress, A.P.; Kim, K.; White, J.L.; Cassidy, J.J.; Li, X.; Lubell, K.; Lim, D.-H.; Cho, I.S.; et al. Silencing by small RNAs is linked to endosomal trafficking. Nat. Cell Biol. 2009, 11, 1150–1156. [Google Scholar] [CrossRef] [Green Version]
- Skog, J.; Würdinger, T.; Van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Nik Mohamed Kama, N.N.S.B.; Shahidan, W.N.S. Non-Exosomal and Exosomal Circulatory MicroRNAs: Which Are More Valid as Biomarkers? Front. Pharmacol. 2020, 10, 1500. [Google Scholar] [CrossRef] [Green Version]
- Mateescu, B.; Kowal, E.J.K.; Van Balkom, B.W.M.; Bartel, S.; Bhattacharyya, S.N.; Buzás, E.I.; Buck, A.H.; de Candia, P.; Chow, F.W.N.; Das, S.; et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA—An ISEV position paper. J. Extracell. Vesicles 2017, 6, 1286095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bobrie, A.; Colombo, M.; Krumeich, S.; Raposo, G.; Théry, C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J. Extracell. Vesicles 2012, 1, 18397. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Takada, R.; Noda, C.; Kobayashi, S.; Takada, S. Different populations of Wnt-containing vesicles are individually released from polarized epithelial cells. Sci. Rep. 2016, 6, 35562. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wilkinson, R.; Kildey, K.; Ungerer, J.P.J.; Hill, M.M.; Shah, A.K.; Mohamed, A.; Dutt, M.; Molendijk, J.; Healy, H.; et al. Molecular and functional profiling of apical versus basolateral small extracellular vesicles derived from primary human proximal tubular epithelial cells under inflammatory conditions. J. Extracell. Vesicles 2021, 10, e12064. [Google Scholar] [CrossRef] [PubMed]
- Van Deun, J.; Mestdagh, P.; Sormunen, R.; Cocquyt, V.; Vermaelen, K.; Vandesompele, J.; Bracke, M.; De Wever, O.; Hendrix, A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 2014, 3, 24858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohzawa, H.; Kumagai, Y.; Yamaguchi, H.; Miyato, H.; Sakuma, Y.; Horie, H.; Hosoya, Y.; Lefor, A.K.; Sata, N.; Kitayama, J. Exosomal microRNA in peritoneal fluid as a biomarker of peritoneal metastases from gastric cancer. Ann. Gastroenterol. Surg. 2020, 4, 84–93. [Google Scholar] [CrossRef]
- Tokuhisa, M.; Ichikawa, Y.; Kosaka, N.; Ochiya, T.; Yashiro, M.; Hirakawa, K.; Kosaka, T.; Makino, H.; Akiyama, H.; Kunisaki, C.; et al. Exosomal miRNAs from Peritoneum Lavage Fluid as Potential Prognostic Biomarkers of Peritoneal Metastasis in Gastric Cancer. PLoS ONE 2015, 10, e0130472. [Google Scholar] [CrossRef]
- Nazri, H.M.; Imran, M.; Fischer, R.; Heilig, R.; Manek, S.; Dragovic, R.A.; Kessler, B.; Zondervan, K.T.; Tapmeier, T.T.; Becker, C.M. Characterization of exosomes in peritoneal fluid of endometriosis patients. Fertil. Steril. 2020, 113, 364–373.e2. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, C.M.; Oakes, M.L.; Murakami, T.; Muto, M.G.; Berkowitz, R.S.; Ng, S.-W. Comparison of benign peritoneal fluid- and ovarian cancer ascites-derived extracellular vesicle RNA biomarkers. J. Ovarian Res. 2018, 11, 20. [Google Scholar] [CrossRef] [Green Version]
- Ng, Y.H.; Rome, S.; Jalabert, A.; Forterre, A.; Singh, H.; Hincks, C.L.; Salamonsen, L.A. Endometrial Exosomes/Microvesicles in the Uterine Microenvironment: A New Paradigm for Embryo-Endometrial Cross Talk at Implantation. PLoS ONE 2013, 8, e58502. [Google Scholar] [CrossRef] [Green Version]
- Lheureux, S.; Msc, M.B.; Oza, A.M. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J. Clin. 2019, 69, 280–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, A.; Yoshioka, Y.; Hirakawa, A.; Yamamoto, Y.; Ishikawa, M.; Ikeda, S.-I.; Kato, T.; Niimi, K.; Kajiyama, H.; Kikkawa, F.; et al. A combination of circulating miRNAs for the early detection of ovarian cancer. Oncotarget 2017, 8, 89811–89823. [Google Scholar] [CrossRef] [Green Version]
- Takashina, T.; Ito, E.; Kudo, R. Cytologic diagnosis of primary tubal cancer. Acta Cytol. 1985, 29, 367–372. [Google Scholar]
- Whiteside, T.L. Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv. Clin. Chem. 2016, 74, 103–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.-M.; Gaur, A.B.; Lengyel, E.; Peter, M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008, 22, 894–907. [Google Scholar] [CrossRef] [Green Version]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef]
- Yang, J.; Zhou, Y.; Junzheng, Y.; Huang, K.-C.; Ni, X.; Choi, P.-W.; Hasselblatt, K.; Muto, M.G.; Welch, W.R.; Berkowitz, R.S. Characterization of MicroRNA-200 pathway in ovarian cancer and serous intraepithelial carcinoma of fallopian tube. BMC Cancer 2017, 17, 422. [Google Scholar] [CrossRef]
- Hales, K.H.; Speckman, S.C.; Kurrey, N.K.; Hales, D.B. Uncovering molecular events associated with the chemosuppressive effects of flaxseed: A microarray analysis of the laying hen model of ovarian cancer. BMC Genom. 2014, 15, 709. [Google Scholar] [CrossRef] [Green Version]
- Choi, P.-W.; So, W.W.; Yang, J.; Liu, S.; Tong, K.K.; Kwan, K.M.; Kwok, J.S.-L.; Tsui, S.K.W.; Ng, S.-K.; Hales, K.H.; et al. MicroRNA-200 family governs ovarian inclusion cyst formation and mode of ovarian cancer spread. Oncogene 2020, 39, 4045–4060. [Google Scholar] [CrossRef]
- Choi, P.W.; Ng, S.W. The Functions of MicroRNA-200 Family in Ovarian Cancer: Beyond Epithelial-Mesenchymal Transition. Int. J. Mol. Sci. 2017, 18, 1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savolainen, K.; Scaravilli, M.; Ilvesmäki, A.; Staff, S.; Tolonen, T.; Mäenpää, J.U.; Visakorpi, T.; Auranen, A. Expression of the miR-200 family in tumor tissue, plasma and urine of epithelial ovarian cancer patients in comparison to benign counterparts. BMC Res. Notes 2020, 13, 311. [Google Scholar] [CrossRef]
- Choi, P.-W.; Bahrampour, A.; Ng, S.-K.; Liu, S.K.; Qiu, W.; Xie, F.; Kuo, W.P.; Kwong, J.; Hales, K.H.; Hales, D.B.; et al. Characterization of miR-200 family members as blood biomarkers for human and laying hen ovarian cancer. Sci. Rep. 2020, 10, 20071. [Google Scholar] [CrossRef] [PubMed]
- Kan, C.W.S.; Hahn, M.A.; Gard, G.B.; Maidens, J.; Huh, J.Y.; Marsh, D.J.; Howell, V.M. Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer. BMC Cancer 2012, 12, 627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuberi, M.; Mir, A.R.; Das, J.; Ahmad, I.; Javid, J.; Yadav, P.; Masroor, M.; Ahmad, S.; Ray, P.C.; Saxena, A. Expression of serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin. Transl. Oncol. 2015, 17, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.-C.; Wu, J. MicroRNA-200c and microRNA-141 as potential diagnostic and prognostic biomarkers for ovarian cancer. Tumor Biol. 2015, 36, 4843–4850. [Google Scholar] [CrossRef]
- Zheng, H.; Zhang, L.; Zhao, Y.; Yang, D.; Song, F.; Wen, Y.; Hao, Q.; Hu, Z.; Zhang, W.; Chen, K. Plasma miRNAs as Diagnostic and Prognostic Biomarkers for Ovarian Cancer. PLoS ONE 2013, 8, e77853. [Google Scholar] [CrossRef]
- Pendlebury, A.; Hannan, N.J.; Binder, N.; Beard, S.; McGauran, M.; Grant, P.; Tong, S.; Whitehead, C.L. The circulating microRNA-200 family in whole blood are potential biomarkers for high-grade serous epithelial ovarian cancer. Biomed. Rep. 2017, 6, 319–322. [Google Scholar] [CrossRef] [Green Version]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef]
- Xu, Y.-F.; Hannafon, B.N.; Khatri, U.; Gin, A.; Ding, W.-Q. The origin of exosomal miR-1246 in human cancer cells. RNA Biol. 2019, 16, 770–784. [Google Scholar] [CrossRef]
- Cha, S.Y.; Choi, Y.H.; Hwang, S.; Jeong, J.-Y.; An, H.J. Clinical Impact of microRNAs Associated With Cancer Stem Cells as a Prognostic Factor in Ovarian Carcinoma. J. Cancer 2017, 8, 3538–3547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishige, F.; Hoshino, I.; Iwatate, Y.; Chiba, S.; Arimitsu, H.; Yanagibashi, H.; Nagase, H.; Takayama, W. MIR1246 in body fluids as a biomarker for pancreatic cancer. Sci. Rep. 2020, 10, 8723. [Google Scholar] [CrossRef] [PubMed]
- Salah, M.; Shaheen, I.; El-Shanawany, P.; Saad, N.E.; Saad, R.; El Guibaly, M.; Momen, N. Detection of miR-1246, miR-23a and miR-451 in sera of colorectal carcinoma patients: A case-control study in Cairo University hospital. Afr. Health Sci. 2020, 20, 1283–1291. [Google Scholar] [CrossRef] [PubMed]
- Todeschini, P.; Salviato, E.; Paracchini, L.; Ferracin, M.; Petrillo, M.; Zanotti, L.; Tognon, G.; Gambino, A.; Calura, E.; Caratti, G.; et al. Circulating miRNA landscape identifies miR-1246 as promising diagnostic biomarker in high-grade serous ovarian carcinoma: A validation across two independent cohorts. Cancer Lett. 2017, 388, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Ren, Z.J.; Tang, J.H.; Yu, Q. Exosomal MicroRNA MiR-1246 Promotes Cell Proliferation, Invasion and Drug Resistance by Targeting CCNG2 in Breast Cancer. Cell. Physiol. Biochem. 2017, 44, 1741–1748. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Li, Y.; Huang, K.; Li, G.; He, M. Exosomal miR-1246 in body fluids is a potential biomarker for gastrointestinal cancer. Biomark. Med. 2018, 12, 1185–1196. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wang, Z.; Zhu, X.; Chen, L.; Ma, Y.; Wang, J.; Yang, X.; Liu, Z. Exosomal miR-1246 in serum as a potential biomarker for early diagnosis of gastric cancer. Int. J. Clin. Oncol. 2019, 25, 89–99. [Google Scholar] [CrossRef]
- Bai, H.; Wu, S. miR-451: A Novel Biomarker and Potential Therapeutic Target for Cancer. OncoTargets Ther. 2019, 12, 11069–11082. [Google Scholar] [CrossRef] [Green Version]
- Ling, S.; Ruiqin, M.; Guohong, Z.; Ying, W. Expression and prognostic significance of microRNA-451 in human epithelial ovarian cancer. Eur. J. Gynaecol. Oncol. 2015, 36, 463–468. [Google Scholar]
- Wu, R.L.; Ali, S.; Bandyopadhyay, S.; Alosh, B.; Hayek, K.; Daaboul, M.F.; Winer, I.; Sarkar, F.H.; Ali-Fehmi, R. Comparative Analysis of Differentially Expressed miRNAs and their Downstream mRNAs in Ovarian Cancer and its Associated Endometriosis. J. Cancer Sci. Ther. 2015, 7, 258–265. [Google Scholar] [CrossRef] [Green Version]
- Ji, T.; Zheng, Z.-G.; Wang, F.-M.; Xu, L.-J.; Li, L.-F.; Cheng, Q.-H.; Guo, J.-F.; Ding, X.-F. Differential microRNA expression by Solexa sequencing in the sera of ovarian cancer patients. Asian Pac. J. Cancer Prev. 2014, 15, 1739–1743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, D.N.P.; Carlsen, A.L.; Heegaard, N.H.H.; Prahm, K.P.; Christensen, I.J.; Høgdall, C.K.; Høgdall, E.V. Diagnostic plasma miRNA-profiles for ovarian cancer in patients with pelvic mass. PLoS ONE 2019, 14, e0225249. [Google Scholar] [CrossRef]
- Chen, S.F.; Liu, Z.; Chaurasiya, S.; Dellinger, T.; Lu, J.; Wu, X.; Qin, H.; Wang, J.; Fong, Y.; Yuan, Y.-C. Identification of core aberrantly expressed microRNAs in serous ovarian carcinoma. Oncotarget 2018, 9, 20451–20466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Jing, Y.; Li, W.; Qian, X.; Xu, Q.; Li, F.-S.; Liu, L.-Z.; Jiang, B.-H.; Jiang, Y. Roles and Mechanism of miR-199a and miR-125b in Tumor Angiogenesis. PLoS ONE 2013, 8, e56647. [Google Scholar] [CrossRef]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.-G.; Alder, H.; et al. MicroRNA Signatures in Human Ovarian Cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, E.J.; Yoon, H.; Kim, S.W.; Kim, H.; Kim, Y.T.; Kim, J.H.; Kim, J.W.; Kim, S. MicroRNA Expression Profiles in Serous Ovarian Carcinoma. Clin. Cancer Res. 2008, 14, 2690–2695. [Google Scholar] [CrossRef] [Green Version]




| Sample | TNM | Grade | Stage (FIGO) | Histological Subtype |
|---|---|---|---|---|
| T1 | T3cN0M1 | High (G3) | IV | serous adenocarcinoma |
| T2 | T3cN1M0 | High (G3) | IIIC | serous papillary adenocarcinoma |
| T3 | T3cN1M0 | High (G3) | IIIC | serous papillary adenocarcinoma |
| T4 | T1cN0M0 | High (G2) | IC | serous adenocarcinoma |
| T5 | T3cN0M1 | High (G3) | IVA | serous papillary adenocarcinoma |
| Gene ID | logFC | logCPM | F | p Value | FDR | Fold |
|---|---|---|---|---|---|---|
| hsa-miR-451a | −4.2758589 | 12.4477224 | 36.9140415 | 1.29 × 10−9 | 3.42 × 10−6 | 0.052 |
| hsa-miR-542-3p | −3.6525866 | 9.5709767 | 23.3039465 | 0.0000014 | 0.001709212 | 0.080 |
| hsa-miR-1246 | 3.8004008 | 9.0879369 | 22.5883317 | 0.0000024 | 0.001709212 | 13.933 |
| hsa-miR-375-3p | 3.2136512 | 11.7583475 | 22.0561752 | 0.0000027 | 0.001709212 | 9.277 |
| hsa-miR-125a-5p | 2.9175311 | 13.6969615 | 21.0651975 | 0.0000045 | 0.001709212 | 7.556 |
| hsa-miR-320b | 2.9883874 | 12.9093771 | 20.9288553 | 0.0000048 | 0.001709212 | 7.936 |
| hsa-miR-200c-3p | 2.9497286 | 13.1576673 | 20.7891165 | 0.0000052 | 0.001709212 | 7.726 |
| hsa-miR-424-5p | −3.2575984 | 10.0135020 | 20.1278828 | 0.0000073 | 0.002145565 | 0.105 |
| hsa-miR-200b-5p | 3.3101791 | 9.7988733 | 18.3239500 | 0.0000187 | 0.004512432 | 9.919 |
| hsa-miR-183-5p | 3.0544179 | 9.9486138 | 17.8818248 | 0.0000238 | 0.005116818 | 8.308 |
| hsa-miR-200b-3p | 2.5691068 | 14.7872804 | 17.7661256 | 0.0000251 | 0.005116818 | 5.934 |
| hsa-miR-320c | 2.8136239 | 11.4890222 | 17.2009005 | 0.0000338 | 0.005592499 | 7.030 |
| hsa-miR-224-5p | 2.9448739 | 10.2072139 | 17.1046125 | 0.0000358 | 0.005592499 | 7.700 |
| hsa-miR-450b-5p | −3.0021235 | 9.3416476 | 16.4403970 | 0.0000508 | 0.006731828 | 0.125 |
| hsa-miR-320a-3p | 3.0742866 | 9.1541706 | 16.2224191 | 0.0000565 | 0.006813182 | 8.423 |
| hsa-let-7b-5p | 2.3190987 | 14.6406996 | 14.6399033 | 0.0001311 | 0.012859111 | 4.990 |
| hsa-miR-125b-5p | 2.3615198 | 13.7812621 | 14.4991659 | 0.0001406 | 0.012859111 | 5.139 |
| hsa-miR-182-5p | 2.5967790 | 10.9627996 | 14.4123220 | 0.0001484 | 0.01311557 | 6.049 |
| hsa-miR-143-3p | −2.1783923 | 15.8799851 | 13.8648943 | 0.0001978 | 0.01604251 | 0.221 |
| hsa-miR-196b-5p | −2.4547336 | 11.9317925 | 13.8508279 | 0.0001996 | 0.01604251 | 0.182 |
| hsa-miR-449c-5p | −2.7414632 | 9.7370987 | 13.1831605 | 0.0002831 | 0.020700409 | 0.150 |
| hsa-miR-9-5p | 2.5062128 | 10.4029623 | 13.0367742 | 0.0003083 | 0.020962645 | 5.681 |
| hsa-miR-19b-3p | −2.6217121 | 9.0047643 | 12.4768255 | 0.0004129 | 0.026072094 | 0.162 |
| hsa-miR-411-5p | −2.7085572 | 9.3188322 | 12.2746304 | 0.0004601 | 0.028203095 | 0.153 |
| hsa-miR-10a-5p | 2.0485626 | 14.9248514 | 11.8254458 | 0.0005854 | 0.033030914 | 4.137 |
| hsa-miR-141-3p | 1.9032256 | 9.1714047 | 10.9735792 | 0.0067931 | 0.040527411 | 3.740 |
| hsa-miR-429 | 2.0748839 | 9.8455066 | 10.4926311 | 0.0006894 | 0.041667954 | 4.213 |
| hsa-miR-23b-3p | −2.0119478 | 14.1412192 | 10.3749023 | 0.0009300 | 0.046277078 | 0.248 |
| hsa-miR-199a-3p | −2.0198928 | 14.8878751 | 9.9660032 | 0.0011188 | 0.049553691 | 0.247 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skryabin, G.O.; Komelkov, A.V.; Zhordania, K.I.; Bagrov, D.V.; Vinokurova, S.V.; Galetsky, S.A.; Elkina, N.V.; Denisova, D.A.; Enikeev, A.D.; Tchevkina, E.M. Extracellular Vesicles from Uterine Aspirates Represent a Promising Source for Screening Markers of Gynecologic Cancers. Cells 2022, 11, 1064. https://doi.org/10.3390/cells11071064
Skryabin GO, Komelkov AV, Zhordania KI, Bagrov DV, Vinokurova SV, Galetsky SA, Elkina NV, Denisova DA, Enikeev AD, Tchevkina EM. Extracellular Vesicles from Uterine Aspirates Represent a Promising Source for Screening Markers of Gynecologic Cancers. Cells. 2022; 11(7):1064. https://doi.org/10.3390/cells11071064
Chicago/Turabian StyleSkryabin, Gleb O., Andrey V. Komelkov, Kirill I. Zhordania, Dmitry V. Bagrov, Svetlana V. Vinokurova, Sergey A. Galetsky, Nadezhda V. Elkina, Darya A. Denisova, Adel D. Enikeev, and Elena M. Tchevkina. 2022. "Extracellular Vesicles from Uterine Aspirates Represent a Promising Source for Screening Markers of Gynecologic Cancers" Cells 11, no. 7: 1064. https://doi.org/10.3390/cells11071064