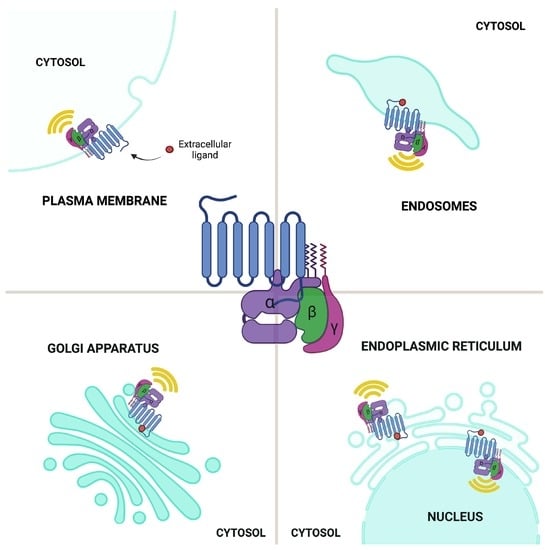
Endomembrane-Based Signaling by GPCRs and G-Proteins
Abstract
:1. Introduction
1.1. GPCR Classes
1.2. G-Protein Signaling Complexes
2. GPCR Activation-Signal Amplification and Inactivation-Desensitization Add Further Regulatory Complexities
2.1. Mechanisms of Ligand-Induced Activation
2.2. Molecular Players in GPCR Desensitization: GRKs, Arrestins and More
2.3. GPCR-Independent Mechanisms for G-Protein Activation: RGS and AGS Proteins
2.4. GPCR and G Proteins in the Secretory Pathway
2.4.1. G-Protein Localization in the Endomembranes
2.4.2. Regulation in GPCR Transport to the Plasma Membrane by ER–Golgi Proteins
2.5. GPCR Signaling: Conventional Plasma Membrane vs. Endomembrane Signaling
2.5.1. GPCR Signaling: ER–Golgi Pool
2.5.2. GPCR Signaling: Endosomal Pool
2.5.3. GPCR Signaling: Nuclear Envelope Pool
3. Conclusion and Perspective: GPCR and G-Protein Signaling in Control Organization and Cell Reprogramming
Author Contributions
Funding
Conflicts of Interest
References
- Fredriksson, R.; Schiöth, H.B. The Repertoire of G-Protein–Coupled Receptors in Fully Sequenced Genomes. Mol. Pharmacol. 2005, 67, 1414–1425. [Google Scholar] [CrossRef] [PubMed]
- Lappano, R.; Maggiolini, M. G protein-coupled receptors: Novel targets for drug discovery in cancer. Nat. Rev. Drug Discov. 2010, 10, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Davenport, A.P.; Scully, C.C.G.; De Graaf, C.; Brown, A.J.H.; Maguire, J.J. Advances in therapeutic peptides targeting G protein-coupled receptors. Nat. Rev. Drug Discov. 2020, 19, 389–413. [Google Scholar] [CrossRef] [PubMed]
- Luttrell, L.M.; Roudabush, F.L.; Choy, E.W.; Miller, W.; Field, M.E.; Pierce, K.L.; Lefkowitz, R.J. Activation and targeting of extracellular signal-regulated kinases by -arrestin scaffolds. Proc. Natl. Acad. Sci. USA 2001, 98, 2449–2454. [Google Scholar] [CrossRef] [Green Version]
- Hanyaloglu, A.C.; von Zastrow, M. Regulation of GPCRs by Endocytic Membrane Trafficking and Its Potential Implications. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 537–568. [Google Scholar] [CrossRef] [Green Version]
- Kolakowski, L.F. GCRDb: A G-protein-coupled receptor database. Recept. Channels 1994, 2. [Google Scholar]
- Attwood, T.K.; Findlay, J.B.C. Fingerprinting G-protein-coupled receptors. Protein Eng. Des. Sel. 1994, 7, 195–203. [Google Scholar] [CrossRef]
- Schiöth, H.B.; Fredriksson, R. The GRAFS classification system of G-protein coupled receptors in comparative perspective. Gen. Comp. Endocrinol. 2005, 142, 94–101. [Google Scholar] [CrossRef]
- Ha, T.S.; Smith, D.P. Insect Odorant Receptors: Channeling Scent. Cell 2008, 133, 761–763. [Google Scholar] [CrossRef] [Green Version]
- DeMaria, S.; Ngai, J. The cell biology of smell. J. Cell Biol. 2010, 191, 443–452. [Google Scholar] [CrossRef]
- Leal, W.S. Odorant Reception in Insects: Roles of Receptors, Binding Proteins, and Degrading Enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Wicher, D.; Schäfer, R.; Bauernfeind, R.; Stensmyr, M.; Heller, R.; Heinemann, S.H.; Hansson, B.S. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 2008, 452, 1007–1011. [Google Scholar] [CrossRef]
- Fredriksson, R.; Lagerström, M.C.; Lundin, L.G.; Schiöth, H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003, 63, 1256–1272. [Google Scholar] [CrossRef] [Green Version]
- Watkins, H.A.; Au, M.; Hay, D.L. The structure of secretin family GPCR peptide ligands: Implications for receptor pharmacology and drug development. Drug Discov. Today 2012, 17, 1006–1014. [Google Scholar] [CrossRef]
- Harmar, A.J. Family-B G-protein-coupled receptors. Genome Biol. 2001, 2, reviews3013.1. [Google Scholar] [CrossRef] [Green Version]
- McKnight, A.J.; Gordon, S. The EGF-TM7 family: Unusual structures at the leukocyte surface. J. Leukoc. Biol. 1998, 63, 271–280. [Google Scholar] [CrossRef]
- Chun, L.; Zhang, W.-H.; Liu, J.-F. Structure and ligand recognition of class C GPCRs. Acta Pharmacol. Sin. 2012, 33, 312–323. [Google Scholar] [CrossRef] [Green Version]
- Kunishima, N.; Shimada, Y.; Tsuji, Y.; Sato, T.; Yamamoto, M.; Kumasaka, T.; Nakanishi, S.; Jingami, H.; Morikawa, K. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature 2000, 407, 971–977. [Google Scholar] [CrossRef]
- Bessis, A.-S.; Rondard, P.; Gaven, F.; Brabet, I.; Triballeau, N.; Prézeau, L.; Acher, F.; Pin, J.-P. Closure of the Venus flytrap module of mGlu8 receptor and the activation process: Insights from mutations converting antagonists into agonists. Proc. Natl. Acad. Sci. USA 2002, 99, 11097–11102. [Google Scholar] [CrossRef] [Green Version]
- Schulte, G.; Kozielewicz, P. Structural insight into Class F receptors—What have we learnt regarding agonist-induced activation? Basic Clin. Pharmacol. Toxicol. 2019, 126 (Suppl. S6), 17–24. [Google Scholar] [CrossRef]
- Kilander, M.B.C.; Petersen, J.; Andressen, K.W.; Ganji, R.S.; Levy, F.O.; Schuster, J.; Dahl, N.; Bryja, V.; Schulte, G. Disheveled regulates precoupling of heterotrimeric G proteins to Frizzled 6. FASEB J. 2014, 28, 2293–2305. [Google Scholar] [CrossRef] [PubMed]
- Wright, S.C.; Kozielewicz, P.; Kowalski-Jahn, M.; Petersen, J.; Bowin, C.-F.; Slodkowicz, G.; Marti-Solano, M.; Rodríguez, D.; Hot, B.; Okashah, N.; et al. A conserved molecular switch in Class F receptors regulates receptor activation and pathway selection. Nat. Commun. 2019, 10, 667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kilander, M.B.; Dijksterhuis, J.P.; Ganji, R.S.; Bryja, V.; Schulte, G. WNT-5A stimulates the GDP/GTP exchange at pertussis toxin-sensitive heterotrimeric G proteins. Cell. Signal. 2011, 23, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Riobo, N.A.; Saucy, B.; DiLizio, C.; Manning, D.R. Activation of heterotrimeric G proteins by Smoothened. Proc. Natl. Acad. Sci. USA 2006, 103, 12607–12612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Medarde, A.; De Las Rivas, J.; Santos, E. 40 Years of RAS—A Historic Overview. Genes 2021, 12, 681. [Google Scholar] [CrossRef] [PubMed]
- Goitre, L.; Trapani, E.; Trabalzini, L.; Retta, S.F. The Ras Superfamily of Small GTPases: The Unlocked Secrets. Methods Mol. Biol. 2013, 1120, 1–18. [Google Scholar] [CrossRef]
- Vögler, O.; Barceló, J.M.; Ribas, C.; Escribá, P.V. Membrane interactions of G proteins and other related proteins. Biochim. Biophys. Acta 2008, 1778, 1640–1652. [Google Scholar] [CrossRef] [Green Version]
- Weis, W.I.; Kobilka, B.K. The Molecular Basis of G Protein–Coupled Receptor Activation. Annu. Rev. Biochem. 2018, 87, 897–919. [Google Scholar] [CrossRef]
- Wettschureck, N.; Offermanns, S. Mammalian G Proteins and Their Cell Type Specific Functions. Physiol. Rev. 2005, 85, 1159–1204. [Google Scholar] [CrossRef] [Green Version]
- Sadana, R.; Dessauer, C.W. Physiological Roles for G Protein-Regulated Adenylyl Cyclase Isoforms: Insights from Knockout and Overexpression Studies. Neurosignals 2009, 17, 5–22. [Google Scholar] [CrossRef]
- Cerione, R.A.; Staniszewski, C.; Benovic, J.L.; Lefkowitz, R.J.; Caron, M.G.; Gierschik, P.; Somers, R.; Spiegel, A.M.; Codina, J.; Birnbaumer, L. Specificity of the functional interactions of the beta-adrenergic receptor and rhodopsin with guanine nucleotide regulatory proteins reconstituted in phospholipid vesicles. J. Biol. Chem. 1985, 260, 1493–1500. [Google Scholar] [CrossRef]
- Dawaliby, R.; Trubbia, C.; Delporte, C.; Masureel, M.; Van Antwerpen, P.; Kobilka, B.B.K.; Govaerts, C. Allosteric regulation of G protein–coupled receptor activity by phospholipids. Nat. Chem. Biol. 2015, 12, 35–39. [Google Scholar] [CrossRef]
- Strohman, M.J.; Maeda, S.; Hilger, D.; Masureel, M.; Du, Y.; Kobilka, B.K. Local membrane charge regulates β2 adrenergic receptor coupling to Gi3. Nat. Commun. 2019, 10, 2234. [Google Scholar] [CrossRef]
- Dingus, J.; Wells, C.A.; Campbell, L.; Cleator, J.H.; Robinson, K.; Hildebrandt, J.D. G Protein βγ Dimer Formation: Gβ and Gγ Differentially Determine Efficiency of in Vitro Dimer Formation. Biochemistry 2005, 44, 11882–11890. [Google Scholar] [CrossRef]
- Masuho, I.; Skamangas, N.K.; Muntean, B.S.; Martemyanov, K.A. Diversity of the Gβγ complexes defines spatial and temporal bias of GPCR signaling. Cell Syst. 2021, 12, 324–337.e5. [Google Scholar] [CrossRef]
- Zhou, Q.; Yang, D.; Wu, M.; Guo, Y.; Guo, W.; Zhong, L.; Cai, X.; Dai, A.; Jang, W.; Shakhnovich, E.I.; et al. Common activation mechanism of class A GPCRs. eLife 2019, 8. [Google Scholar] [CrossRef]
- Nygaard, R.; Frimurer, T.M.; Holst, B.; Rosenkilde, M.M.; Schwartz, T.W. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol. Sci. 2009, 30, 249–259. [Google Scholar] [CrossRef]
- Hoare, S.R.J. Mechanisms of peptide and nonpeptide ligand binding to Class B G-protein-coupled receptors. Drug Discov. Today 2005, 10, 417–427. [Google Scholar] [CrossRef]
- Lee, S.-M.; Hay, D.L.; Pioszak, A.A. Calcitonin and amylin receptor peptide interaction mechanisms. Insights into peptide-binding modes and allosteric modulation of the calcitonin receptor by receptor activity-modifying proteins. J. Biol. Chem. 2016, 291, 16416. [Google Scholar] [CrossRef] [Green Version]
- Geng, Y.; Bush, M.; Mosyak, L.; Wang, F.; Fan, Q.R. Structural mechanism of ligand activation in human GABAB receptor. Nature 2013, 504, 254–259. [Google Scholar] [CrossRef] [Green Version]
- Dijksterhuis, J.P.; Petersen, J.; Schulte, G. WNT/Frizzled signalling: Receptor-ligand selectivity with focus on FZD-G protein signalling and its physiological relevance: IUPHAR Review 3. Br. J. Pharmacol 2014, 171, 1195–1209. [Google Scholar] [CrossRef] [Green Version]
- Dann, C.E.; Hsieh, J.-C.; Rattner, A.; Sharma, D.; Nathans, J.; Leahy, D.J. Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature 2001, 412, 86–90. [Google Scholar] [CrossRef]
- Ross, E.M. G Protein-coupled receptors: Multi-turnover GDP/GTP exchange catalysis on heterotrimeric G proteins. Cell. Logist. 2014, 4, e29391. [Google Scholar] [CrossRef] [Green Version]
- Benovic, J.L.; Strasser, R.H.; Caron, M.G.; Lefkowitz, R.J. Beta-adrenergic receptor kinase: Identification of a novel protein kinase that phosphorylates the agonist-occupied form of the receptor. Proc. Natl. Acad. Sci. USA 1986, 83, 2797–2801. [Google Scholar] [CrossRef] [Green Version]
- Strasser, R.H.; Sibley, D.R.; Lefkowitz, R.J. A novel catecholamine-activated adenosine cyclic 3′,5′-phosphate independent pathway for β-adrenergic receptor phosphorylation in wild-type and mutant S49 lymphoma cells: Mechanism of homologous desensitization of adenylate cyclase. Biochemistry 1986, 25, 1371–1377. [Google Scholar] [CrossRef]
- Yang, Z.; Yang, F.; Zhang, D.; Liu, Z.; Lin, A.; Liu, C.; Xiao, P.; Yu, X.; Sun, J.-P. Phosphorylation of G Protein-Coupled Receptors: From the Barcode Hypothesis to the Flute Model. Mol. Pharmacol. 2017, 92, 201–210. [Google Scholar] [CrossRef]
- Shen, A.; Nieves-Cintron, M.; Deng, Y.; Shi, Q.; Chowdhury, D.; Qi, J.; Hell, J.W.; Navedo, M.F.; Xiang, Y.K. Functionally distinct and selectively phosphorylated GPCR subpopulations co-exist in a single cell. Nat. Commun. 2018, 9, 1050. [Google Scholar] [CrossRef] [Green Version]
- Clark, R.B.; Kunkel, M.W.; Friedman, J.; Goka, T.J.; Johnson, J.A. Activation of cAMP-dependent protein kinase is required for heterologous desensitization of adenylyl cyclase in S49 wild-type lymphoma cells. Proc. Natl. Acad. Sci. USA 1988, 85, 1442–1446. [Google Scholar] [CrossRef] [Green Version]
- Kelly, E.; Bailey, C.P.; Henderson, G. Agonist-selective mechanisms of GPCR desensitization. J. Cereb. Blood Flow Metab. 2008, 153 (Suppl. S1), S379–S388. [Google Scholar] [CrossRef] [Green Version]
- Yao, L.; Ji, S.; Liu, X.; Li, S.; Shen, L.; Li, F.; Wang, J.; Han, J. Ph domain of G protein-coupled receptor kinase-2 binds to protein kinase C PKC and negatively regulates activity of PKC kinase. Front. Biosci. 2003, 8, a34–a39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitcher, J.A.; Freedman, N.J.; Lefkowitz, R.J. G protein–coupled receptor kinases. Annu. Rev. Biochem. 1998, 67, 653–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurevich, V.V.; Gurevich, E.V. GPCR Signaling Regulation: The Role of GRKs and Arrestins. Front. Pharmacol. 2019, 10, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krupnick, J.G.; Benovic, J.L. The role of receptor kinases and arrestins in g protein–coupled receptor regulation. Annu. Rev. Pharmacol. Toxicol. 1998, 38, 289–319. [Google Scholar] [CrossRef]
- Magalhaes, A.C.; Dunn, H.; Ferguson, S.S. Regulation of GPCR activity, trafficking and localization by GPCR-interacting proteins. Br. J. Pharmacol. 2012, 165, 1717–1736. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.-C.; Tesmer, J.J. Recognition in the Face of Diversity: Interactions of Heterotrimeric G proteins and G Protein-coupled Receptor (GPCR) Kinases with Activated GPCRs. J. Biol. Chem. 2011, 286, 7715–7721. [Google Scholar] [CrossRef] [Green Version]
- Gurevich, E.V.; Tesmer, J.J.; Mushegian, A.; Gurevich, V.V. G protein-coupled receptor kinases: More than just kinases and not only for GPCRs. Pharmacol. Ther. 2012, 133, 40–69. [Google Scholar] [CrossRef] [Green Version]
- Lohse, M.J.; Benovic, J.L.; Codina, J.; Caron, M.G.; Lefkowitz, R.J. β-Arrestin: A Protein that Regulates β-adrenergic Receptor Function. Science 1990, 248, 1547–1550. [Google Scholar] [CrossRef]
- Zhou, X.E.; He, Y.; de Waal, P.; Gao, X.; Kang, Y.; Van Eps, N.; Yin, Y.; Pal, K.; Goswami, D.; White, T.A.; et al. Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors. Cell 2017, 170, 457–469.e13. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.; Zhou, X.E.; Gao, X.; He, Y.; Liu, W.; Ishchenko, A.; Barty, A.; White, T.A.; Yefanov, O.; Han, G.W.; et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 2015, 523, 561–567. [Google Scholar] [CrossRef] [Green Version]
- Kang, D.S.; Tian, X.; Benovic, J.L. Role of β-arrestins and arrestin domain-containing proteins in G protein-coupled receptor trafficking. Curr. Opin. Cell Biol. 2013, 27, 63–71. [Google Scholar] [CrossRef] [Green Version]
- Bagnato, A.; Rosanò, L. New Routes in GPCR/β-Arrestin-Driven Signaling in Cancer Progression and Metastasis. Front. Pharmacol. 2019, 10, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, A.K.; Dwivedi-Agnihotri, H. Structure and function of β-arrestins, their emerging role in breast cancer, and potential opportunities for therapeutic manipulation. Adv. Cancer Res. 2020, 145, 139–156. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, S.; Shenoy, S.K. GPCR desensitization: Acute and prolonged phases. Cell. Signal. 2017, 41, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Zaccolo, M.; Zerio, A.; Lobo, M.J. Subcellular Organization of the cAMP Signaling Pathway. Pharmacol. Rev. 2020, 73, 278–309. [Google Scholar] [CrossRef]
- Laubach, W. Who earns how much? Krankenpflege 1989, 43, 647–648. [Google Scholar]
- Brandt, H.; Capulong, Z.L.; Lee, E.Y. Purification and properties of rabbit liver phosphorylase phosphatase. J. Biol. Chem. 1975, 250, 8038–8044. [Google Scholar] [CrossRef]
- Ingebritsen, T.S.; Cohen, P. Protein Phosphatases: Properties and Role in Cellular Regulation. Science 1983, 221, 331–338. [Google Scholar] [CrossRef]
- Terrin, A.; Di Benedetto, G.; Pertegato, V.; Cheung, Y.-F.; Baillie, G.; Lynch, M.J.; Elvassore, N.; Prinz, A.; Herberg, F.W.; Houslay, M.; et al. PGE1 stimulation of HEK293 cells generates multiple contiguous domains with different [cAMP]: Role of compartmentalized phosphodiesterases. J. Cell Biol. 2006, 175, 441–451. [Google Scholar] [CrossRef]
- Leroy, J.; Abi-Gerges, A.; Nikolaev, V.O.; Richter, W.; Lechêne, P.; Mazet, J.-L.; Conti, M.; Fischmeister, R.; Vandecasteele, G. Spatiotemporal Dynamics of β-Adrenergic cAMP Signals and L-Type Ca 2+ Channel Regulation in Adult Rat Ventricular Myocytes. Circ. Res. 2008, 102, 1091–1100. [Google Scholar] [CrossRef] [Green Version]
- Dohlman, H.G.; Thorner, J. RGS Proteins and Signaling by Heterotrimeric G Proteins. J. Biol. Chem. 1997, 272, 3871–3874. [Google Scholar] [CrossRef] [Green Version]
- Tesmer, J.J.G. Structure and Function of Regulator of G Protein Signaling Homology Domains. Prog. Mol. Biol. Transl. Sci. 2009, 86, 75–113. [Google Scholar] [CrossRef]
- Ross, E.M. Coordinating Speed and Amplitude in G-Protein Signaling. Curr. Biol. 2008, 18, R777–R783. [Google Scholar] [CrossRef] [Green Version]
- Hollinger, S. Cellular Regulation of RGS Proteins: Modulators and Integrators of G Protein Signaling. Pharmacol. Rev. 2002, 54, 527–559. [Google Scholar] [CrossRef] [Green Version]
- Masuho, I.; Balaji, S.; Muntean, B.S.; Skamangas, N.K.; Chavali, S.; Tesmer, J.J.; Babu, M.M.; Martemyanov, K.A. A Global Map of G Protein Signaling Regulation by RGS Proteins. Cell 2020, 183, 503–521.e19. [Google Scholar] [CrossRef]
- Chatterjee, T.K.; Fisher, R. Cytoplasmic, Nuclear, and Golgi Localization of RGS Proteins. Evidence for N-terminal and RGS domain sequences as intracellular targeting motifs. J. Biol. Chem. 2000, 275, 24013–24021. [Google Scholar] [CrossRef] [Green Version]
- Bastin, G.; Singh, K.; Dissanayake, K.; Mighiu, A.S.; Nurmohamed, A.; Heximer, S.P. Amino-terminal Cysteine Residues Differentially Influence RGS4 Protein Plasma Membrane Targeting, Intracellular Trafficking, and Function. J. Biol. Chem. 2012, 287, 28966–28974. [Google Scholar] [CrossRef] [Green Version]
- Bastin, G.; Yang, J.Y.; Heximer, S.P. Gαi3-Dependent Inhibition of JNK Activity on Intracellular Membranes. Front. Bioeng. Biotechnol. 2015, 3, 128. [Google Scholar] [CrossRef] [Green Version]
- Wylie, F.; Heimann, K.; Le, T.L.; Brown, D.; Rabnott, G.; Stow, J.L. GAIP, a Gαi-3-binding protein, is associated with Golgi-derived vesicles and protein trafficking. Am. J. Physiol. Physiol. 1999, 276, C497–C506. [Google Scholar] [CrossRef]
- Fischer, T.; Elenko, E.; Wan, L.; Thomas, G.; Farquhar, M.G. Membrane-associated GAIP is a phosphoprotein and can be phosphorylated by clathrin-coated vesicles. Proc. Natl. Acad. Sci. USA 2000, 97, 4040–4045. [Google Scholar] [CrossRef] [Green Version]
- Cismowski, M.J.; Takesono, A.; Ma, C.; Lizano, J.S.; Xie, X.; Fuernkranz, H.; Lanier, S.; Duzic, E. Genetic screens in yeast to identify mammalian nonreceptor modulators of G-protein signaling. Nat. Biotechnol. 1999, 17, 878–883. [Google Scholar] [CrossRef]
- Takesono, A.; Cismowski, M.J.; Ribas, C.; Bernard, M.; Chung, P.; Hazard, S.; Duzic, E.; Lanier, S. Receptor-independent Activators of Heterotrimeric G-protein Signaling Pathways. J. Biol. Chem. 1999, 274, 33202–33205. [Google Scholar] [CrossRef] [Green Version]
- Blumer, J.B.; Lanier, S.M. Activators of G Protein Signaling Exhibit Broad Functionality and Define a Distinct Core Signaling Triad. Mol. Pharmacol. 2013, 85, 388–396. [Google Scholar] [CrossRef] [Green Version]
- Sato, M.; Hiraoka, M.; Suzuki, H.; Bai, Y.; Kurotani, R.; Yokoyama, U.; Okumura, S.; Cismowski, M.J.; Lanier, S.M.; Ishikawa, Y. Identification of Transcription Factor E3 (TFE3) as a Receptor-independent Activator of Gα16: Gene regulation by nuclear Gα subunit and its activator. J. Biol. Chem. 2011, 286, 17766–17776. [Google Scholar] [CrossRef] [Green Version]
- Lo, I.-C.; Gupta, V.; Midde, K.K.; Taupin, V.; Lopez-Sanchez, I.; Kufareva, I.; Abagyan, R.; Randazzo, P.A.; Farquhar, M.G.; Ghosh, P. Activation of Gαi at the Golgi by GIV/Girdin Imposes Finiteness in Arf1 Signaling. Dev. Cell 2015, 33, 189–203. [Google Scholar] [CrossRef] [Green Version]
- Oner, S.S.; Vural, A.; Lanier, S.M. Translocation of Activator of G-protein Signaling 3 to the Golgi Apparatus in Response to Receptor Activation and Its Effect on the trans-Golgi Network. J. Biol. Chem. 2013, 288, 24091–24103. [Google Scholar] [CrossRef] [Green Version]
- Gupta, V.; Bhandari, D.; Leyme, A.; Aznar, N.; Midde, K.K.; Lo, I.-C.; Ear, J.; Niesman, I.; López-Sánchez, I.; Blanco-Canosa, J.B.; et al. GIV/Girdin activates Gαi and inhibits Gαs via the same motif. Proc. Natl. Acad. Sci. USA 2016, 113, E5721–E5730. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, P.; Rangamani, P.; Kufareva, I. The GAPs, GEFs, GDIs and…now, GEMs: New kids on the heterotrimeric G protein signaling block. Cell Cycle 2017, 16, 607–612. [Google Scholar] [CrossRef] [Green Version]
- De Vries, L.; Fischer, T.; Tronchère, H.; Brothers, G.M.; Strockbine, B.; Siderovski, D.; Farquhar, M.G. Activator of G protein signaling 3 is a guanine dissociation inhibitor for Galpha i subunits. Proc. Natl. Acad. Sci. USA 2000, 97, 14364–14369. [Google Scholar] [CrossRef] [Green Version]
- Groves, B.; Gong, Q.; Xu, Z.; Huntsman, C.; Nguyen, C.; Li, D.; Ma, D. A specific role of AGS3 in the surface expression of plasma membrane proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 18103–18108. [Google Scholar] [CrossRef] [Green Version]
- Nie, Z.-W.; Niu, Y.-J.; Zhou, W.; Zhou, D.-J.; Kim, J.-Y.; Cui, X.-S. AGS3-dependent trans-Golgi network membrane trafficking is essential for compaction in mouse embryos. J. Cell Sci. 2020. [Google Scholar] [CrossRef]
- Baschieri, F.; Farhan, H. Crosstalk of small GTPases at the Golgi apparatus. Small GTPases 2012, 3, 80–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, A.; Capalbo, A.; Iyengar, N.R.; Rizzo, R.; di Campli, A.; Di Martino, R.; Monte, M.L.; Beccari, A.R.; Yerudkar, A.; Del Vecchio, C.; et al. Auto-regulation of Secretory Flux by Sensing and Responding to the Folded Cargo Protein Load in the Endoplasmic Reticulum. Cell 2019, 176, 1461–1476.e23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denker, S.P.; McCaffery, J.M.; Palade, G.E.; Insel, P.A.; Farquhar, M.G. Differential distribution of alpha subunits and beta gamma subunits of heterotrimeric G proteins on Golgi membranes of the exocrine pancreas. J. Cell Biol. 1996, 133, 1027–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stow, J.; De Almeida, J.B.; Narula, N.; Holtzman, E.J.; Ercolani, L.; Ausiello, D.A. A heterotrimeric G protein, G alpha i-3, on Golgi membranes regulates the secretion of a heparan sulfate proteoglycan in LLC-PK1 epithelial cells. J. Cell Biol. 1991, 114, 1113–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stow, J.L.; Heimann, K. Vesicle budding on Golgi membranes: Regulation by G proteins and myosin motors. Biochim. Biophys. Acta 1998, 1404, 161–171. [Google Scholar] [CrossRef] [Green Version]
- Cancino, J.; Capalbo, A.; DI Campli, A.; Giannotta, M.; Rizzo, R.; Jung, J.E.; Di Martino, R.; Persico, M.; Heinklein, P.; Sallese, M.; et al. Control Systems of Membrane Transport at the Interface between the Endoplasmic Reticulum and the Golgi. Dev. Cell 2014, 30, 280–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giannotta, M.; Ruggiero, C.; Grossi, M.; Cancino, J.; Capitani, M.; Pulvirenti, T.; Consoli, G.M.L.; Geraci, C.; Fanelli, F.; Luini, A.; et al. The KDEL receptor couples to Gαq/11to activate Src kinases and regulate transport through the Golgi. EMBO J. 2012, 31, 2869–2881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solis, G.P.; Bilousov, O.; Koval, A.; Lüchtenborg, A.-M.; Lin, C.; Katanaev, V.L. Golgi-Resident Gαo Promotes Protrusive Membrane Dynamics. Cell 2017, 170, 939–955.e24. [Google Scholar] [CrossRef] [Green Version]
- Pulvirenti, T.; Giannotta, M.; Capestrano, M.; Capitani, M.; Pisanu, A.; Polishchuk, R.; Pietro, E.S.; Beznoussenko, G.V.; Mironov, A.A.; Turacchio, G.; et al. A traffic-activated Golgi-based signalling circuit coordinates the secretory pathway. Nat. Cell Biol. 2008, 10, 912–922. [Google Scholar] [CrossRef]
- Di Martino, R.; Capalbo, A.; Sticco, L.; Varavallo, A.; Kunnathully, V.; De Luca, V.; Iyengar, N.R.; Monte, M.L.; Henklein, P.; Cancino, J.; et al. Autoregulatory circuit regulating basolateral cargo export from the TGN: Role of the orphan receptor GPRC5A in PKD signaling and cell polarity. bioRxiv 2020. [Google Scholar] [CrossRef]
- Jamora, C.; Yamanouye, N.; Van Lint, J.; Laudenslager, J.; Vandenheede, J.R.; Faulkner, D.; Malhotra, V. Gβγ-Mediated Regulation of Golgi Organization Is through the Direct Activation of Protein Kinase D. Cell 1999, 98, 59–68. [Google Scholar] [CrossRef] [Green Version]
- Irannejad, R.; Wedegaertner, P.B. Regulation of Constitutive Cargo Transport from the trans-Golgi Network to Plasma Membrane by Golgi-localized G Protein βγ Subunits. J. Biol. Chem. 2010, 285, 32393–32404. [Google Scholar] [CrossRef] [Green Version]
- Khater, M.; Bryant, C.N.; Wu, G. Gβγ translocation to the Golgi apparatus activates ARF1 to spatiotemporally regulate G protein–coupled receptor signaling to MAPK. J. Biol. Chem. 2021, 296, 100805. [Google Scholar] [CrossRef]
- Akgoz, M.; Kalyanaraman, V.; Gautam, N.; Yew, K.-H.; Hembree, M.; Prasadan, K.; Preuett, B.; McFall, C.; Benjes, C.; Crowley, A.; et al. Receptor-mediated Reversible Translocation of the G Protein βγ Complex from the Plasma Membrane to the Golgi Complex. J. Biol. Chem. 2004, 279, 51541–51544. [Google Scholar] [CrossRef] [Green Version]
- Saini, D.K.; Karunarathne, W.K.A.; Angaswamy, N.; Cho, J.-H.; Kalyanaraman, V.; Gautam, N. Regulation of Golgi structure and secretion by receptor-induced G protein complex translocation. Proc. Natl. Acad. Sci. USA 2010, 107, 11417–11422. [Google Scholar] [CrossRef] [Green Version]
- Bechler, M.E.; Brown, W.J. Gβ1γ2 activates phospholipase A2-dependent Golgi membrane tubule formation. Front. Cell Dev. Biol. 2014, 2, 4. [Google Scholar] [CrossRef] [Green Version]
- Conn, P.M.; Ulloa-Aguirre, A.; Ito, J.; Janovick, J.A. G Protein-Coupled Receptor Trafficking in Health and Disease: Lessons Learned to Prepare for Therapeutic Mutant Rescue in Vivo. Pharmacol. Rev. 2007, 59, 225–250. [Google Scholar] [CrossRef] [Green Version]
- Dong, C.; Filipeanu, C.M.; Duvernay, M.T.; Wu, G. Regulation of G protein-coupled receptor export trafficking. Biochim. Biophys. Acta 2007, 1768, 853–870. [Google Scholar] [CrossRef] [Green Version]
- Ulloa-Aguirre, A.; Janovick, J.A.; Miranda, A.L.; Conn, P.M. G-Protein-Coupled Receptor Trafficking: Understanding the Chemical Basis of Health and Disease. ACS Chem. Biol. 2006, 1, 631–638. [Google Scholar] [CrossRef]
- Ulloa-Aguirre, A.; Janovick, J.A.; Leaños-Miranda, A.; Conn, P.M. Misrouted cell surface GnRH receptors as a disease aetiology for congenital isolated hypogonadotrophic hypogonadism. Hum. Reprod. Update 2004, 10, 177–192. [Google Scholar] [CrossRef] [Green Version]
- Hicks, S.W.; Horn, T.A.; McCaffery, J.M.; Zuckerman, D.M.; Machamer, C.E. Golgin-160 Promotes Cell Surface Expression of the Beta-1 Adrenergic Receptor. Traffic 2006, 7, 1666–1677. [Google Scholar] [CrossRef]
- Zhang, M.; Davis, J.E.; Li, C.; Gao, J.; Huang, W.; Lambert, N.A.; Terry, A.; Wu, G. GGA3 Interacts with a G Protein-Coupled Receptor and Modulates Its Cell Surface Export. Mol. Cell. Biol. 2016, 36, 1152–1163. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Huang, W.; Gao, J.; Terry, A.V.; Wu, G. Regulation of α2B-Adrenergic Receptor Cell Surface Transport by GGA1 and GGA2. Sci. Rep. 2016, 6, 37921. [Google Scholar] [CrossRef]
- Wu, G.; Davis, J.E.; Zhang, M. Regulation of α2B-Adrenerigc Receptor Export Trafficking by Specific Motifs. Prog. Mol. Biol. Transl. Sci. 2015, 132, 227–244. [Google Scholar] [CrossRef] [Green Version]
- Feinstein, T.N.; Wehbi, V.L.; Ardura, J.A.; Wheeler, D.S.; Ferrandon, S.; Gardella, T.J.; Vilardaga, J.-P. Retromer terminates the generation of cAMP by internalized PTH receptors. Nat. Chem. Biol. 2011, 7, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Godbole, A.; Lyga, S.; Lohse, M.J.; Calebiro, D. Internalized TSH receptors en route to the TGN induce local Gs-protein signaling and gene transcription. Nat. Commun. 2017, 8, 443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nezhady, M.A.M.; Rivera, J.C.; Chemtob, S. Location Bias as Emerging Paradigm in GPCR Biology and Drug Discovery. iScience 2020, 23, 101643. [Google Scholar] [CrossRef]
- Steyaert, J.; Kobilka, B.K. Nanobody stabilization of G protein-coupled receptor conformational states. Curr. Opin. Struct. Biol. 2011, 21, 567–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irannejad, R.; Pessino, V.; Mika, D.; Huang, B.; Wedegaertner, P.B.; Conti, M.; Von Zastrow, M. Functional selectivity of GPCR-directed drug action through location bias. Nat. Chem. Biol. 2017, 13, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Bathe-Peters, M.; Gmach, P.; Boltz, H.-H.; Einsiedel, J.; Gotthardt, M.; Hübner, H.; Gmeiner, P.; Lohse, M.J.; Annibale, P. Visualization of β-adrenergic receptor dynamics and differential localization in cardiomyocytes. Proc. Natl. Acad. Sci. USA 2021, 118, e2101119118. [Google Scholar] [CrossRef] [PubMed]
- Christopoulos, A.; Christopoulos, G.; Morfis, M.; Udawela, M.; Laburthe, M.; Couvineau, A.; Kuwasako, K.; Tilakaratne, N.; Sexton, P.M. Novel Receptor Partners and Function of Receptor Activity-modifying Proteins. J. Biol. Chem. 2003, 278, 3293–3297. [Google Scholar] [CrossRef] [Green Version]
- Chan, L.F.; Webb, T.R.; Chung, T.-T.; Meimaridou, E.; Cooray, S.N.; Guasti, L.; Chapple, J.P.; Egertová, M.; Elphick, M.R.; Cheetham, M.E.; et al. MRAP and MRAP2 are bidirectional regulators of the melanocortin receptor family. Proc. Natl. Acad. Sci. USA 2009, 106, 6146–6151. [Google Scholar] [CrossRef] [Green Version]
- Saito, H.; Kubota, M.; Roberts, R.W.; Chi, Q.; Matsunami, H. RTP Family Members Induce Functional Expression of Mammalian Odorant Receptors. Cell 2004, 119, 679–691. [Google Scholar] [CrossRef] [Green Version]
- Jong, Y.-J.I.; Kumar, V.; O’Malley, K.L. Intracellular Metabotropic Glutamate Receptor 5 (mGluR5) Activates Signaling Cascades Distinct from Cell Surface Counterparts. J. Biol. Chem. 2009, 284, 35827–35838. [Google Scholar] [CrossRef] [Green Version]
- Puri, N.M.; Romano, G.R.; Lin, T.-Y.; Mai, Q.N.; Irannejad, R. The OCT2 Transporter Regulates Dopamine D1 Receptor Signaling at the Golgi Apparatus. bioRxiv 2021. [Google Scholar] [CrossRef]
- Gasser, P.J.; Hurley, M.M.; Chan, J.; Pickel, V.M. Organic cation transporter 3 (OCT3) is localized to intracellular and surface membranes in select glial and neuronal cells within the basolateral amygdaloid complex of both rats and mice. Brain Struct. Funct. 2016, 222, 1913–1928. [Google Scholar] [CrossRef]
- Zwart, R.; Verhaagh, S.; Buitelaar, M.; Popp-Snijders, C.; Barlow, D.P. Impaired Activity of the Extraneuronal Monoamine Transporter System Known as Uptake-2 in Orct3/Slc22a3-Deficient Mice. Mol. Cell. Biol. 2001, 21, 4188–4196. [Google Scholar] [CrossRef] [Green Version]
- Revankar, C.M.; Cimino, D.F.; Sklar, L.A.; Arterburn, J.B.; Prossnitz, E.R. A Transmembrane Intracellular Estrogen Receptor Mediates Rapid Cell Signaling. Science 2005, 307, 1625–1630. [Google Scholar] [CrossRef] [Green Version]
- Purgert, C.A.; Izumi, Y.; Jong, Y.-J.I.; Kumar, V.; Zorumski, C.F.; O’Malley, K.L. Intracellular mGluR5 Can Mediate Synaptic Plasticity in the Hippocampus. J. Neurosci. 2014, 34, 4589–4598. [Google Scholar] [CrossRef] [Green Version]
- Cunningham, M.R.; McIntosh, K.A.; Pediani, J.; Robben, J.; Cooke, A.E.; Nilsson, M.; Gould, G.W.; Mundell, S.; Milligan, G.; Plevin, R. Novel Role for Proteinase-activated Receptor 2 (PAR2) in Membrane Trafficking of Proteinase-activated Receptor 4 (PAR4). J. Biol. Chem. 2012, 287, 16656–16669. [Google Scholar] [CrossRef] [Green Version]
- Stoeber, M.; Jullié, D.; Lobingier, B.T.; Laeremans, T.; Steyaert, J.; Schiller, P.W.; Manglik, A.; von Zastrow, M. A Genetically Encoded Biosensor Reveals Location Bias of Opioid Drug Action. Neuron 2018, 98, 963–976.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Rigoutsos, I. The emerging roles of GPRC5A in diseases. Oncoscience 2014, 1, 765–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadara, H.; Fujimoto, J.; Men, T.; Ye, X.; Lotan, D.; Lee, J.-S.; Lotan, R. A Gprc5a Tumor Suppressor Loss of Expression Signature Is Conserved, Prevalent, and Associated with Survival in Human Lung Adenocarcinomas. Neoplasia 2010, 12, 499-IN8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, Q.; Fujimoto, J.; Men, T.; Ye, X.; Deng, J.; Lacroix, L.; Clifford, J.L.; Mao, L.; Van Pelt, C.S.; Lee, J.J.; et al. Identification of the Retinoic Acid-Inducible Gprc5a as a New Lung Tumor Suppressor Gene. J. Natl. Cancer Inst. 2007, 99, 1668–1682. [Google Scholar] [CrossRef]
- Kurtenbach, S.; Mayer, C.; Pelz, T.; Hatt, H.; Leese, F.; Neuhaus, E.M. Molecular evolution of a chordate specific family of G protein-coupled receptors. BMC Evol. Biol. 2011, 11, 234. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Greimel, P.; Hirabayashi, Y. GPRC5B-Mediated Sphingomyelin Synthase 2 Phosphorylation Plays a Critical Role in Insulin Resistance. iScience 2018, 8, 250–266. [Google Scholar] [CrossRef]
- Watkins, L.R.; Orlandi, C. Orphan G Protein Coupled Receptors in Affective Disorders. Genes 2020, 11, 694. [Google Scholar] [CrossRef]
- Kim, K.A.; von Zastrow, M. Neurotrophin-regulated sorting of opioid receptors in the biosynthetic pathway of neurosecretory cells. J. Neurosci. 2003, 23, 2075–2085. [Google Scholar] [CrossRef]
- Shiwarski, D.J.; Tipton, A.; Giraldo, M.D.; Schmidt, B.F.; Gold, M.S.; Pradhan, A.A.; Puthenveedu, M.A. A PTEN-Regulated Checkpoint Controls Surface Delivery of δ Opioid Receptors. J. Neurosci. 2017, 37, 3741–3752. [Google Scholar] [CrossRef] [Green Version]
- Laporte, S.A.; Oakley, R.H.; Zhang, J.; Holt, J.A.; Ferguson, S.S.G.; Caron, M.G.; Barak, L.S. The 2-adrenergic receptor/ arrestin complex recruits the clathrin adaptor AP-2 during endocytosis. Proc. Natl. Acad. Sci. USA 1999, 96, 3712–3717. [Google Scholar] [CrossRef] [Green Version]
- Pierce, K.L.; Premont, R.T.; Lefkowitz, R.J. Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 2002, 3, 639–650. [Google Scholar] [CrossRef]
- Drake, M.T.; Shenoy, S.K.; Lefkowitz, R.J. Trafficking of G Protein–Coupled Receptors. Circ. Res. 2006, 99, 570–582. [Google Scholar] [CrossRef]
- DeWire, S.M.; Ahn, S.; Lefkowitz, R.J.; Shenoy, S.K. β-Arrestins and Cell Signaling. Annu. Rev. Physiol. 2007, 69, 483–510. [Google Scholar] [CrossRef] [Green Version]
- Sorkin, A.; von Zastrow, M. Endocytosis and signalling: Intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 2009, 10, 609–622. [Google Scholar] [CrossRef] [Green Version]
- Cahill, T.J.; Thomsen, A.; Tarrasch, J.T.; Plouffe, B.; Nguyen, A.; Yang, F.; Huang, L.-Y.; Kahsai, A.W.; Bassoni, D.L.; Gavino, B.J.; et al. Distinct conformations of GPCR–β-arrestin complexes mediate desensitization, signaling, and endocytosis. Proc. Natl. Acad. Sci. USA 2017, 114, 2562–2567. [Google Scholar] [CrossRef] [Green Version]
- Irannejad, R.; Tsvetanova, N.G.; Lobingier, B.T.; von Zastrow, M. Effects of endocytosis on receptor-mediated signaling. Curr. Opin. Cell Biol. 2015, 35, 137–143. [Google Scholar] [CrossRef] [Green Version]
- Ferrandon, S.; Feinstein, T.; Castro, M.; Wang, B.; Bouley, R.; Potts, J.T.; Gardella, T.J.; Vilardaga, J.-P. Sustained cyclic AMP production by parathyroid hormone receptor endocytosis. Nat. Chem. Biol. 2009, 5, 734–742. [Google Scholar] [CrossRef]
- Jensen, D.D.; Lieu, T.; Halls, M.L.; Veldhuis, N.A.; Imlach, W.L.; Mai, Q.N.; Poole, D.P.; Quach, T.; Aurelio, L.; Conner, J.; et al. Neurokinin 1 receptor signaling in endosomes mediates sustained nociception and is a viable therapeutic target for prolonged pain relief. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef]
- Irannejad, R.; Tomshine, J.C.; Tomshine, J.R.; Chevalier, M.; Mahoney, J.P.; Steyaert, J.; Rasmussen, S.; Sunahara, R.K.; El-Samad, H.; Huang, B.; et al. Conformational biosensors reveal GPCR signalling from endosomes. Nature 2013, 495, 534–538. [Google Scholar] [CrossRef] [Green Version]
- Kotowski, S.J.; Hopf, F.W.; Seif, T.; Bonci, A.; von Zastrow, M. Endocytosis Promotes Rapid Dopaminergic Signaling. Neuron 2011, 71, 278–290. [Google Scholar] [CrossRef] [Green Version]
- Calebiro, D.; Nikolaev, V.O.; Gagliani, M.C.; de Filippis, T.; Dees, C.; Tacchetti, C.; Persani, L.; Lohse, M.J. Persistent cAMP-Signals Triggered by Internalized G-Protein–Coupled Receptors. PLOS Biol. 2009, 7, e1000172. [Google Scholar] [CrossRef] [Green Version]
- Bowman, S.L.; Shiwarski, D.; Puthenveedu, M.A. Distinct G protein–coupled receptor recycling pathways allow spatial control of downstream G protein signaling. J. Cell Biol. 2016, 214, 797–806. [Google Scholar] [CrossRef] [Green Version]
- Feinstein, T.N.; Yui, N.; Webber, M.; Wehbi, V.L.; Stevenson, H.P.; King, J.D.; Hallows, K.R.; Brown, D.; Bouley, R.; Vilardaga, J.-P. Noncanonical Control of Vasopressin Receptor Type 2 Signaling by Retromer and Arrestin. J. Biol. Chem. 2013, 288, 27849–27860. [Google Scholar] [CrossRef] [Green Version]
- Thomsen, A.; Plouffe, B.; Cahill, T.J.; Shukla, A.; Tarrasch, J.T.; Dosey, A.M.; Kahsai, A.W.; Strachan, R.T.; Pani, B.; Mahoney, J.P.; et al. GPCR-G Protein-β-Arrestin Super-Complex Mediates Sustained G Protein Signaling. Cell 2016, 166, 907–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, A.; Thomsen, A.; Cahill, T.J.; Huang, R.; Huang, L.-Y.; Marcink, T.; Clarke, O.B.; Heissel, S.; Masoudi, A.; Ben-Hail, D.; et al. Structure of an endosomal signaling GPCR–G protein–β-arrestin megacomplex. Nat. Struct. Mol. Biol. 2019, 26, 1123–1131. [Google Scholar] [CrossRef]
- Koshimizu, T.-A.; Nakamura, K.; Egashira, N.; Hiroyama, M.; Nonoguchi, H.; Tanoue, A. Vasopressin V1a and V1b Receptors: From Molecules to Physiological Systems. Physiol. Rev. 2012, 92, 1813–1864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juul, K.V.; Bichet, D.-G.; Nielsen, S.; Nørgaard, J.P. The physiological and pathophysiological functions of renal and extrarenal vasopressin V2 receptors. Am. J. Physiol. Physiol. 2014, 306, F931–F940. [Google Scholar] [CrossRef]
- Jonassen, T.E.N.; Nielsen, S.; Christensen, S.; Petersen, J.S. Decreased vasopressin-mediated renal water reabsorption in rats with compensated liver cirrhosis. Am. J. Physiol. Content 1998, 275, F216–F225. [Google Scholar] [CrossRef]
- Bankir, L.; Bichet, D.G.; Bouby, N. Vasopressin V2 receptors, ENaC, and sodium reabsorption: A risk factor for hypertension? Am. J. Physiol. Physiol. 2010, 299, F917–F928. [Google Scholar] [CrossRef] [Green Version]
- Henderson, K.K.; Byron, K.L. Vasopressin-induced vasoconstriction: Two concentration-dependent signaling pathways. J. Appl. Physiol. 2007, 102, 1402–1409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina, P.; Acuña, A.; Martínez-León, J.B.; Otero, E.; Vila, J.M.; Aldasoro, M.; Lluch, S. Arginine Vasopressin Enhances Sympathetic Constriction Through the V 1 Vasopressin Receptor in Human Saphenous Vein. Circulation 1998, 97, 865–870. [Google Scholar] [CrossRef] [Green Version]
- Holmes, C.L.; Landry, D.W.; Granton, J.T. Science Review: Vasopressin and the cardiovascular system part 1—Receptor physiology. Crit. Care 2003, 7, 427–434. [Google Scholar] [CrossRef] [Green Version]
- Goldsmith, S.R. Arginine vasopressin antagonism in heart failure: Current status and possible new directions. J. Cardiol. 2019, 74, 49–52. [Google Scholar] [CrossRef]
- Schweiger, T.A.; Zdanowicz, M.M. Vasopressin-receptor antagonists in heart failure. Am. J. Health Pharm. 2008, 65, 807–817. [Google Scholar] [CrossRef] [Green Version]
- Wasilewski, M.A.; Myers, V.D.; Recchia, F.A.; Feldman, A.M.; Tilley, D.G. Arginine vasopressin receptor signaling and functional outcomes in heart failure. Cell. Signal. 2015, 28, 224–233. [Google Scholar] [CrossRef] [Green Version]
- Manglik, A.; Kobilka, B.K.; Steyaert, J. Nanobodies to Study G Protein–Coupled Receptor Structure and Function. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 19–37. [Google Scholar] [CrossRef] [Green Version]
- Westfield, G.H.; Rasmussen, S.G.F.; Su, M.; Dutta, S.; DeVree, B.T.; Chung, K.Y.; Calinski, D.; Velez-Ruiz, G.; Oleskie, A.N.; Pardon, E.; et al. Structural flexibility of the G s -helical domain in the 2-adrenoceptor Gs complex. Proc. Natl. Acad. Sci. USA 2011, 108, 16086–16091. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, S.; Choi, H.-J.; Fung, J.J.; Pardon, E.; Casarosa, P.; Chae, P.S.; DeVree, B.; Rosenbaum, D.M.; Thian, F.S.; Kobilka, T.S.; et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 2011, 469, 175–180. [Google Scholar] [CrossRef] [Green Version]
- Sposini, S.; Jean-Alphonse, F.; Ayoub, M.A.; Oqua, A.; West, C.; Lavery, S.; Brosens, J.; Reiter, E.; Hanyaloglu, A.C. Integration of GPCR Signaling and Sorting from Very Early Endosomes via Opposing APPL1 Mechanisms. Cell Rep. 2017, 21, 2855–2867. [Google Scholar] [CrossRef] [Green Version]
- Gorvin, C.; Rogers, A.; Hastoy, B.; Tarasov, A.; Frost, M.; Sposini, S.; Inoue, A.; Whyte, M.P.; Rorsman, P.; Hanyaloglu, A.C.; et al. AP2σ Mutations Impair Calcium-Sensing Receptor Trafficking and Signaling, and Show an Endosomal Pathway to Spatially Direct G-Protein Selectivity. Cell Rep. 2018, 22, 1054–1066. [Google Scholar] [CrossRef] [Green Version]
- Jimenez-Vargas, N.N.; Gong, J.; Wisdom, M.J.; Jensen, D.D.; Latorre, R.; Hegron, A.; Teng, S.; DiCello, J.J.; Rajasekhar, P.; Veldhuis, N.A.; et al. Endosomal signaling of delta opioid receptors is an endogenous mechanism and therapeutic target for relief from inflammatory pain. Proc. Natl. Acad. Sci. USA 2020, 117, 15281–15292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-H.; Barr, V.A.; Mo, Y.; Rojkova, A.M.; Liu, S.; Simonds, W.F. Nuclear Localization of G Protein β5 and Regulator of G Protein Signaling 7 in Neurons and Brain. J. Biol. Chem. 2001, 276, 10284–10289. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, S.; Kawamura, K.; James, T.N. Intracellular distribution of adenylate cyclase in human cardiocytes determined by electron microscopic cytochemistry. Microsc. Res. Tech. 1998, 40, 479–487. [Google Scholar] [CrossRef]
- Schulze, W.; Buchwalow, I.B. Adenylyl cyclase in the heart: An enzymocytochemical and immunocytochemical approach. Microsc. Res. Tech. 1998, 40, 473–478. [Google Scholar] [CrossRef]
- Schievella, A.R.; Regier, M.K.; Smith, W.L.; Lin, L.-L. Calcium-mediated Translocation of Cytosolic Phospholipase A2 to the Nuclear Envelope and Endoplasmic Reticulum. J. Biol. Chem. 1995, 270, 30749–30754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.G.; Park, D.; Rhee, S.G. The Role of Carboxyl-terminal Basic Amino Acids in Gqα-dependent Activation, Particulate Association, and Nuclear Localization of Phospholipase C-β1. J. Biol. Chem. 1996, 271, 21187–21192. [Google Scholar] [CrossRef] [Green Version]
- Pendergrass, K.D.; Gwathmey, T.M.; Michalek, R.D.; Grayson, J.M.; Chappell, M.C. The angiotensin II–AT1 receptor stimulates reactive oxygen species within the cell nucleus. Biochem. Biophys. Res. Commun. 2009, 384, 149–154. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Yang, H.; Shaw, G.; Raizada, M.K. Angiotensin II-Induced Nuclear Targeting of the Angiotensin Type 1 (AT1) Receptor in Brain Neurons*. Endocrinology 1998, 139, 365–375. [Google Scholar] [CrossRef]
- Gwathmey, T.M.; Shaltout, H.A.; Pendergrass, K.D.; Pirro, N.T.; Figueroa, J.P.; Rose, J.C.; Diz, D.I.; Chappell, M.C. Nuclear angiotensin II type 2 (AT2) receptors are functionally linked to nitric oxide production. Am. J. Physiol. Physiol. 2009, 296, F1484–F1493. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, M.; Peri, K.G.; Almazan, G.; Ribeiro-Da-Silva, A.; Shichi, H.; Durocher, Y.; Abramovitz, M.; Hou, X.; Varma, D.R.; Chemtob, S. Nuclear localization of prostaglandin E2 receptors. Proc. Natl. Acad. Sci. USA 1998, 95, 15792–15797. [Google Scholar] [CrossRef] [Green Version]
- Bhosle, V.K.; Rivera, J.C.; Zhou, T.; Omri, S.; Sanchez, M.; Hamel, D.; Zhu, T.; Rouget, R.; Al Rabea, A.; Hou, X.; et al. Nuclear localization of platelet-activating factor receptor controls retinal neovascularization. Cell Discov. 2016, 2, 16017. [Google Scholar] [CrossRef] [Green Version]
- Wright, C.D.; Chen, Q.; Baye, N.L.; Huang, Y.; Healy, C.L.; Kasinathan, S.; O’Connell, T.D. Nuclear α1-Adrenergic Receptors Signal Activated ERK Localization to Caveolae in Adult Cardiac Myocytes. Circ. Res. 2008, 103, 992–1000. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.C.; O’connell, T.D. Nuclear Compartmentalization of α1-Adrenergic Receptor Signaling in Adult Cardiac Myocytes. J. Cardiovasc. Pharmacol. 2015, 65, 91–100. [Google Scholar] [CrossRef]
- Vincent, K.; Cornea, V.M.; Jong, Y.-J.I.; Laferrière, A.; Kumar, N.; Mickeviciute, A.; Fung, J.S.T.; Bandegi, P.; Ribeiro-Da-Silva, A.; O’Malley, K.L.; et al. Intracellular mGluR5 plays a critical role in neuropathic pain. Nat. Commun. 2016, 7, 10604. [Google Scholar] [CrossRef] [Green Version]
- Um, J.W.; Kaufman, A.C.; Kostylev, M.; Heiss, J.K.; Stagi, M.; Takahashi, H.; Kerrisk, M.E.; Vortmeyer, A.; Wisniewski, T.; Koleske, A.J.; et al. Metabotropic Glutamate Receptor 5 Is a Coreceptor for Alzheimer Aβ Oligomer Bound to Cellular Prion Protein. Neuron 2013, 79, 887–902. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, A.; Esseltine, J.L.; DeVries, R.A.; Cregan, S.P.; Ferguson, S.S.G. Metabotropic glutamate receptor 5 knockout reduces cognitive impairment and pathogenesis in a mouse model of Alzheimer’s disease. Mol. Brain 2014, 7, 40. [Google Scholar] [CrossRef] [Green Version]
- Boivin, B.; Lavoie, C.; Vaniotis, G.; Baragli, A.; Villeneuve, L.-R.; Ethier, N.; Trieu, P.; Allen, B.G.; Hébert, T.E. Functional β-adrenergic receptor signalling on nuclear membranes in adult rat and mouse ventricular cardiomyocytes. Cardiovasc. Res. 2006, 71, 69–78. [Google Scholar] [CrossRef]
- Vaniotis, G.; Del Duca, D.; Trieu, P.; Rohlicek, C.V.; Hébert, T.E.; Allen, B.G. Nuclear β-adrenergic receptors modulate gene expression in adult rat heart. Cell. Signal. 2011, 23, 89–98. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liccardo, F.; Luini, A.; Di Martino, R. Endomembrane-Based Signaling by GPCRs and G-Proteins. Cells 2022, 11, 528. https://doi.org/10.3390/cells11030528
Liccardo F, Luini A, Di Martino R. Endomembrane-Based Signaling by GPCRs and G-Proteins. Cells. 2022; 11(3):528. https://doi.org/10.3390/cells11030528
Chicago/Turabian StyleLiccardo, Federica, Alberto Luini, and Rosaria Di Martino. 2022. "Endomembrane-Based Signaling by GPCRs and G-Proteins" Cells 11, no. 3: 528. https://doi.org/10.3390/cells11030528