Structural Aspects of LIMK Regulation and Pharmacology
Abstract
:1. LIMKs Regulate Actin Dynamics
2. The Conformational Space of the LIMK Kinase Domain
2.1. Basic Features of the Kinase Fold
2.2. The Active LIMK Kinase Conformation
2.3. Inactive LIMK Kinase Conformations
2.4. Kinase Activation by T508 Phosphorylation
3. The Unusual LIMK Catalytic Mechanism
3.1. Catalytic Mechanism and Fidelity Control in Conventional Kinases
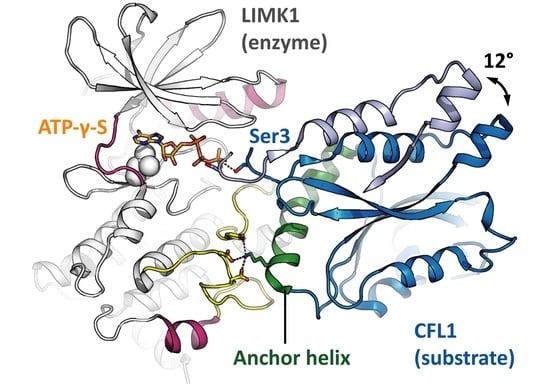
3.2. LIMK Substrate Recognition and ‘Rock-and-Poke’ Mechanism
3.3. Role of the LIM Domains and the PDZ Domain
4. Pharmacological Targeting of LIMKs
4.1. LIMKs Are Involved in Human Disease
4.2. LIMK Inhibitors Binding to the Active Site
4.3. Inhibitors Interfering with Substrate Recognition
4.4. PROTACs to Induce LIMK Degradation
4.5. Outlook—Isoform-Specific LIMK Inhibitors
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blanchoin, L.; Boujemaa-Paterski, R.; Sykes, C.; Plastino, J. Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 2014, 94, 235–263. [Google Scholar] [CrossRef] [Green Version]
- Kanellos, G.; Frame, M.C. Cellular functions of the ADF/cofilin family at a glance. J. Cell Sci. 2016, 129, 3211–3218. [Google Scholar] [CrossRef] [Green Version]
- Yang, N.; Higuchi, O.; Ohashi, K.; Nagata, K.; Wada, A.; Kangawa, K.; Nishida, E.; Mizuno, K. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 1998, 393, 809–812. [Google Scholar] [CrossRef]
- Niwa, R.; Nagata-Ohashi, K.; Takeichi, M.; Mizuno, K.; Uemura, T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell 2002, 108, 233–246. [Google Scholar] [CrossRef] [Green Version]
- Ben Zablah, Y.; Zhang, H.; Gugustea, R.; Jia, Z. LIM-Kinases in Synaptic Plasticity, Memory, and Brain Diseases. Cells 2021, 10, 2079. [Google Scholar] [CrossRef] [PubMed]
- Vallée, B.; Cuberos, H.; Doudeau, M.; Godin, F.; Gosset, D.; Vourc’h, P.; Andres, C.R.; Bénédetti, H. LIMK2-1, a new isoform of human LIMK2, regulates actin cytoskeleton remodeling via a different signaling pathway than that of its two homologs, LIMK2a and LIMK2b. Biochem. J. 2018, 475, 3745–3761. [Google Scholar] [CrossRef] [PubMed]
- McClendon, C.L.; Kornev, A.P.; Gilson, M.K.; Taylor, S.S. Dynamic architecture of a protein kinase. Proc. Natl. Acad. Sci. USA 2014, 111, E4623–E4631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodwin, N.C.; Cianchetta, G.; Burgoon, H.A.; Healy, J.; Mabon, R.; Strobel, E.D.; Allen, J.; Wang, S.; Hamman, B.D.; Rawlins, D.B. Discovery of a Type III Inhibitor of LIM Kinase 2 That Binds in a DFG-Out Conformation. ACS Med. Chem. Lett. 2015, 6, 53–57. [Google Scholar] [CrossRef] [Green Version]
- Hamill, S.; Lou, H.J.; Turk, B.E.; Boggon, T.J. Structural Basis for Noncanonical Substrate Recognition of Cofilin/ADF Proteins by LIM Kinases. Mol. Cell 2016, 62, 397–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salah, E.; Chatterjee, D.; Beltrami, A.; Tumber, A.; Preuss, F.; Canning, P.; Chaikuad, A.; Knaus, P.; Knapp, S.; Bullock, A.N.; et al. Lessons from LIMK1 enzymology and their impact on inhibitor design. Biochem. J. 2019, 476, 3197–3209. [Google Scholar] [CrossRef] [PubMed]
- Kornev, A.P.; Taylor, S.S. Dynamics-Driven Allostery in Protein Kinases. Trends Biochem. Sci. 2015, 40, 628–647. [Google Scholar] [CrossRef] [Green Version]
- Bernard, O. Lim kinases, regulators of actin dynamics. Int. J. Biochem. Cell Biol. 2007, 39, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.W.; Olson, M.F. LIM kinases: Function, regulation and association with human disease. J. Mol. Med. 2007, 85, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Edwards, D.C.; Sanders, L.C.; Bokoch, G.M.; Gill, G.N. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat. Cell Biol. 1999, 1, 253–259. [Google Scholar] [CrossRef]
- Maekawa, M.; Ishizaki, T.; Boku, S.; Watanabe, N.; Fujita, A.; Iwamatsu, A.; Obinata, T.; Ohashi, K.; Mizuno, K.; Narumiya, S. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science 1999, 285, 895–898. [Google Scholar] [CrossRef] [PubMed]
- Sumi, T.; Matsumoto, K.; Shibuya, A.; Nakamura, T. Activation of LIM kinases by myotonic dystrophy kinase-related Cdc42-binding kinase alpha. J. Biol. Chem. 2001, 276, 23092–23096. [Google Scholar] [CrossRef] [Green Version]
- Ohashi, K.; Nagata, K.; Maekawa, M.; Ishizaki, T.; Narumiya, S.; Mizuno, K. Rho-associated kinase ROCK activates LIM-kinase 1 by phosphorylation at threonine 508 within the activation loop. J. Biol. Chem. 2000, 275, 3577–3582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chetty, A.K.; Sexton, J.A.; Ha, B.H.; Turk, B.E.; Boggon, T.J. Recognition of physiological phosphorylation sites by p21-activated kinase 4. J. Struct. Biol. 2020, 211, 107553. [Google Scholar] [CrossRef]
- Miller, C.J.; Turk, B.E. Homing in: Mechanisms of Substrate Targeting by Protein Kinases. Trends Biochem. Sci. 2018, 43, 380–394. [Google Scholar] [CrossRef]
- Yang, J.; Cron, P.; Good, V.M.; Thompson, V.; Hemmings, B.A.; Barford, D. Crystal structure of an activated Akt/protein kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat. Struct. Biol. 2002, 9, 940–944. [Google Scholar] [CrossRef]
- Miller, M.L.; Jensen, L.J.; Diella, F.; Jørgensen, C.; Tinti, M.; Li, L.; Hsiung, M.; Parker, S.A.; Bordeaux, J.; Sicheritz-Ponten, T.; et al. Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal. 2008, 1, ra2. [Google Scholar] [CrossRef]
- Bradley, D.; Beltrao, P. Evolution of protein kinase substrate recognition at the active site. PLoS Biol. 2019, 17, e3000341. [Google Scholar] [CrossRef] [Green Version]
- Pröschel, C.; Blouin, M.J.; Gutowski, N.J.; Ludwig, R.; Noble, M. Limk1 is predominantly expressed in neural tissues and phosphorylates serine, threonine and tyrosine residues in vitro. Oncogene 1995, 11, 1271–1281. [Google Scholar] [PubMed]
- Chen, C.; Ha, B.H.; Thévenin, A.F.; Lou, H.J.; Zhang, R.; Yip, K.Y.; Peterson, J.R.; Gerstein, M.; Kim, P.M.; Filippakopoulos, P.; et al. Identification of a major determinant for serine-threonine kinase phosphoacceptor specificity. Mol. Cell 2014, 53, 140–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphreys, J.M.; Piala, A.T.; Akella, R.; He, H.; Goldsmith, E.J. Precisely ordered phosphorylation reactions in the p38 mitogen-activated protein (MAP) kinase cascade. J. Biol. Chem. 2013, 288, 23322–23330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagoutte, E.; Villeneuve, C.; Lafanechère, L.; Wells, C.M.; Jones, G.E.; Chavrier, P.; Rossé, C. LIMK Regulates Tumor-Cell Invasion and Matrix Degradation Through Tyrosine Phosphorylation of MT1-MMP. Sci. Rep. 2016, 6, 24925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huehn, A.R.; Bibeau, J.P.; Schramm, A.C.; Cao, W.; de La Cruz, E.M.; Sindelar, C.V. Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments. Proc. Natl. Acad. Sci. USA 2020, 117, 1478–1484. [Google Scholar] [CrossRef]
- Anderson, C.A.; Kovar, D.R.; Gardel, M.L.; Winkelman, J.D. LIM domain proteins in cell mechanobiology. Cytoskeleton 2021, 78, 303–311. [Google Scholar] [CrossRef]
- Sun, X.; Phua, D.Y.Z.; Axiotakis, L.; Smith, M.A.; Blankman, E.; Gong, R.; Cail, R.C.; Espinosa de Los Reyes, S.; Beckerle, M.C.; Waterman, C.M.; et al. Mechanosensing through Direct Binding of Tensed F-Actin by LIM Domains. Dev. Cell 2020, 55, 468–482.e7. [Google Scholar] [CrossRef]
- Chen, X.; Macara, I.G. Par-3 mediates the inhibition of LIM kinase 2 to regulate cofilin phosphorylation and tight junction assembly. J. Cell Biol. 2006, 172, 671–678. [Google Scholar] [CrossRef]
- Sacchetti, P.; Carpentier, R.; Ségard, P.; Olivé-Cren, C.; Lefebvre, P. Multiple signaling pathways regulate the transcriptional activity of the orphan nuclear receptor NURR1. Nucleic Acids Res. 2006, 34, 5515–5527. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-J.; Zheng, J.J. PDZ domains and their binding partners: Structure, specificity, and modification. Cell Commun. Signal. CCS 2010, 8, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amacher, J.F.; Brooks, L.; Hampton, T.H.; Madden, D.R. Specificity in PDZ-peptide interaction networks: Computational analysis and review. J. Struct. Biol. X 2020, 4, 100022. [Google Scholar] [CrossRef] [PubMed]
- Erlendsson, S.; Thorsen, T.S.; Vauquelin, G.; Ammendrup-Johnsen, I.; Wirth, V.; Martinez, K.L.; Teilum, K.; Gether, U.; Madsen, K.L. Mechanisms of PDZ domain scaffold assembly illuminated by use of supported cell membrane sheets. eLife 2019, 8, e39180. [Google Scholar] [CrossRef]
- Rademacher, N.; Kuropka, B.; Kunde, S.-A.; Wahl, M.C.; Freund, C.; Shoichet, S.A. Intramolecular domain dynamics regulate synaptic MAGUK protein interactions. eLife 2019, 8, e41299. [Google Scholar] [CrossRef]
- Vallée, B.; Doudeau, M.; Godin, F.; Gombault, A.; Tchalikian, A.; de Tauzia, M.-L.; Bénédetti, H. Nf1 RasGAP inhibition of LIMK2 mediates a new cross-talk between Ras and Rho pathways. PLoS ONE 2012, 7, e47283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Milosavljevic, T.; Alahari, S.K. Nischarin inhibits LIM kinase to regulate cofilin phosphorylation and cell invasion. Mol. Cell. Biol. 2008, 28, 3742–3756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorrell, F.J.; Kilian, L.M.; Elkins, J.M. Solution structures and biophysical analysis of full-length group A PAKs reveal they are monomeric and auto-inhibited in cis. Biochem. J. 2019, 476, 1037–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caballe, A.; Wenzel, D.M.; Agromayor, M.; Alam, S.L.; Skalicky, J.J.; Kloc, M.; Carlton, J.G.; Labrador, L.; Sundquist, W.I.; Martin-Serrano, J. ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins. eLife 2015, 4, e06547. [Google Scholar] [CrossRef] [Green Version]
- Mathea, S.; Salah, E.; Tallant, C.; Chatterjee, D.; Berger, B.-T.; Konietzny, R.; Müller, S.; Kessler, B.M.; Knapp, S. Conformational plasticity of the ULK3 kinase domain. Biochem. J. 2021, 478, 2811–2823. [Google Scholar] [CrossRef]
- Hiraoka, J.; Okano, I.; Higuchi, O.; Yang, N.; Mizuno, K. Self-association of LIM-kinase 1 mediated by the interaction between an N-terminal LIM domain and a C-terminal kinase domain. FEBS Lett. 1996, 399, 117–121. [Google Scholar] [CrossRef] [Green Version]
- Edwards, D.C.; Gill, G.N. Structural features of LIM kinase that control effects on the actin cytoskeleton. J. Biol. Chem. 1999, 274, 11352–11361. [Google Scholar] [CrossRef] [Green Version]
- Nagata, K.; Ohashi, K.; Yang, N.; Mizuno, K. The N-terminal LIM domain negatively regulates the kinase activity of LIM-kinase 1. Biochem. J. 1999, 343 Pt 1, 99–105. [Google Scholar] [CrossRef]
- Kobayashi, M.; Nishita, M.; Mishima, T.; Ohashi, K.; Mizuno, K. MAPKAPK-2-mediated LIM-kinase activation is critical for VEGF-induced actin remodeling and cell migration. EMBO J. 2006, 25, 713–726. [Google Scholar] [CrossRef] [Green Version]
- Ritchey, L.; Ottman, R.; Roumanos, M.; Chakrabarti, R. A functional cooperativity between Aurora A kinase and LIM kinase1: Implication in the mitotic process. Cell Cycle (Georget. Tex.) 2012, 11, 296–309. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Soosairajah, J.; Harari, D.; Citri, A.; Price, J.; Ng, H.L.; Morton, C.J.; Parker, M.W.; Yarden, Y.; Bernard, O. Hsp90 increases LIM kinase activity by promoting its homo-dimerization. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2006, 20, 1218–1220. [Google Scholar] [CrossRef] [PubMed]
- Sivadasan, R.; Hornburg, D.; Drepper, C.; Frank, N.; Jablonka, S.; Hansel, A.; Lojewski, X.; Sterneckert, J.; Hermann, A.; Shaw, P.J.; et al. C9ORF72 interaction with cofilin modulates actin dynamics in motor neurons. Nat. Neurosci. 2016, 19, 1610–1618. [Google Scholar] [CrossRef]
- Kashima, R.; Roy, S.; Ascano, M.; Martinez-Cerdeno, V.; Ariza-Torres, J.; Kim, S.; Louie, J.; Lu, Y.; Leyton, P.; Bloch, K.D.; et al. Augmented noncanonical BMP type II receptor signaling mediates the synaptic abnormality of fragile X syndrome. Sci. Signal. 2016, 9, ra58. [Google Scholar] [CrossRef] [Green Version]
- Petrilli, A.; Copik, A.; Posadas, M.; Chang, L.-S.; Welling, D.B.; Giovannini, M.; Fernández-Valle, C. LIM domain kinases as potential therapeutic targets for neurofibromatosis type 2. Oncogene 2014, 33, 3571–3582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sousa-Squiavinato, A.C.M.; Vasconcelos, R.I.; Gehren, A.S.; Fernandes, P.V.; de Oliveira, I.M.; Boroni, M.; Morgado-Díaz, J.A. Cofilin-1, LIMK1 and SSH1 are differentially expressed in locally advanced colorectal cancer and according to consensus molecular subtypes. Cancer Cell Int. 2021, 21, 69. [Google Scholar] [CrossRef]
- Nikhil, K.; Chang, L.; Viccaro, K.; Jacobsen, M.; McGuire, C.; Satapathy, S.R.; Tandiary, M.; Broman, M.M.; Cresswell, G.; He, Y.J.; et al. Identification of LIMK2 as a therapeutic target in castration resistant prostate cancer. Cancer Lett. 2019, 448, 182–196. [Google Scholar] [CrossRef] [Green Version]
- Attwood, M.M.; Fabbro, D.; Sokolov, A.V.; Knapp, S.; Schiöth, H.B. Trends in kinase drug discovery: Targets, indications and inhibitor design. Nat. Rev. Drug Discov. 2021, 20, 839–861. [Google Scholar] [CrossRef] [PubMed]
- Kanev, G.K.; de Graaf, C.; Westerman, B.A.; de Esch, I.J.P.; Kooistra, A.J. KLIFS: An overhaul after the first 5 years of supporting kinase research. Nucleic Acids Res. 2021, 49, D562–D569. [Google Scholar] [CrossRef] [PubMed]
- Ross-Macdonald, P.; de Silva, H.; Guo, Q.; Xiao, H.; Hung, C.-Y.; Penhallow, B.; Markwalder, J.; He, L.; Attar, R.M.; Lin, T.; et al. Identification of a nonkinase target mediating cytotoxicity of novel kinase inhibitors. Mol. Cancer Ther. 2008, 7, 3490–3498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, Z.M.; Real, A.M.; Marsiglia, W.M.; Chow, A.; Duffy, M.E.; Yerabolu, J.R.; Scopton, A.P.; Dar, A.C. Structural basis for the action of the drug trametinib at KSR-bound MEK. Nature 2020, 588, 509–514. [Google Scholar] [CrossRef]
- Donovan, K.A.; Ferguson, F.M.; Bushman, J.W.; Eleuteri, N.A.; Bhunia, D.; Ryu, S.; Tan, L.; Shi, K.; Yue, H.; Liu, X.; et al. Mapping the Degradable Kinome Provides a Resource for Expedited Degrader Development. Cell 2020, 183, 1714–1731.e10. [Google Scholar] [CrossRef]
- Christensen, N.R.; Čalyševa, J.; Fernandes, E.F.A.; Lüchow, S.; Clemmensen, L.S.; Haugaard-Kedström, L.M.; Strømgaard, K. PDZ Domains as Drug Targets. Adv. Ther. 2019, 2, 1800143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amano, T.; Tanabe, K.; Eto, T.; Narumiya, S.; Mizuno, K. LIM-kinase 2 induces formation of stress fibres, focal adhesions and membrane blebs, dependent on its activation by Rho-associated kinase-catalysed phosphorylation at threonine-505. Biochem. J. 2001, 354, 149–159. [Google Scholar] [CrossRef]
- Chaikuad, A.; Koch, P.; Laufer, S.A.; Knapp, S. The Cysteinome of Protein Kinases as a Target in Drug Development. Angewandte Chemie (Int. Ed. Engl.) 2018, 57, 4372–4385. [Google Scholar]
- Meng, Y.; Takahashi, H.; Meng, J.; Zhang, Y.; Lu, G.; Asrar, S.; Nakamura, T.; Jia, Z. Regulation of ADF/cofilin phosphorylation and synaptic function by LIM-kinase. Neuropharmacology 2004, 47, 746–754. [Google Scholar] [CrossRef]
- Todorovski, Z.; Asrar, S.; Liu, J.; Saw, N.M.N.; Joshi, K.; Cortez, M.A.; Snead, O.C.; Xie, W.; Jia, Z. LIMK1 regulates long-term memory and synaptic plasticity via the transcriptional factor CREB. Mol. Cell. Biol. 2015, 35, 1316–1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, Y.; Zhang, Y.; Tregoubov, V.; Janus, C.; Cruz, L.; Jackson, M.; Lu, W.Y.; MacDonald, J.F.; Wang, J.Y.; Falls, D.L.; et al. Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron 2002, 35, 121–133. [Google Scholar] [CrossRef] [Green Version]
- Estevez, B.; Stojanovic-Terpo, A.; Delaney, M.K.; O’Brien, K.A.; Berndt, M.C.; Ruan, C.; Du, X. LIM kinase-1 selectively promotes glycoprotein Ib-IX-mediated TXA2 synthesis, platelet activation, and thrombosis. Blood 2013, 121, 4586–4594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawano, T.; Zhu, M.; Troiano, N.; Horowitz, M.; Bian, J.; Gundberg, C.; Kolodziejczak, K.; Insogna, K. LIM kinase 1 deficient mice have reduced bone mass. Bone 2013, 52, 70–82. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, H.; Koshimizu, U.; Miyazaki, J.; Nakamura, T. Impaired spermatogenic ability of testicular germ cells in mice deficient in the LIM-kinase 2 gene. Dev. Biol. 2002, 241, 259–272. [Google Scholar] [CrossRef] [Green Version]
- Antonipillai, J.; Mittelstaedt, K.; Rigby, S.; Bassler, N.; Bernard, O. LIM kinase 2 (LIMK2) may play an essential role in platelet function. Exp. Cell Res. 2020, 388, 111822. [Google Scholar] [CrossRef]



| Protein | Boundaries | Ligand | Ligand Type | Space Group | PDB ID |
|---|---|---|---|---|---|
| LIMK1 | 330–637 | Staurosporine | Type-1 | C 2 2 21 | 3S95 [10] |
| LIMK1 | 330–637 | PF477736 | Type-1 | C 2 2 21 | 5NXC [10] |
| LIMK1 | 330–637 | LIJTF500127 | Type-1 | P 61 2 2 | 7ATS |
| LIMK1 | 330–637 | LIJTF500025 | Type-3 | P 21 | 7ATU |
| LIMK2 | 330–632 | Ligand 22 | Type-3 | P 21 | 4TPT [8] |
| LIMK2 | 330–632 | TH300 | Type-3 | P 21 | 5NXD |
| LIMK1 D460N | 329–638 | AMP-PNP | Nucleotide | P 21 | 5HVJ [9] |
| LIMK1 D460N | 329–638 | ADP | Nucleotide | P 21 21 21 | 5HVK [9] |
| LIMK1 | 330–637 | ATP-γ-S | Nucleotide | P 32 2 1 | 5L6W [10] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chatterjee, D.; Preuss, F.; Dederer, V.; Knapp, S.; Mathea, S. Structural Aspects of LIMK Regulation and Pharmacology. Cells 2022, 11, 142. https://doi.org/10.3390/cells11010142
Chatterjee D, Preuss F, Dederer V, Knapp S, Mathea S. Structural Aspects of LIMK Regulation and Pharmacology. Cells. 2022; 11(1):142. https://doi.org/10.3390/cells11010142
Chicago/Turabian StyleChatterjee, Deep, Franziska Preuss, Verena Dederer, Stefan Knapp, and Sebastian Mathea. 2022. "Structural Aspects of LIMK Regulation and Pharmacology" Cells 11, no. 1: 142. https://doi.org/10.3390/cells11010142