Label-Free and Quantitative Dry Mass Monitoring for Single Cells during In Situ Culture
Abstract
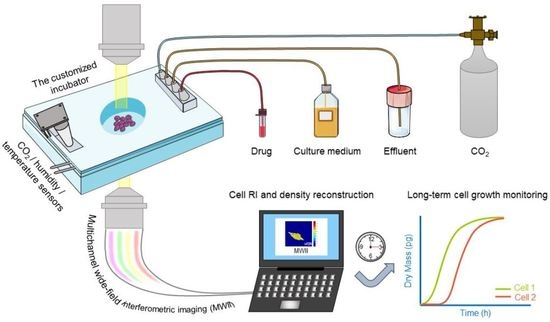
:1. Introduction
2. Materials and Methods
2.1. Quantitative Dry Mass Imaging
2.2. Image Segmentation
2.3. Preparation
3. Results
3.1. Principle Verification
3.2. Quantitative Dry Mass Measurement of Living Cells at the Ultrastructural Level
3.3. Long-Term Dry Mass Monitoring of HeLa Cells and HCerEpiC during Cell Growth
3.4. Quantitative Analysis of Single-Cell Heterogeneity in Dry Mass
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Conlon, I.J.; Dunn, G.A.; Mudge, A.W.; Raff, M.C. Extracellular control of cell size. Nat. Cell Biol. 2001, 3, 918–921. [Google Scholar] [CrossRef]
- Wang, R.; Wang, Z.; Leigh, J.; Sobh, N.; Millet, L.; Gillette, M.U.; Levine, A.J.; Popescu, G. One-dimensional deterministic transport in neurons measured by dispersion-relation phase spectroscopy. J. Phys. Condens. Matter 2011, 23, 374107. [Google Scholar] [CrossRef] [Green Version]
- Alberts, D.S.; Einspahr, J.G.; Krouse, R.S.; Prasad, A.; Ranger-Moore, J.; Hamilton, P.; Ismail, A.; Lance, P.; Goldschmid, S.; Hess, L.M.; et al. Karyometry of the Colonic Mucosa. Cancer Epidemiol. Biomark. Prev. 2007, 16, 2704–2716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, H.; Roy, H.K.; Pradhan, P.; Goldberg, M.J.; Muldoon, J.; Brand, R.E.; Sturgis, C.; Hensing, T.; Ray, D.; Bogojevic, A.; et al. Nanoscale Cellular Changes in Field Carcinogenesis Detected by Partial Wave Spectroscopy. Cancer Res. 2009, 69, 5357–5363. [Google Scholar] [CrossRef] [Green Version]
- Almassalha, L.M.; Bauer, G.M.; Wu, W.; Cherkezyan, L.; Zhang, D.; Kendra, A.; Gladstein, S.; Chandler, J.E.; VanDerway, D.; Seagle, B.-L.L.; et al. Macrogenomic engineering via modulation of the scaling of chromatin packing density. Nat. Biomed. Eng. 2017, 1, 902–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godin, M.; Delgado, F.F.; Son, S.; Grover, W.H.; Bryan, A.K.; Tzur, A.; Jorgensen, P.; Payer, K.; Grossman, A.D.; Kirschner, M.W.; et al. Using buoyant mass to measure the growth of single cells. Nat. Methods 2010, 7, 387–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, K.; Millet, L.; Kim, N.; Li, H.; Jin, X.; Popescu, G.; Aluru, N.R.; Hsia, K.J.; Bashir, R. Measurement of adherent cell mass and growth. Proc. Natl. Acad. Sci. USA 2010, 107, 20691–20696. [Google Scholar] [CrossRef] [Green Version]
- Mir, M.; Wang, Z.; Shen, Z.; Bednarz, M.; Bashir, R.; Golding, I.; Prasanth, S.G.; Popescu, G. Optical measurement of cycle-dependent cell growth. Proc. Natl. Acad. Sci. USA 2011, 108, 13124–13129. [Google Scholar] [CrossRef] [Green Version]
- Popescu, G.; Park, Y.; Lue, N.; Best-Popescu, C.; DeFlores, L.; Dasari, R.R.; Feld, M.S.; Badizadegan, K. Optical imaging of cell mass and growth dynamics. Am. J. Physiol. Physiol. 2008, 295, C538–C544. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Amidi, E.; Nandy, S.; Mostafa, A.; Zhu, Q. Optimized light delivery probe using ball lenses for co-registered photoacoustic and ultrasound endo-cavity subsurface imaging. Photoacoustics 2019, 13, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.; Bao, Y.; Chen, R.; Zhou, Q.; Hu, S. High-speed wide-field multi-parametric photoacoustic microscopy. Opt. Lett. 2020, 45, 2756–2759. [Google Scholar] [CrossRef] [PubMed]
- Cherkezyan, L.; Zhang, D.; Subramanian, H.; Capoglu, I.; Taflove, A.; Backman, V. Review of interferometric spectroscopy of scattered light for the quantification of subdiffractional structure of biomaterials. J. Biomed. Opt. 2017, 22, 030901. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Li, Q.; Wang, R.; Xue, N.; Lin, X.; Su, Y.; Jiang, K.; Jin, X.; Lin, R.; Gan, W.; et al. An interferometric imaging biosensor using weighted spectrum analysis to confirm DNA monolayer films with attogram sensitivity. Talanta 2018, 181, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Popescu, G. Quantitative Phase Imaging (QPI) in Neuroscience. IEEE J. Sel. Top. Quantum Electron. 2018, 25, 1–9. [Google Scholar] [CrossRef]
- Chowdhury, S.; Eldridge, W.J.; Wax, A.; Izatt, J.A. Structured illumination multimodal 3D-resolved quantitative phase and fluorescence sub-diffraction microscopy. Biomed. Opt. Express 2017, 8, 2496–2518. [Google Scholar] [CrossRef] [Green Version]
- Fu, R.; Su, Y.; Wang, R.; Lin, X.; Jiang, K.; Jin, X.; Yang, H.; Ma, L.; Luo, X.; Lu, Y.; et al. Label-free tomography of living cellular nanoarchitecture using hyperspectral self-interference microscopy. Biomed. Opt. Express 2019, 10, 2757–2767. [Google Scholar] [CrossRef]
- Li, Q.; Fu, R.; Zhang, J.; Wang, R.; Ye, J.; Xue, N.; Lin, X.; Su, Y.; Gan, W.; Lu, Y.; et al. Label-Free Method Using a Weighted-Phase Algorithm To Quantitate Nanoscale Interactions between Molecules on DNA Microarrays. Anal. Chem. 2017, 89, 3501–3507. [Google Scholar] [CrossRef] [PubMed]
- Sevenler, D.; Avci, O.; Ünlü, M.S. Quantitative interferometric reflectance imaging for the detection and measurement of biological nanoparticles. Biomed. Opt. Express 2017, 8, 2976–2989. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Kandel, M.E.; Hu, C.; Lee, Y.J.; Popescu, G. Wolf phase tomography (WPT) of transparent structures using partially coherent illumination. Light. Sci. Appl. 2020, 9, 1–9. [Google Scholar] [CrossRef]
- Steelman, Z.A.; Eldridge, W.J.; Weintraub, J.B.; Wax, A. Is the nuclear refractive index lower than cytoplasm? Validation of phase measurements and implications for light scattering technologies. J. Biophotonics 2017, 10, 1714–1722. [Google Scholar] [CrossRef]
- Mohsin, A.S.M.; Salim, M.B. Probing the intracellular refractive index and molecular interaction of gold nanoparticle in HeLa cells using single particle spectroscopy. Int. J. Nanomed. 2018, 13, 6019–6028. [Google Scholar] [CrossRef] [Green Version]
- Firdaus, E.R.; Park, J.-H.; Lee, S.-K.; Park, Y.; Cha, G.-H.; Han, E.-T. 3D morphological and biophysical changes in a single tachyzoite and its infected cells using three-dimensional quantitative phase imaging. J. Biophotonics 2020, 13. [Google Scholar] [CrossRef]
- Zhang, J.; Fu, R.; Xie, L.; Li, Q.; Zhou, W.; Wang, R.; Ye, J.; Wang, D.; Xue, N.; Lin, X.; et al. A smart device for label-free and real-time detection of gene point mutations based on the high dark phase contrast of vapor condensation. Lab. A Chip 2015, 15, 3891–3896. [Google Scholar] [CrossRef]
- Cherkezyan, L.; Capoglu, I.; Subramanian, H.; Rogers, J.D.; Damania, D.; Taflove, A.; Backman, V. Interferometric spectroscopy of scattered light can quantify the statistics of subdiffractional refractive-index fluctuations. Phys. Rev. Lett. 2013, 111, 033903. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, A.; Takigawa, K.; Kurishita, Y.; Kuwata, K.; Ishida, M.; Shimoda, Y.; Hamachi, I.; Tsukiji, S. Hoechst tagging: A modular strategy to design synthetic fluorescent probes for live-cell nucleus imaging. Chem. Commun. 2014, 50, 6149–6152. [Google Scholar] [CrossRef]
- Jing, G.; Perry, S.F.; Tatic-Lucic, S. Precise cell patterning using cytophobic self-assembled monolayer deposited on top of semi-transparent gold. Biomed. Microdevices 2010, 12, 935–948. [Google Scholar] [CrossRef]
- Gatti, R.; Belletti, S.; Orlandini, G.; Bussolati, O.; Dall’Asta, V.; Gazzola, G.C. Comparison of annexin V and calcein-AM as early vital markers of apoptosis in adherent cells by confocal laser microscopy. J. Histochem. Cytochem. 1998, 46, 895–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.W.T.; Xie, L.X.L.; Huang, H.H.H.; Li, X.L.X.; Wang, R.W.R.; Yang, G.Y.G.; Du, Y.D.Y.; Huang, G.H.G. Label-free biomolecular imaging using scanning spectral interferometry. Chin. Opt. Lett. 2013, 11, 111102–111105. [Google Scholar] [CrossRef]
- Cheng, Y.-Y.; Wyant, J.C. Two-wavelength phase shifting interferometry. Appl. Opt. 1984, 23, 4539–4543. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Su, Y.; Wang, R.; Lin, X.; Jin, X.; Yang, H.; Du, W.; Shan, X.; Lv, W.; Huang, G. Single cell capture, isolation, and long-term in-situ imaging using quantitative self-interference spectroscopy. Cytom. A 2021, 99, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Lam, V.K.; Nguyen, T.C.; Bui, V.; Chung, B.M.; Chang, L.-C.; Nehmetallah, G.; Raub, C.B. Quantitative scoring of epithelial and mesenchymal qualities of cancer cells using machine learning and quantitative phase imaging. J. Biomed. Opt. 2020, 25, 026002-17. [Google Scholar] [CrossRef]
- Iyer, R.R.; Zurauskas, M.; Cui, Q.; Gao, L.; Smith, R.T.; Boppart, S.A. Full-field spectral domain optical interferometryfor snapshot three-dimensional microscopy. Biomed. Opt. Express 2020, 11, 5903–5919. [Google Scholar] [CrossRef]
- Ling, T.; Boyle, K.C.; Zuckerman, V.; Flores, T.; Ramakrishnan, C.; Deisseroth, K.; Palanker, D. High-speed interferometric imaging reveals dynamics of neuronal deformation during the action potential. Proc. Natl. Acad. Sci. USA 2020, 117, 10278–10285. [Google Scholar] [CrossRef] [PubMed]
- Nava, M.; Miroshnikova, Y.A.; Biggs, L.C.; Whitefield, D.B.; Metge, F.; Boucas, J.; Vihinen, H.; Jokitalo, E.; Li, X.; Arcos, J.M.G.; et al. Heterochromatin-Driven Nuclear Softening Protects the Genome against Mechanical Stress-Induced Damage. Cell 2020, 181, 800–817.e22. [Google Scholar] [CrossRef] [PubMed]
- Felkl, M.; Tomas, K.; Smid, M.; Mattes, J.; Windoffer, R.; Leube, R.E. Monitoring the Cytoskeletal EGF Response in Live Gastric Carcinoma Cells. PLoS ONE 2012, 7, e45280. [Google Scholar] [CrossRef] [Green Version]
- Leal-Esteban, L.C.; Fajas, L. Cell cycle regulators in cancer cell metabolism. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2020, 1866, 165715. [Google Scholar] [CrossRef]
- Jackman, M.; Lindon, C.; Nigg, E.A.; Pines, J. Active cyclin B1–Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 2003, 5, 143–148. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, Y.; Fu, R.; Du, W.; Yang, H.; Ma, L.; Luo, X.; Wang, R.; Lin, X.; Jin, X.; Shan, X.; et al. Label-Free and Quantitative Dry Mass Monitoring for Single Cells during In Situ Culture. Cells 2021, 10, 1635. https://doi.org/10.3390/cells10071635
Su Y, Fu R, Du W, Yang H, Ma L, Luo X, Wang R, Lin X, Jin X, Shan X, et al. Label-Free and Quantitative Dry Mass Monitoring for Single Cells during In Situ Culture. Cells. 2021; 10(7):1635. https://doi.org/10.3390/cells10071635
Chicago/Turabian StyleSu, Ya, Rongxin Fu, Wenli Du, Han Yang, Li Ma, Xianbo Luo, Ruliang Wang, Xue Lin, Xiangyu Jin, Xiaohui Shan, and et al. 2021. "Label-Free and Quantitative Dry Mass Monitoring for Single Cells during In Situ Culture" Cells 10, no. 7: 1635. https://doi.org/10.3390/cells10071635