Development and External Validation of 18F-FDG PET-Based Radiomic Model for Predicting Pathologic Complete Response after Neoadjuvant Chemotherapy in Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
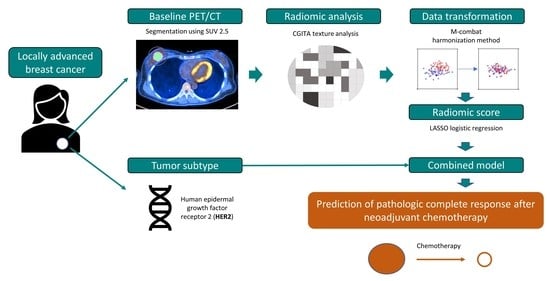
2. Patients and Methods
2.1. Study Population
2.2. Data Collection
2.3. 18F-FDG PET/CT Image Acquisition and Analysis
2.4. 18F-FDG Radiomic Feature Extraction
2.5. 18F-FDG Radiomic Feature Selection and Model Construction
2.6. Statistical Analysis
3. Results
3.1. Baseline Characteristics of Patients
3.2. Comparison of Clinical Variables and Conventional PET Parameters According to pCR
3.3. 18F-FDG Radiomic Feature Selection and Model Construction
3.4. Model Performance and Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Killelea, B.K.; Yang, V.Q.; Mougalian, S.; Horowitz, N.R.; Pusztai, L.; Chagpar, A.B.; Lannin, D.R. Neoadjuvant chemotherapy for breast cancer increases the rate of breast conservation: Results from the National Cancer Database. J. Am. Coll. Surg. 2015, 220, 1063–1069. [Google Scholar] [CrossRef] [PubMed]
- Pilewskie, M.; Morrow, M. Axillary nodal management following neoadjuvant chemotherapy: A review. JAMA Oncol. 2017, 3, 549–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orsaria, P.; Grasso, A.; Ippolito, E.; Pantano, F.; Sammarra, M.; Altomare, C.; Cagli, B.; Costa, F.; Perrone, G.; Soponaru, G. Clinical outcomes among major breast cancer subtypes after neoadjuvant chemotherapy: Impact on breast cancer recurrence and survival. Anticancer Res. 2021, 41, 2697–2709. [Google Scholar] [CrossRef] [PubMed]
- Spring, L.M.; Fell, G.; Arfe, A.; Sharma, C.; Greenup, R.; Reynolds, K.L.; Smith, B.L.; Alexander, B.; Moy, B.; Isakoff, S.J.; et al. Pathologic Complete Response after Neoadjuvant Chemotherapy and Impact on Breast Cancer Recurrence and Survival: A Comprehensive Meta-analysis. Clin. Cancer Res. 2020, 26, 2838–2848. [Google Scholar]
- Derks, M.G.; van de Velde, C.J. Neoadjuvant chemotherapy in breast cancer: More than just downsizing. Lancet Oncol. 2018, 19, 2–3. [Google Scholar] [CrossRef] [Green Version]
- Sener, S.F.; Sargent, R.E.; Lee, C.; Manchandia, T.; Le-Tran, V.; Olimpiadi, Y.; Zaremba, N.; Alabd, A.; Nelson, M.; Lang, J.E. MRI does not predict pathologic complete response after neoadjuvant chemotherapy for breast cancer. J. Surg. Oncol. 2019, 120, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Schaefgen, B.; Mati, M.; Sinn, H.P.; Golatta, M.; Stieber, A.; Rauch, G.; Hennigs, A.; Richter, H.; Domschke, C.; Schuetz, F.; et al. Can Routine Imaging After Neoadjuvant Chemotherapy in Breast Cancer Predict Pathologic Complete Response? Ann. Surg. Oncol. 2016, 23, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Groheux, D.; Cochet, A.; Humbert, O.; Alberini, J.-L.; Hindié, E.; Mankoff, D. 18F-FDG PET/CT for staging and restaging of breast cancer. J. Nucl. Med. 2016, 57, 17S–26S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jadvar, H.; Alavi, A.; Gambhir, S.S. 18F-FDG uptake in lung, breast, and colon cancers: Molecular biology correlates and disease characterization. J. Nucl. Med. 2009, 50, 1820–1827. [Google Scholar] [CrossRef] [Green Version]
- Basu, S.; Kumar, R.; Mavi, A.; Alavi, A. Exploring Tumor Biology with Fluorodeoxyglucose–Positron Emission Tomography Imaging in Breast Carcinoma. PET Clin. 2009, 4, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Kwee, T.C.; Basu, S.; Saboury, B.; Ambrosini, V.; Torigian, D.A.; Alavi, A. A new dimension of FDG-PET interpretation: Assessment of tumor biology. Eur. J. Nucl. Med. Mol. Imaging 2011, 38, 1158–1170. [Google Scholar] [CrossRef] [PubMed]
- Alberini, J.L.; Lerebours, F.; Wartski, M.; Fourme, E.; Le Stanc, E.; Gontier, E.; Madar, O.; Cherel, P.; Pecking, A. 18F-fluorodeoxyglucose positron emission tomography/computed tomography (FDG-PET/CT) imaging in the staging and prognosis of inflammatory breast cancer. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2009, 115, 5038–5047. [Google Scholar] [CrossRef] [PubMed]
- Groheux, D.; Sanna, A.; Majdoub, M.; de Cremoux, P.; Giacchetti, S.; Teixeira, L.; Espié, M.; Merlet, P.; de Roquancourt, A.; Visvikis, D. Baseline tumor 18F-FDG uptake and modifications after 2 cycles of neoadjuvant chemotherapy are prognostic of outcome in ER+/HER2− breast cancer. J. Nucl. Med. 2015, 56, 824–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reginelli, A.; Nardone, V.; Giacobbe, G.; Belfiore, M.P.; Grassi, R.; Schettino, F.; Del Canto, M.; Grassi, R.; Cappabianca, S. Radiomics as a New Frontier of Imaging for Cancer Prognosis: A Narrative Review. Diagnostics 2021, 11, 1796. [Google Scholar] [CrossRef]
- Li, P.; Wang, X.; Xu, C.; Liu, C.; Zheng, C.; Fulham, M.J.; Feng, D.; Wang, L.; Song, S.; Huang, G. 18 F-FDG PET/CT radiomic predictors of pathologic complete response (pCR) to neoadjuvant chemotherapy in breast cancer patients. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1116–1126. [Google Scholar] [CrossRef]
- Antunovic, L.; De Sanctis, R.; Cozzi, L.; Kirienko, M.; Sagona, A.; Torrisi, R.; Tinterri, C.; Santoro, A.; Chiti, A.; Zelic, R.; et al. PET/CT radiomics in breast cancer: Promising tool for prediction of pathological response to neoadjuvant chemotherapy. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 1468–1477. [Google Scholar] [CrossRef]
- Lee, H.; Lee, D.E.; Park, S.; Kim, T.S.; Jung, S.Y.; Lee, S.; Kang, H.S.; Lee, E.S.; Sim, S.H.; Park, I.H.; et al. Predicting Response to Neoadjuvant Chemotherapy in Patients With Breast Cancer: Combined Statistical Modeling Using Clinicopathological Factors and FDG PET/CT Texture Parameters. Clin. Nucl. Med. 2019, 44, 21–29. [Google Scholar] [CrossRef]
- Park, J.E.; Park, S.Y.; Kim, H.J.; Kim, H.S. Reproducibility and Generalizability in Radiomics Modeling: Possible Strategies in Radiologic and Statistical Perspectives. Korean J. Radiol. 2019, 20, 1124–1137. [Google Scholar] [CrossRef]
- Heinze, G.; Wallisch, C.; Dunkler, D. Variable selection–a review and recommendations for the practicing statistician. Biom. J. 2018, 60, 431–449. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.H.; Lin, C.Y.; Shih, M.J.; Wang, H.M.; Ho, T.Y.; Liao, C.T.; Yen, T.C. Development and evaluation of an open-source software package "CGITA" for quantifying tumor heterogeneity with molecular images. BioMed Res. Int. 2014, 2014, 248505. [Google Scholar] [CrossRef] [Green Version]
- Stein, C.K.; Qu, P.; Epstein, J.; Buros, A.; Rosenthal, A.; Crowley, J.; Morgan, G.; Barlogie, B. Removing batch effects from purified plasma cell gene expression microarrays with modified ComBat. BMC Bioinform. 2015, 16, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da-Ano, R.; Masson, I.; Lucia, F.; Doré, M.; Robin, P.; Alfieri, J.; Rousseau, C.; Mervoyer, A.; Reinhold, C.; Castelli, J. Performance comparison of modified ComBat for harmonization of radiomic features for multicenter studies. Sci. Rep. 2020, 10, 10248. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Vasquez, M.M.; Hu, C.; Roe, D.J.; Chen, Z.; Halonen, M.; Guerra, S. Least absolute shrinkage and selection operator type methods for the identification of serum biomarkers of overweight and obesity: Simulation and application. BMC Med. Res. Methodol. 2016, 16, 154. [Google Scholar]
- Zhou, J.; Zou, S.; Kuang, D.; Yan, J.; Zhao, J.; Zhu, X. A novel approach using FDG-PET/CT-Based radiomics to assess tumor immune phenotypes in patients with non-small cell lung cancer. Front. Oncol. 2021, 11, 769272. [Google Scholar] [CrossRef]
- Lv, L.; Xin, B.; Hao, Y.; Yang, Z.; Xu, J.; Wang, L.; Wang, X.; Song, S.; Guo, X. Radiomic analysis for predicting prognosis of colorectal cancer from preoperative 18F-FDG PET/CT. J. Transl. Med. 2022, 20, 66. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. Ann. Intern. Med. 2015, 162, 55–63. [Google Scholar] [CrossRef] [Green Version]
- Pak, K.; Cheon, G.J.; Nam, H.-Y.; Kim, S.-J.; Kang, K.W.; Chung, J.-K.; Kim, E.E.; Lee, D.S. Prognostic value of metabolic tumor volume and total lesion glycolysis in head and neck cancer: A systematic review and meta-analysis. J. Nucl. Med. 2014, 55, 884–890. [Google Scholar] [CrossRef] [Green Version]
- Wen, W.; Xuan, D.; Hu, Y.; Li, X.; Liu, L.; Xu, D. Prognostic value of maximum standard uptake value, metabolic tumor volume, and total lesion glycolysis of positron emission tomography/computed tomography in patients with breast cancer: A systematic review and meta-analysis. PLoS ONE 2019, 14, e0225959. [Google Scholar] [CrossRef] [Green Version]
- Im, H.-J.; Pak, K.; Cheon, G.J.; Kang, K.W.; Kim, S.-J.; Kim, I.-J.; Chung, J.-K.; Kim, E.E.; Lee, D.S. Prognostic value of volumetric parameters of 18 F-FDG PET in non-small-cell lung cancer: A meta-analysis. Eur. J. Nucl. Med. Mol. Imaging 2015, 42, 241–251. [Google Scholar] [CrossRef]
- Lemarignier, C.; Martineau, A.; Teixeira, L.; Vercellino, L.; Espié, M.; Merlet, P.; Groheux, D. Correlation between tumour characteristics, SUV measurements, metabolic tumour volume, TLG and textural features assessed with 18F-FDG PET in a large cohort of oestrogen receptor-positive breast cancer patients. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 1145–1154. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.H.; Lee, S.J.; Lee, J.; Jung, J.H.; Park, H.Y.; Jeong, S.Y.; Lee, S.-w.; Chae, Y.S. Utility of 18F-FDG PET/CT for predicting pathologic complete response in hormone receptor-positive, HER2-negative breast cancer patients receiving neoadjuvant chemotherapy. BMC Cancer 2020, 20, 1106. [Google Scholar] [CrossRef] [PubMed]
- Martelotto, L.G.; Ng, C.K.Y.; Piscuoglio, S.; Weigelt, B.; Reis-Filho, J.S. Breast cancer intra-tumor heterogeneity. Breast Cancer Res. 2014, 16, 210. [Google Scholar] [CrossRef]
- Hirata, K.; Tamaki, N. Quantitative FDG PET assessment for oncology therapy. Cancers 2021, 13, 869. [Google Scholar] [CrossRef]
- Denisov, E.V.; Litviakov, N.V.; Zavyalova, M.V.; Perelmuter, V.M.; Vtorushin, S.V.; Tsyganov, M.M.; Gerashchenko, T.S.; Garbukov, E.Y.; Slonimskaya, E.M.; Cherdyntseva, N.V. Intratumoral morphological heterogeneity of breast cancer: Neoadjuvant chemotherapy efficiency and multidrug resistance gene expression. Sci. Rep. 2014, 4, 4709. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Jiang, Y.-C.; Sun, C.-K.; Chen, Q.-M. Role of the tumor microenvironment in tumor progression and the clinical applications. Oncol. Rep. 2016, 35, 2499–2515. [Google Scholar] [CrossRef] [Green Version]
- Hatt, M.; Tixier, F.; Pierce, L.; Kinahan, P.E.; Le Rest, C.C.; Visvikis, D. Characterization of PET/CT images using texture analysis: The past, the present… any future? Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 151–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnstone, I.M.; Titterington, D.M. Statistical Challenges of High-Dimensional Data; The Royal Society Publishing: London, UK, 2009; Volume 367, pp. 4237–4253. [Google Scholar]
- Ha, S.; Choi, H.; Paeng, J.C.; Cheon, G.J. Radiomics in Oncological PET/CT: A Methodological Overview. Nucl. Med. Mol. Imaging 2019, 53, 14–29. [Google Scholar] [CrossRef] [PubMed]
- Nestle, U.; Kremp, S.; Schaefer-Schuler, A.; Sebastian-Welsch, C.; Hellwig, D.; Rübe, C.; Kirsch, C.-M. Comparison of different methods for delineation of 18F-FDG PET–positive tissue for target volume definition in radiotherapy of patients with non–small cell lung cancer. J. Nucl. Med. 2005, 46, 1342–1348. [Google Scholar] [PubMed]
- Ha, S.; Park, S.; Bang, J.-I.; Kim, E.-K.; Lee, H.-Y. Metabolic radiomics for pretreatment 18F-FDG PET/CT to characterize locally advanced breast cancer: Histopathologic characteristics, response to neoadjuvant chemotherapy, and prognosis. Sci. Rep. 2017, 7, 1556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilyas, H.; Mikhaeel, N.G.; Dunn, J.T.; Rahman, F.; Møller, H.; Smith, D.; Barrington, S.F. Defining the optimal method for measuring baseline metabolic tumour volume in diffuse large B cell lymphoma. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 1142–1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hatt, M.; Tixier, F.; Cheze Le Rest, C.; Pradier, O.; Visvikis, D. Robustness of intratumour 18F-FDG PET uptake heterogeneity quantification for therapy response prediction in oesophageal carcinoma. Eur. J. Nucl. Med. Mol. Imaging 2013, 40, 1662–1671. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Chu-Shern, J.L.; Loi, H.Y.; Khor, L.K.; Sinha, A.K.; Quek, S.T.; Tham, I.W.; Townsend, D. Impact of image reconstruction settings on texture features in 18F-FDG PET. J. Nucl. Med. 2015, 56, 1667–1673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orlhac, F.; Eertink, J.J.; Cottereau, A.-S.; Zijlstra, J.M.; Thieblemont, C.; Meignan, M.; Boellaard, R.; Buvat, I. A guide to ComBat harmonization of imaging biomarkers in multicenter studies. J. Nucl. Med. 2022, 63, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.; Lee, J.; Cheon, M.; Woo, S.-K.; Ahn, M.-J.; Pyo, H.R.; Choi, Y.S.; Han, J.H.; Choi, J.Y. Predictive value of 18F-FDG PET/CT using machine learning for pathological response to neoadjuvant concurrent chemoradiotherapy in patients with stage iii non-small cell lung cancer. Cancers 2022, 14, 1987. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.-J.; Kim, Y.; Kim, B.S. Intratumoral metabolic heterogeneity predicts invasive components in breast ductal carcinoma in situ. Eur. Radiol. 2015, 25, 3648–3658. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-C.; Chen, C.-J.; Hsu, W.-L.; Chang, S.-M.; Huang, Y.-F.; Tyan, Y.-C. Prognostic significance of metabolic parameters and textural features on 18F-FDG PET/CT in invasive ductal carcinoma of breast. Sci. Rep. 2019, 9, 10946. [Google Scholar] [CrossRef] [Green Version]
- Leijenaar, R.T.; Carvalho, S.; Velazquez, E.R.; Van Elmpt, W.J.; Parmar, C.; Hoekstra, O.S.; Hoekstra, C.J.; Boellaard, R.; Dekker, A.L.; Gillies, R.J. Stability of FDG-PET Radiomics features: An integrated analysis of test-retest and inter-observer variability. Acta Oncol. 2013, 52, 1391–1397. [Google Scholar] [CrossRef] [Green Version]
- Yip, S.S.; Aerts, H.J. Applications and limitations of radiomics. Phys. Med. Biol. 2016, 61, R150. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Tham, I.W.; Yan, J. Quantitative accuracy of radiomic features of low-dose 18F-FDG PET imaging. Transl. Cancer Res. 2020, 9, 4646. [Google Scholar] [CrossRef]
- Martens, R.M.; Koopman, T.; Noij, D.P.; Pfaehler, E.; Übelhör, C.; Sharma, S.; Vergeer, M.R.; Leemans, C.R.; Hoekstra, O.S.; Yaqub, M. Predictive value of quantitative 18F-FDG-PET radiomics analysis in patients with head and neck squamous cell carcinoma. EJNMMI Res. 2020, 10, 102. [Google Scholar] [CrossRef] [PubMed]




| Variable | Training (n = 53) | Internal Validation (n = 34) | External Validation (n = 28) | p Value |
|---|---|---|---|---|
| Age | 49.5 ± 10.8 | 49.7 ± 9.6 | 48.7 ± 9.8 | 0.929 |
| Clinical tumor stage | 0.190 | |||
| II | 23 (43.4) | 9 (26.5) | 13 (46.4) | |
| III | 30 (56.6) | 25 (73.5) | 15 (53.6) | |
| Receptor status in histology | 0.812 | |||
| ER-positive | 33 (62.3) | 19 (55.9) | 15 (53.6) | |
| PR-positive | 29 (54.7) | 18 (52.9) | 10 (35.7) | |
| HER2-postive | 16 (30.2) | 7 (20.6) | 7 (25.0) | |
| TNBC | 11 (20.8) | 9 (26.5) | 9 (32.1) | |
| Surgery | 0.239 | |||
| Breast conserving surgery | 12 (22.6) | 13 (38.2) | 10 (35.7) | |
| Mastectomy | 41 (77.4) | 21 (61.8) | 18 (64.3) | |
| Baseline CA15-3 | 9.6 ± 4.5 | 9.7 ± 4.5 | 18.8 ± 11.7 | 0.001 |
| Tumor location | 0.155 | |||
| Right | 28 (52.8) | 11 (32.4) | 14 (50.0) | |
| Left | 25 (47.2) | 23 (67.6) | 14 (50.0) | |
| Response to NAC | 0.821 | |||
| pCR | 10 (18.9) | 5 (14.7) | 4 (14.3) | |
| Non-pCR | 43 (81.1) | 29 (85.3) | 24 (85.7) |
| Variable | pCR (n = 10) | Non-pCR (n = 43) | p Value |
|---|---|---|---|
| Age | 52.8 ± 10.4 | 48.7 ± 10.8 | 0.223 |
| Clinical tumor stage | 0.484 | ||
| II | 3 (30.0) | 20 (46.5) | |
| III | 7 (70.0) | 23 (53.5) | |
| Receptor status in histology | |||
| ER-positive | 5 (50.0) | 28 (65.1) | 0.475 |
| PR-positive | 3 (30.0) | 26 (60.5) | 0.156 |
| HER2-postive | 8 (80.0) | 8 (18.6) | <0.001 |
| TNBC | 1 (10.0) | 10 (23.3) | 0.667 |
| Surgery | 0.207 | ||
| Breast conserving surgery | 4 (40.0) | 8 (18.6) | |
| Mastectomy | 6 (60.0) | 35 (81.4) | |
| Baseline CA15-3 | 8.1 ± 3.5 | 9.9 ± 4.6 | 0.285 |
| Tumor location | 0.999 | ||
| Right | 5 (50.0) | 23 (53.5) | |
| Left | 5 (50.0) | 20 (46.5) | |
| Conventional PET parameter | |||
| SUVmax | 12.2 (6.4–19.6) | 8.3 (5.5–15.1) | 0.328 |
| MTV | 8.2 (4.2–11.4) | 12.3 (6.0–22.7) | 0.112 |
| TLG | 25.1 (11.7–40.5) | 38.3 (18.2–112.9) | 0.228 |
| Variables | Univariate Logistic Analysis | Multivariate Logistic Analysis | ||
|---|---|---|---|---|
| OR (95% CI) | p Value | OR (95% CI) | p Value | |
| Age | 1.04 (0.97–1.11) | 0.273 | - | |
| CA15-3 | 0.90 (0.74–1.08) | 0.799 | - | |
| Stage | 0.49 (0.11–2.16) | 0.335 | - | |
| ER positive | 1.80 (0.45–7.23) | 0.576 | - | |
| PR positive | 3.43 (0.78–15.17) | 0.193 | - | |
| HER2 positive | 8.82 (1.89–41.09) | 0.003 | 433.82 (1.03–182,988.53) | 0.049 |
| TNBC | 0.37 (0.04–3.26) | 0.320 | - | |
| Radiomics score | 9.71 (2.01–46.91) | <0.001 | 38.33 (1.70–866.00) | 0.022 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, C.H.; Choi, J.Y.; Choi, J.H.; Lee, J.-H.; Lee, J.; Lim, C.W.; Kim, Z.; Woo, S.-K.; Park, S.B.; Park, J.M. Development and External Validation of 18F-FDG PET-Based Radiomic Model for Predicting Pathologic Complete Response after Neoadjuvant Chemotherapy in Breast Cancer. Cancers 2023, 15, 3842. https://doi.org/10.3390/cancers15153842
Lim CH, Choi JY, Choi JH, Lee J-H, Lee J, Lim CW, Kim Z, Woo S-K, Park SB, Park JM. Development and External Validation of 18F-FDG PET-Based Radiomic Model for Predicting Pathologic Complete Response after Neoadjuvant Chemotherapy in Breast Cancer. Cancers. 2023; 15(15):3842. https://doi.org/10.3390/cancers15153842
Chicago/Turabian StyleLim, Chae Hong, Joon Young Choi, Joon Ho Choi, Jun-Hee Lee, Jihyoun Lee, Cheol Wan Lim, Zisun Kim, Sang-Keun Woo, Soo Bin Park, and Jung Mi Park. 2023. "Development and External Validation of 18F-FDG PET-Based Radiomic Model for Predicting Pathologic Complete Response after Neoadjuvant Chemotherapy in Breast Cancer" Cancers 15, no. 15: 3842. https://doi.org/10.3390/cancers15153842