Functional and Clinical Significance of Dysregulated microRNAs in Liver Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Pathological Characteristics and Clinical Development of HCC
3. The Biosynthesis and Action Mechanisms of MicroRNAs
4. The Important Roles and Functions of MicroRNAs in HCC
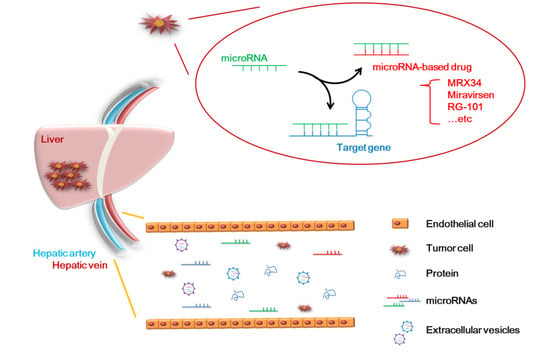
5. Application of MicroRNAs to Clinical Technology
6. The Advantages of MicroRNAs for the Diagnosis and Treatment of HCC
7. The Disadvantages of MicroRNAs in the Diagnosis and Treatment of HCC
8. The Current Clinical Application of MicroRNAs in HCC
| Name | microRNA/Role | Phase Status | Diseases | Therapeutic Agent | Regulation Gene | Side Effect | Reference |
|---|---|---|---|---|---|---|---|
| MRX34 | miR-34/Suppressor | Phase1 (terminated) | HCC | miR-34 mimic | SMAD4, SATB2, PDGFR, c-MET, Axl | Immune-mediated toxicity | [130,131,132,133,134,135] |
| Miravirsen (SPC3649) | miR-122/Oncogene | Phase2 (suspended) | HCV | Anti-miR-122 | Ago2, DCAF1, CAT-1, NIK, LPL, NS5B | No clear side effects | [27,136,137,138,139,140,141] |
| RG-101 | miR-122/Oncogene | Phase2 (discontinued) | HCV | Anti-miR-122 | Jaundice | [142,143] | |
| RG-125 (AZD4076) | miR-103/Oncogene miR-107/Oncogene | Phase1 | NASH | Anti-miR-103/ 107 | HMGB1, P120, ZO-1, LATS2, RGS4, HMGCS2 | Hyperbilirubinemia | [144,145,146,147,148,149,150] |
9. Challenges and Solutions for the Clinical Application of microRNAs
10. Discussion
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Fornari, F.; Gramantieri, L.; Callegari, E.; Shankaraiah, R.C.; Piscaglia, F.; Negrini, M.; Giovannini, C. MicroRNAs in Animal Models of HCC. Cancers 2019, 11, 1961. [Google Scholar] [CrossRef] [Green Version]
- Ghouri, Y.A.; Mian, I.; Rowe, J.H. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J. Carcinog. 2017, 16, 1. [Google Scholar]
- Schwabe, R.F.; Greten, T.F. Gut microbiome in HCC—Mechanisms, diagnosis and therapy. J. Hepatol. 2020, 72, 230–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Reviews Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Han, Q.; Zhao, H.; Zhang, J. The Mechanisms of HBV-Induced Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2021, 8, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Teufel, A.; Staib, F.; Kanzler, S.; Weinmann, A.; Schulze-Bergkamen, H.; Galle, P.R. Genetics of hepatocellular carcinoma. World J. Gastroenterol. 2007, 13, 2271–2282. [Google Scholar] [CrossRef] [PubMed]
- Buendia, M.A. Genetics of hepatocellular carcinoma. Semin. Cancer Biol. 2000, 10, 185–200. [Google Scholar] [CrossRef]
- Laurent-Puig, P.; Zucman-Rossi, J. Genetics of hepatocellular tumors. Oncogene 2006, 25, 3778–3786. [Google Scholar] [CrossRef] [Green Version]
- Tellapuri, S.; Sutphin, P.D.; Beg, M.S.; Singal, A.G.; Kalva, S.P. Staging systems of hepatocellular carcinoma: A review. Indian J. Gastroenterol. Off. J. Indian Soc. Gastroenterol. 2018, 37, 481–491. [Google Scholar] [CrossRef]
- Shao, Y.Y.; Wang, S.Y.; Lin, S.M.; Diagnosis, G.; Systemic Therapy, G. Management consensus guideline for hepatocellular carcinoma: 2020 update on surveillance, diagnosis, and systemic treatment by the Taiwan Liver Cancer Association and the Gastroenterological Society of Taiwan. J. Formos. Med. Assoc. 2021, 120, 1051–1060. [Google Scholar] [CrossRef]
- Lee, A.; Lee, F.C. Medical oncology management of advanced hepatocellular carcinoma 2019: A reality check. Front. Med. 2020, 14, 273–283. [Google Scholar] [CrossRef] [Green Version]
- Mazzaferro, V.; Regalia, E.; Doci, R.; Andreola, S.; Pulvirenti, A.; Bozzetti, F.; Montalto, F.; Ammatuna, M.; Morabito, A.; Gennari, L. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N. Engl. J. Med. 1996, 334, 693–699. [Google Scholar] [CrossRef]
- Yuan, J.N.; Chao, Y.; Lee, W.P.; Li, C.P.; Lee, R.C.; Chang, F.Y.; Yen, S.H.; Lee, S.D.; Whang-Peng, J. Chemotherapy with etoposide, doxorubicin, cisplatin, 5-fluorouracil, and leucovorin for patients with advanced hepatocellular carcinoma. Med. Oncol. 2008, 25, 201–206. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Bandara, K.V.; Michael, M.Z.; Gleadle, J.M. MicroRNA Biogenesis in Hypoxia. MicroRNA 2017, 6, 80–96. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bofill-De Ros, X.; Rovira-Rigau, M.; Fillat, C. Implications of MicroRNAs in Oncolytic Virotherapy. Front. Oncol. 2017, 7, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sempere, L.F.; Azmi, A.S.; Moore, A. microRNA-based diagnostic and therapeutic applications in cancer medicine. Wiley Interdiscip. Rev. RNA 2021, 12, e1662. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Sun, W.; Guo, Z.; Zhang, J.; Yu, H.; Liu, B. Mechanisms of lncRNA/microRNA interactions in angiogenesis. Life Sci. 2020, 254, 116900. [Google Scholar] [CrossRef]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Reviews Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.K.; Wang, Z.; Li, G. MicroRNA-125 in immunity and cancer. Cancer Lett. 2019, 454, 134–145. [Google Scholar] [CrossRef]
- Gmerek, L.; Martyniak, K.; Horbacka, K.; Krokowicz, P.; Scierski, W.; Golusinski, P.; Golusinski, W.; Schneider, A.; Masternak, M.M. MicroRNA regulation in colorectal cancer tissue and serum. PLoS ONE 2019, 14, e0222013. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.F.; Li, N.; Lin, D.D.; Sun, L.B. Circulating MicroRNA-122 for the Diagnosis of Hepatocellular Carcinoma: A Meta-Analysis. BioMed Res. Int. 2020, 2020, 5353695. [Google Scholar] [CrossRef] [Green Version]
- Mahmoudian-Sani, M.R.; Asgharzade, S.; Alghasi, A.; Saeedi-Boroujeni, A.; Adnani Sadati, S.J.; Moradi, M.T. MicroRNA-122 in patients with hepatitis B and hepatitis B virus-associated hepatocellular carcinoma. J. Gastrointest. Oncol. 2019, 10, 789–796. [Google Scholar] [CrossRef]
- Ono, C.; Fukuhara, T.; Li, S.; Wang, J.; Sato, A.; Izumi, T.; Fauzyah, Y.; Yamamoto, T.; Morioka, Y.; Dokholyan, N.V.; et al. Various miRNAs compensate the role of miR-122 on HCV replication. PLoS Pathog. 2020, 16, e1008308. [Google Scholar] [CrossRef]
- Yan, Y.; Li, C.; Sun, B.; Yang, R. DCAF1 is involved in HCV replication through regulation of miR-122. Arch. Virol. 2018, 163, 977–985. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Huang, M.; Tang, C.; Yue, Y.; Liu, X.; Zheng, Z.; Dong, H.; Liu, D. Dietary daidzein inhibits hepatitis C virus replication by decreasing microRNA-122 levels. Virus Res. 2021, 298, 198404. [Google Scholar] [CrossRef] [PubMed]
- Mata, M.; Neben, S.; Majzoub, K.; Carette, J.; Ramanathan, M.; Khavari, P.A.; Sarnow, P. Impact of a patient-derived hepatitis C viral RNA genome with a mutated microRNA binding site. PLoS Pathog. 2019, 15, e1007467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamontagne, J.; Steel, L.F.; Bouchard, M.J. Hepatitis B virus and microRNAs: Complex interactions affecting hepatitis B virus replication and hepatitis B virus-associated diseases. World J. Gastroenterol. 2015, 21, 7375–7399. [Google Scholar] [CrossRef]
- Caviglia, G.P.; Abate, M.L.; Gaia, S.; Petrini, E.; Bosco, C.; Olivero, A.; Rosso, C.; Ciancio, A.; Pellicano, R.; Saracco, G.M.; et al. Risk of hepatocellular carcinoma in HBV cirrhotic patients assessed by the combination of miR-122, AFP and PIVKA-II. Panminerva Med. 2017, 59, 283–289. [Google Scholar] [CrossRef]
- Bharali, D.; Banerjee, B.D.; Bharadwaj, M.; Husain, S.A.; Kar, P. Expression Analysis of MicroRNA-21 and MicroRNA-122 in Hepatocellular Carcinoma. J. Clin. Exp. Hepatol. 2019, 9, 294–301. [Google Scholar] [CrossRef]
- Diaz, G.; Melis, M.; Tice, A.; Kleiner, D.E.; Mishra, L.; Zamboni, F.; Farci, P. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int. J. Cancer 2013, 133, 816–824. [Google Scholar] [CrossRef] [Green Version]
- Lou, Z.; Gong, Y.Q.; Zhou, X.; Hu, G.H. Low expression of miR-199 in hepatocellular carcinoma contributes to tumor cell hyper-proliferation by negatively suppressing XBP1. Oncol. Lett. 2018, 16, 6531–6539. [Google Scholar] [CrossRef] [Green Version]
- Zhan, Y.; Zheng, N.; Teng, F.; Bao, L.; Liu, F.; Zhang, M.; Guo, M.; Guo, W.; Ding, G.; Wang, Q. MiR-199a/b-5p inhibits hepatocellular carcinoma progression by post-transcriptionally suppressing ROCK1. Oncotarget 2017, 8, 67169–67180. [Google Scholar] [CrossRef]
- Giovannini, C.; Fornari, F.; Dallo, R.; Gagliardi, M.; Nipoti, E.; Vasuri, F.; Coada, C.A.; Ravaioli, M.; Bolondi, L.; Gramantieri, L. MiR-199-3p replacement affects E-cadherin expression through Notch1 targeting in hepatocellular carcinoma. Acta Histochem. 2018, 120, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Qian, S.; Yang, G.; Zhu, L.; Zhou, B.; Wang, J.; Liu, R.; Yan, Z.; Qu, X. MicroRNA-199 suppresses cell proliferation, migration and invasion by downregulating RGS17 in hepatocellular carcinoma. Gene 2018, 659, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.Y.; Yeh, K.Y.; Lin, C.Y.; Hsieh, Y.W.; Lai, H.H.; Chen, J.R.; Hsu, C.C.; Her, G.M. MicroRNA-21 Plays Multiple Oncometabolic Roles in the Process of NAFLD-Related Hepatocellular Carcinoma via PI3K/AKT, TGF-beta, and STAT3 Signaling. Cancers 2021, 13, 940. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Ye, M.; Wang, F.; Fang, J.; Wang, C.; Luo, J.; Liu, J.; Liu, J.; Liu, L.; Zhao, Q.; et al. MiR-21-3p Promotes Hepatocellular Carcinoma Progression via SMAD7/YAP1 Regulation. Front. Oncol. 2021, 11, 642030. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ren, H.; Wang, J.; Zhang, P.; Shi, X. Extracellular HMGB1 promotes CD44 expression in hepatocellular carcinoma via regulating miR-21. Aging 2021, 13, 8380–8395. [Google Scholar] [CrossRef]
- Yu, Q.; Zhou, J.; Jian, Y.; Xiu, Z.; Xiang, L.; Yang, D.; Zeng, W. MicroRNA-214 suppresses cell proliferation and migration and cell metabolism by targeting PDK2 and PHF6 in hepatocellular carcinoma. Cell Biol. Int. 2020, 44, 117–126. [Google Scholar] [CrossRef]
- Huang, P.S.; Lin, Y.H.; Chi, H.C.; Chen, P.Y.; Huang, Y.H.; Yeh, C.T.; Wang, C.S.; Lin, K.H. Thyroid hormone inhibits growth of hepatoma cells through induction of miR-214. Sci. Rep. 2017, 7, 14868. [Google Scholar] [CrossRef]
- Karimkhanloo, H.; Mohammadi-Yeganeh, S.; Hadavi, R.; Koochaki, A.; Paryan, M. Potential role of miR-214 in beta-catenin gene expression within hepatocellular carcinoma. Mol. Biol. Rep. 2020, 47, 7429–7437. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Yuan, J.; Zhang, N.; Zheng, Y.; Liu, J.; Yang, M. Silencing of lncRNA PVT1 by miR-214 inhibits the oncogenic GDF15 signaling and suppresses hepatocarcinogenesis. Biochem. Biophys. Res. Commun. 2020, 521, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Gong, X.; Yang, J.; Ouyang, L.; Xiao, R.; You, X.; Ouyang, Y. Suppressive role of microRNA-29 in hepatocellular carcinoma via targeting IGF2BP1. Int. J. Clin. Exp. Pathol. 2018, 11, 1175–1185. [Google Scholar]
- Jampoka, K.; Muangpaisarn, P.; Khongnomnan, K.; Treeprasertsuk, S.; Tangkijvanich, P.; Payungporn, S. Serum miR-29a and miR-122 as Potential Biomarkers for Non-Alcoholic Fatty Liver Disease (NAFLD). MicroRNA 2018, 7, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.S.; Dai, G.L.; Liu, S.J. MicroRNA-29 family functions as a tumor suppressor by targeting RPS15A and regulating cell cycle in hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2017, 10, 8031–8042. [Google Scholar]
- Zhou, Y.; Li, K.; Dai, T.; Wang, H.; Hua, Z.; Bian, W.; Wang, H.; Chen, F.; Ai, X. Long non-coding RNA HCP5 functions as a sponge of miR-29b-3p and promotes cell growth and metastasis in hepatocellular carcinoma through upregulating DNMT3A. Aging 2021, 13, 16267–16286. [Google Scholar] [CrossRef]
- Xie, S.C.; Zhang, J.Q.; Jiang, X.L.; Hua, Y.Y.; Xie, S.W.; Qin, Y.A.; Yang, Y.J. LncRNA CRNDE facilitates epigenetic suppression of CELF2 and LATS2 to promote proliferation, migration and chemoresistance in hepatocellular carcinoma. Cell Death Dis. 2020, 11, 676. [Google Scholar] [CrossRef]
- Lin, C.; Xiang, Y.; Sheng, J.; Liu, S.; Cui, M.; Zhang, X. Long non-coding RNA CRNDE promotes malignant progression of hepatocellular carcinoma through the miR-33a-5p/CDK6 axis. J. Physiol. Biochem. 2020, 76, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Gamaev, L.; Mizrahi, L.; Friehmann, T.; Rosenberg, N.; Pappo, O.; Olam, D.; Zeira, E.; Bahar Halpern, K.; Caruso, S.; Zucman-Rossi, J.; et al. The pro-oncogenic effect of the lncRNA H19 in the development of chronic inflammation-mediated hepatocellular carcinoma. Oncogene 2021, 40, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, Y.; Li, Z.; Li, H.; Li, X.; Yan, L.; Mao, J.; Shen, J.; Chen, W.; Xue, F. Long non-coding RNA H19 is involved in sorafenib resistance in hepatocellular carcinoma by upregulating miR-675. Oncol. Rep. 2020, 44, 165–173. [Google Scholar] [CrossRef]
- Wong, C.M.; Wei, L.; Au, S.L.; Fan, D.N.; Zhou, Y.; Tsang, F.H.; Law, C.T.; Lee, J.M.; He, X.; Shi, J.; et al. MiR-200b/200c/429 subfamily negatively regulates Rho/ROCK signaling pathway to suppress hepatocellular carcinoma metastasis. Oncotarget 2015, 6, 13658–13670. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; Chu, L.; Liang, J.; Tan, X.; Wang, Y.; Wen, J.; Chen, J.; Wu, Y.; Liu, S.; Liao, J.; et al. H19 Promotes HCC Bone Metastasis Through Reducing Osteoprotegerin Expression in a Protein Phosphatase 1 Catalytic Subunit Alpha/p38 Mitogen-Activated Protein Kinase-Dependent Manner and Sponging microRNA 200b-3p. Hepatology 2021, 74, 214–232. [Google Scholar] [CrossRef]
- Wei, L.; Wang, X.; Lv, L.; Liu, J.; Xing, H.; Song, Y.; Xie, M.; Lei, T.; Zhang, N.; Yang, M. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol. Cancer 2019, 18, 147. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, M.; Dureja, H. Sorafenib for hepatocellular carcinoma: Potential molecular targets and resistance mechanisms. J. Chemother. 2021, 1–16. [Google Scholar] [CrossRef]
- Ding, B.; Lou, W.; Xu, L.; Fan, W. Non-coding RNA in drug resistance of hepatocellular carcinoma. Biosci. Rep. 2018, 38, BSR20180915. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qin, C.; Yang, G.; Zhao, B.; Wang, W. The role of autophagy in pancreatic cancer progression. Biochim. Biophys. Acta. Rev. Cancer 2021, 1876, 188592. [Google Scholar] [CrossRef]
- Xia, Y.H.; Ren, L.; Li, J.Z.; Gao, F. Role of miR-541-3p/TMPRSS4 in the metastasis and EMT of hepatocellular carcinoma. Eur. Rev. Med Pharmacol. Sci. 2019, 23, 10721–10728. [Google Scholar]
- Xu, W.P.; Liu, J.P.; Feng, J.F.; Zhu, C.P.; Yang, Y.; Zhou, W.P.; Ding, J.; Huang, C.K.; Cui, Y.L.; Ding, C.H.; et al. miR-541 potentiates the response of human hepatocellular carcinoma to sorafenib treatment by inhibiting autophagy. Gut 2020, 69, 1309–1321. [Google Scholar] [CrossRef]
- Han, W.; Li, N.; Liu, J.; Sun, Y.; Yang, X.; Wang, Y. MicroRNA-26b-5p enhances T cell responses by targeting PIM-2 in hepatocellular carcinoma. Cell. Signal. 2019, 59, 182–190. [Google Scholar] [CrossRef]
- Hu, X.; Wang, R.; Ren, Z.; Liu, X.; Gu, J.; Cui, G.; Li, Q. MiR-26b suppresses hepatocellular carcinoma development by negatively regulating ZNRD1 and Wnt/β-catenin signaling. Cancer Med. 2019, 8, 7359–7371. [Google Scholar] [CrossRef]
- Chen, E.; Li, E.; Liu, H.; Zhou, Y.; Wen, L.; Wang, J.; Wang, Y.; Ye, L.; Liang, T. miR-26b enhances the sensitivity of hepatocellular carcinoma to Doxorubicin via USP9X-dependent degradation of p53 and regulation of autophagy. Int. J. Biol. Sci. 2021, 17, 781–795. [Google Scholar] [CrossRef] [PubMed]
- Pratedrat, P.; Chuaypen, N.; Nimsamer, P.; Payungporn, S.; Pinjaroen, N.; Sirichindakul, B.; Tangkijvanich, P. Diagnostic and prognostic roles of circulating miRNA-223-3p in hepatitis B virus-related hepatocellular carcinoma. PLoS ONE 2020, 15, e0232211. [Google Scholar]
- Zhou, Y.; Chen, E.; Tang, Y.; Mao, J.; Shen, J.; Zheng, X.; Xie, S.; Zhang, S.; Wu, Y.; Liu, H.; et al. miR-223 overexpression inhibits doxorubicin-induced autophagy by targeting FOXO3a and reverses chemoresistance in hepatocellular carcinoma cells. Cell Death Dis. 2019, 10, 843. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Liang, Q.; Qiao, L.; Xiao, F. MicroRNAs Modulate Drug Resistance-Related Mechanisms in Hepatocellular Carcinoma. Front. Oncol. 2020, 10, 920. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Qin, Z.; Cai, S.; Yu, L.; Hu, H.; Zeng, S. The role of non-coding RNAs in ABC transporters regulation and their clinical implications of multidrug resistance in cancer. Expert Opin. Drug Metab. Toxicol. 2021, 17, 291–306. [Google Scholar] [CrossRef]
- Yahya, S.M.M.; Fathy, S.A.; El-Khayat, Z.A.; El-Toukhy, S.E.; Hamed, A.R.; Hegazy, M.G.A.; Nabih, H.K. Possible Role of microRNA-122 in Modulating Multidrug Resistance of Hepatocellular Carcinoma. Indian J. Clin. Biochem. 2018, 33, 21–30. [Google Scholar] [CrossRef]
- Cao, F.; Yin, L.X. miR-122 enhances sensitivity of hepatocellular carcinoma to oxaliplatin via inhibiting MDR1 by targeting Wnt/beta-catenin pathway. Exp. Mol. Pathol. 2019, 106, 34–43. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Z.; Dou, C.; Han, S.; Li, C.; Tu, K.; Yang, W. [miR-491 inhibits the proliferation, invasion and migration of hepatocellular carcinoma cell via down-regulating TPX2 expression]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi Chin. J. Cell. Mol. Immunol. 2016, 32, 512–517. [Google Scholar]
- Zhao, Y.; Qi, X.; Chen, J.; Wei, W.; Yu, C.; Yan, H.; Pu, M.; Li, Y.; Miao, L.; Li, C.; et al. The miR-491-3p/Sp3/ABCB1 axis attenuates multidrug resistance of hepatocellular carcinoma. Cancer Lett. 2017, 408, 102–111. [Google Scholar] [CrossRef]
- Li, T.T.; Mou, J.; Pan, Y.J.; Huo, F.C.; Du, W.Q.; Liang, J.; Wang, Y.; Zhang, L.S.; Pei, D.S. MicroRNA-138-1-3p sensitizes sorafenib to hepatocellular carcinoma by targeting PAK5 mediated β-catenin/ABCB1 signaling pathway. J. Biomed. Sci. 2021, 28, 56. [Google Scholar] [CrossRef]
- Song, K.A.; Faber, A.C. Epithelial-to-mesenchymal transition and drug resistance: Transitioning away from death. J. Thorac. Dis. 2019, 11, E82–E85. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Powell, R.T.; Yuan, X.; Bae, G.; Roarty, K.P.; Stossi, F.; Strempfl, M.; Toneff, M.J.; Johnson, H.L.; Mani, S.A.; et al. Morphological screening of mesenchymal mammary tumor organoids to identify drugs that reverse epithelial-mesenchymal transition. Nat. Commun. 2021, 12, 4262. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.W.; Zhou, Y.; Wei, T.; Wen, L.; Zhang, Y.B.; Shen, S.C.; Zhang, J.; Ma, T.; Chen, W.; Ni, L.; et al. lncRNA-POIR promotes epithelial-mesenchymal transition and suppresses sorafenib sensitivity simultaneously in hepatocellular carcinoma by sponging miR-182-5p. J. Cell. Biochem. 2021, 122, 130–142. [Google Scholar] [CrossRef]
- Yang, H.; Li, Y.; Zhong, X.; Luo, P.; Luo, P.; Sun, R.; Xie, R.; Fu, D.; Ma, Y.; Cong, X.; et al. Upregulation of microRNA-32 is associated with tumorigenesis and poor prognosis in patients with hepatocellular carcinoma. Oncol. Lett. 2018, 15, 4097–4104. [Google Scholar] [CrossRef]
- Yan, S.Y.; Chen, M.M.; Li, G.M.; Wang, Y.Q.; Fan, J.G. MiR-32 induces cell proliferation, migration, and invasion in hepatocellular carcinoma by targeting PTEN. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2015, 36, 4747–4755. [Google Scholar] [CrossRef]
- Fu, X.; Liu, M.; Qu, S.; Ma, J.; Zhang, Y.; Shi, T.; Wen, H.; Yang, Y.; Wang, S.; Wang, J.; et al. Exosomal microRNA-32-5p induces multidrug resistance in hepatocellular carcinoma via the PI3K/Akt pathway. J. Exp. Clin. Cancer Res. CR 2018, 37, 52. [Google Scholar] [CrossRef] [Green Version]
- Zeng, T.; Luo, L.; Huang, Y.; Ye, X.; Lin, J. Upregulation of miR-138 Increases Sensitivity to Cisplatin in Hepatocellular Carcinoma by Regulating EZH2. BioMed Res. Int. 2021, 2021, 6665918. [Google Scholar] [CrossRef]
- Bao, Y.; Zhang, Y.; Lu, Y.; Guo, H.; Dong, Z.; Chen, Q.; Zhang, X.; Shen, W.; Chen, W.; Wang, X. Overexpression of microRNA-9 enhances cisplatin sensitivity in hepatocellular carcinoma by regulating EIF5A2-mediated epithelial-mesenchymal transition. Int. J. Biol. Sci. 2020, 16, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.G.; Lee, S.H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018, 2018, 5416923. [Google Scholar] [CrossRef] [Green Version]
- Zhong, D.; Lyu, X.; Fu, X.; Xie, P.; Liu, M.; He, F.; Huang, G. Upregulation of miR-124-3p by Liver X Receptor Inhibits the Growth of Hepatocellular Carcinoma Cells Via Suppressing Cyclin D1 and CDK6. Technol. Cancer Res. Treat. 2020, 19, 1533033820967473. [Google Scholar] [CrossRef]
- Feng, Y.; Jiang, W.; Zhao, W.; Lu, Z.; Gu, Y.; Dong, Y. miR-124 regulates liver cancer stem cells expansion and sorafenib resistance. Exp. Cell Res. 2020, 394, 112162. [Google Scholar] [CrossRef]
- Wang, Y.; Tai, Q.; Zhang, J.; Kang, J.; Gao, F.; Zhong, F.; Cai, L.; Fang, F.; Gao, Y. MiRNA-206 inhibits hepatocellular carcinoma cell proliferation and migration but promotes apoptosis by modulating cMET expression. Acta Biochim. Biophys. Sin. 2019, 51, 243–253. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, L.; Zhong, Y.; Lai, L.; Li, X. miR-206 inhibits cell proliferation, invasion, and migration by down-regulating PTP1B in hepatocellular carcinoma. Biosci. Rep. 2019, 39, BSR20181823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Li, J.; Wang, W.; Zhong, X.; Xu, F.; Lu, J. miR-206 inhibits liver cancer stem cell expansion by regulating EGFR expression. Cell Cycle 2020, 19, 1077–1088. [Google Scholar] [CrossRef]
- Ran, R.Z.; Chen, J.; Cui, L.J.; Lin, X.L.; Fan, M.M.; Cong, Z.Z.; Zhang, H.; Tan, W.F.; Zhang, G.Q.; Zhang, Y.J. miR-194 inhibits liver cancer stem cell expansion by regulating RAC1 pathway. Exp. Cell Res. 2019, 378, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Tribolet, L.; Kerr, E.; Cowled, C.; Bean, A.G.D.; Stewart, C.R.; Dearnley, M.; Farr, R.J. MicroRNA Biomarkers for Infectious Diseases: From Basic Research to Biosensing. Front. Microbiol. 2020, 11, 1197. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, T.; Liu, Z.; Han, Z.; Ge, Q. MicroRNA Detection Specificity: Recent Advances and Future Perspective. Anal. Chem. 2019, 91, 3179–3186. [Google Scholar] [CrossRef] [Green Version]
- Dave, V.P.; Ngo, T.A.; Pernestig, A.K.; Tilevik, D.; Kant, K.; Nguyen, T.; Wolff, A.; Bang, D.D. MicroRNA amplification and detection technologies: Opportunities and challenges for point of care diagnostics. Lab. Investig. A J. Tech. Methods Pathol. 2019, 99, 452–469. [Google Scholar] [CrossRef] [PubMed]
- Diener, C.; Galata, V.; Keller, A.; Meese, E. MicroRNA profiling from dried blood samples. Crit. Rev. Clin. Lab. Sci. 2019, 56, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Enguita, F.J. New promising circulating RNA biomarkers for early diagnosis of lung adenocarcinoma. Ann. Transl. Med. 2019, 7 (Suppl. S3), S130. [Google Scholar] [CrossRef]
- Rifai, N.; Gillette, M.A.; Carr, S.A. Protein biomarker discovery and validation: The long and uncertain path to clinical utility. Nat. Biotechnol. 2006, 24, 971–983. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Morine, Y.; Ikemoto, T.; Imura, S.; Higashijima, J.; Iwahashi, S.; Saito, Y.U.; Takasu, C.; Yamada, S.; Ishikawa, D.; et al. Elevated Preoperative Serum CEA Level Is Associated with Poor Prognosis in Patients with Hepatocellular Carcinoma Through the Epithelial-Mesenchymal Transition. Anticancer Res. 2017, 37, 1169–1175. [Google Scholar]
- Edoo, M.I.A.; Chutturghoon, V.K.; Wusu-Ansah, G.K.; Zhu, H.; Zhen, T.Y.; Xie, H.Y.; Zheng, S.S. Serum Biomarkers AFP, CEA and CA19-9 Combined Detection for Early Diagnosis of Hepatocellular Carcinoma. Iran. J. Public Health 2019, 48, 314–322. [Google Scholar]
- Xi, X.; Li, T.; Huang, Y.; Sun, J.; Zhu, Y.; Yang, Y.; Lu, Z.J. RNA Biomarkers: Frontier of Precision Medicine for Cancer. Non-Coding RNA 2017, 3, 9. [Google Scholar] [CrossRef]
- Glinge, C.; Clauss, S.; Boddum, K.; Jabbari, R.; Jabbari, J.; Risgaard, B.; Tomsits, P.; Hildebrand, B.; Kaab, S.; Wakili, R.; et al. Stability of Circulating Blood-Based MicroRNAs - Pre-Analytic Methodological Considerations. PLoS ONE 2017, 12, e0167969. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pal, M.K.; Jaiswar, S.P.; Dwivedi, V.N.; Tripathi, A.K.; Dwivedi, A.; Sankhwar, P. MicroRNA: A new and promising potential biomarker for diagnosis and prognosis of ovarian cancer. Cancer Biol. Med. 2015, 12, 328–341. [Google Scholar]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [Green Version]
- Saliminejad, K.; Khorram Khorshid, H.R.; Ghaffari, S.H. Why have microRNA biomarkers not been translated from bench to clinic? Future Oncol. 2019, 15, 801–803. [Google Scholar] [CrossRef]
- Bajan, S.; Hutvagner, G. RNA-Based Therapeutics: From Antisense Oligonucleotides to miRNAs. Cells 2020, 9, 137. [Google Scholar] [CrossRef] [Green Version]
- Segal, M.; Slack, F.J. Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opin. Drug Discov. 2020, 15, 987–992. [Google Scholar] [CrossRef]
- Gagliardi, M.; Ashizawa, A.T. The Challenges and Strategies of Antisense Oligonucleotide Drug Delivery. Biomedicines 2021, 9, 137. [Google Scholar] [CrossRef]
- Hydbring, P.; Badalian-Very, G. Clinical applications of microRNAs. F1000Research 2013, 2, 136. [Google Scholar] [CrossRef] [PubMed]
- Byrnes, S.A.; Weigl, B.H. Selecting analytical biomarkers for diagnostic applications: A first principles approach. Expert Rev. Mol. Diagn. 2018, 18, 19–26. [Google Scholar] [CrossRef]
- Nielsen, M.M.; Pedersen, J.S. miRNA activity inferred from single cell mRNA expression. Sci. Rep. 2021, 11, 9170. [Google Scholar] [CrossRef]
- Zhou, X.; Fang, S.; Wang, M.; Xiong, A.; Zheng, C.; Wang, J.; Yin, C. Diagnostic value of circulating miRNA-122 for hepatitis B virus and/or hepatitis C virus-associated chronic viral hepatitis. Biosci. Rep. 2019, 39, BSR20190900. [Google Scholar] [CrossRef] [Green Version]
- Trung, N.T.; Hoan, N.X.; Trung, P.Q.; Binh, M.T.; Van Tong, H.; Toan, N.L.; Bang, M.H.; Song, L.H. Clinical significance of combined circulating TERT promoter mutations and miR-122 expression for screening HBV-related hepatocellular carcinoma. Sci. Rep. 2020, 10, 8181. [Google Scholar] [CrossRef] [PubMed]
- Turato, C.; Fornari, F.; Pollutri, D.; Fassan, M.; Quarta, S.; Villano, G.; Ruvoletto, M.; Bolondi, L.; Gramantieri, L.; Pontisso, P. MiR-122 Targets SerpinB3 and Is Involved in Sorafenib Resistance in Hepatocellular Carcinoma. J. Clin. Med. 2019, 8, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Huang, J.; Ma, L.; Shan, J.; Shen, J.; Yang, Z.; Liu, L.; Luo, Y.; Yao, C.; Qian, C. MicroRNA-122 confers sorafenib resistance to hepatocellular carcinoma cells by targeting IGF-1R to regulate RAS/RAF/ERK signaling pathways. Cancer Lett. 2016, 371, 171–181. [Google Scholar] [CrossRef]
- Tat Trung, N.; Duong, D.C.; Tong, H.V.; Hien, T.T.T.; Hoan, P.Q.; Bang, M.H.; Binh, M.T.; Ky, T.D.; Tung, N.L.; Thinh, N.T.; et al. Optimisation of quantitative miRNA panels to consolidate the diagnostic surveillance of HBV-related hepatocellular carcinoma. PLoS ONE 2018, 13, e0196081. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Miu, K.K.; Zhang, X.; Wan, A.T.; Lu, G.; Cheung, H.H.; Lee, H.M.; Kong, A.P.; Chan, J.C.; Chan, W.Y. Hepatic miR-192-3p reactivation alleviates steatosis by targeting glucocorticoid receptor. JHEP Rep. Innov. Hepatol. 2020, 2, 100179. [Google Scholar] [CrossRef]
- Raut, A.; Khanna, A. Enhanced expression of hepatocyte-specific microRNAs in valproic acid mediated hepatic trans-differentiation of human umbilical cord derived mesenchymal stem cells. Exp. Cell Res. 2016, 343, 237–247. [Google Scholar] [CrossRef]
- Ren, F.J.; Yao, Y.; Cai, X.Y.; Fang, G.Y. Emerging Role of MiR-192-5p in Human Diseases. Front. Pharmacol. 2021, 12, 614068. [Google Scholar] [CrossRef] [PubMed]
- Musaddaq, G.; Shahzad, N.; Ashraf, M.A.; Arshad, M.I. Circulating liver-specific microRNAs as noninvasive diagnostic biomarkers of hepatic diseases in human. Biomark. Biochem. Indic. Expo. Response Susceptibility Chem. 2019, 24, 103–109. [Google Scholar] [CrossRef]
- Sorop, A.; Constantinescu, D.; Cojocaru, F.; Dinischiotu, A.; Cucu, D.; Dima, S.O. Exosomal microRNAs as Biomarkers and Therapeutic Targets for Hepatocellular Carcinoma. Int. J. Mol. Sci. 2021, 22, 4997. [Google Scholar] [CrossRef]
- Ge, Q.; Zhou, Y.; Lu, J.; Bai, Y.; Xie, X.; Lu, Z. miRNA in plasma exosome is stable under different storage conditions. Molecules 2014, 19, 1568–1575. [Google Scholar] [CrossRef] [Green Version]
- Suehiro, T.; Miyaaki, H.; Kanda, Y.; Shibata, H.; Honda, T.; Ozawa, E.; Miuma, S.; Taura, N.; Nakao, K. Serum exosomal microRNA-122 and microRNA-21 as predictive biomarkers in transarterial chemoembolization-treated hepatocellular carcinoma patients. Oncol. Lett. 2018, 16, 3267–3273. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, C.; Zhang, P.; Guo, G.; Jiang, T.; Zhao, X.; Jiang, J.; Huang, X.; Tong, H.; Tian, Y. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med. 2018, 7, 1670–1679. [Google Scholar] [CrossRef] [Green Version]
- Fornari, F.; Ferracin, M.; Trere, D.; Milazzo, M.; Marinelli, S.; Galassi, M.; Venerandi, L.; Pollutri, D.; Patrizi, C.; Borghi, A.; et al. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, Identify Cirrhotic Patients with HCC. PLoS ONE 2015, 10, e0141448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zhang, L.; Liu, F.; Xiang, G.; Jiang, D.; Pu, X. Identification of endogenous controls for analyzing serum exosomal miRNA in patients with hepatitis B or hepatocellular carcinoma. Dis. Markers 2015, 2015, 893594. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Hu, J.; Zhou, K.; Chen, F.; Wang, Z.; Liao, B.; Dai, Z.; Cao, Y.; Fan, J.; Zhou, J. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. OncoTargets Ther. 2017, 10, 3843–3851. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.; Jiang, Y.; Yang, L.; Yan, S.; Wang, Y.G.; Lu, X.J. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J. Cell. Biochem. 2018, 119, 4711–4716. [Google Scholar] [CrossRef] [PubMed]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Gebert, L.F.; Rebhan, M.A.; Crivelli, S.E.; Denzler, R.; Stoffel, M.; Hall, J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res. 2014, 42, 609–621. [Google Scholar] [CrossRef] [Green Version]
- Bonneau, E.; Neveu, B.; Kostantin, E.; Tsongalis, G.J.; De Guire, V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. Ejifcc 2019, 30, 114–127. [Google Scholar]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef] [Green Version]
- Rotman, Y.; Sanyal, A.J. Current and upcoming pharmacotherapy for non-alcoholic fatty liver disease. Gut 2017, 66, 180–190. [Google Scholar] [CrossRef] [Green Version]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- Schueller, F.; Roy, S.; Vucur, M.; Trautwein, C.; Luedde, T.; Roderburg, C. The Role of miRNAs in the Pathophysiology of Liver Diseases and Toxicity. Int. J. Mol. Sci. 2018, 19, 261. [Google Scholar] [CrossRef] [Green Version]
- Feili, X.; Wu, S.; Ye, W.; Tu, J.; Lou, L. MicroRNA-34a-5p inhibits liver fibrosis by regulating TGF-beta1/Smad3 pathway in hepatic stellate cells. Cell Biol. Int. 2018, 42, 1370–1376. [Google Scholar] [CrossRef]
- Chen, Q.Y.; Des Marais, T.; Costa, M. Deregulation of SATB2 in carcinogenesis with emphasis on miRNA-mediated control. Carcinogenesis 2019, 40, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, A.A.; Fayyaz, S.; Shatynska-Mytsyk, I.; Javed, Z.; Jabeen, S.; Yaylim, I.; Gasparri, M.L.; Panici, P.B. Is miR-34a a Well-equipped Swordsman to Conquer Temple of Molecular Oncology? Chem. Biol. Drug Des. 2016, 87, 321–334. [Google Scholar] [CrossRef]
- Mudduluru, G.; Ceppi, P.; Kumarswamy, R.; Scagliotti, G.V.; Papotti, M.; Allgayer, H. Regulation of Axl receptor tyrosine kinase expression by miR-34a and miR-199a/b in solid cancer. Oncogene 2011, 30, 2888–2899. [Google Scholar] [CrossRef]
- Van der Ree, M.H.; van der Meer, A.J.; de Bruijne, J.; Maan, R.; van Vliet, A.; Welzel, T.M.; Zeuzem, S.; Lawitz, E.J.; Rodriguez-Torres, M.; Kupcova, V.; et al. Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antivir. Res. 2014, 111, 53–59. [Google Scholar] [CrossRef]
- Goldaracena, N.; Spetzler, V.N.; Echeverri, J.; Kaths, J.M.; Cherepanov, V.; Persson, R.; Hodges, M.R.; Janssen, H.L.; Selzner, N.; Grant, D.R.; et al. Inducing Hepatitis C Virus Resistance After Pig Liver Transplantation-A Proof of Concept of Liver Graft Modification Using Warm Ex Vivo Perfusion. Am. J. Transplant. Off. J. Am. Soc. Transplant. Am. Soc. Transpl. Surg. 2017, 17, 970–978. [Google Scholar] [CrossRef]
- Li, Y.; Wang, L.; Rivera-Serrano, E.E.; Chen, X.; Lemon, S.M. TNRC6 proteins modulate hepatitis C virus replication by spatially regulating the binding of miR-122/Ago2 complexes to viral RNA. Nucleic Acids Res. 2019, 47, 6411–6424. [Google Scholar] [CrossRef]
- Lowey, B.; Hertz, L.; Chiu, S.; Valdez, K.; Li, Q.; Liang, T.J. Hepatitis C Virus Infection Induces Hepatic Expression of NF-kappaB-Inducing Kinase and Lipogenesis by Downregulating miR-122. mBio 2019, 10, e01617-19. [Google Scholar]
- Sidorkiewicz, M.; Grek-Kowalinska, M.; Piekarska, A. The Correlation between miR-122 and Lipoprotein Lipase Expression in Chronic Hepatitis C Patients. Can. J. Gastroenterol. Hepatol. 2018, 2018, 6348948. [Google Scholar] [CrossRef]
- Gerresheim, G.K.; Dunnes, N.; Nieder-Rohrmann, A.; Shalamova, L.A.; Fricke, M.; Hofacker, I.; Honer Zu Siederdissen, C.; Marz, M.; Niepmann, M. microRNA-122 target sites in the hepatitis C virus RNA NS5B coding region and 3′ untranslated region: Function in replication and influence of RNA secondary structure. Cell. Mol. Life Sci. 2017, 74, 747–760. [Google Scholar] [CrossRef]
- van der Ree, M.H.; Stelma, F.; Willemse, S.B.; Brown, A.; Swadling, L.; van der Valk, M.; Sinnige, M.J.; van Nuenen, A.C.; de Vree, J.M.L.; Klenerman, P.; et al. Immune responses in DAA treated chronic hepatitis C patients with and without prior RG-101 dosing. Antivir. Res. 2017, 146, 139–145. [Google Scholar] [CrossRef]
- Deng, Y.; Campbell, F.; Han, K.; Theodore, D.; Deeg, M.; Huang, M.; Hamatake, R.; Lahiri, S.; Chen, S.; Horvath, G.; et al. Randomized clinical trials towards a single-visit cure for chronic hepatitis C: Oral GSK2878175 and injectable RG-101 in chronic hepatitis C patients and long-acting injectable GSK2878175 in healthy participants. J. Viral Hepat. 2020, 27, 699–708. [Google Scholar] [CrossRef]
- Dajani, A.; AbuHammour, A. Treatment of nonalcoholic fatty liver disease: Where do we stand? an overview. Saudi J. Gastroenterol. Off. J. Saudi Gastroenterol. Assoc. 2016, 22, 91–105. [Google Scholar]
- Torres, J.L.; Novo-Veleiro, I.; Manzanedo, L.; Alvela-Suárez, L.; Macías, R.; Laso, F.J.; Marcos, M. Role of microRNAs in alcohol-induced liver disorders and non-alcoholic fatty liver disease. World J. Gastroenterol. 2018, 24, 4104–4118. [Google Scholar] [CrossRef]
- Chen, L.; Lu, Q.; Deng, F.; Peng, S.; Yuan, J.; Liu, C.; Du, X. miR-103a-3p Could Attenuate Sepsis-Induced Liver Injury by Targeting HMGB1. Inflammation 2020, 43, 2075–2086. [Google Scholar] [CrossRef]
- Fang, J.H.; Zhang, Z.J.; Shang, L.R.; Luo, Y.W.; Lin, Y.F.; Yuan, Y.; Zhuang, S.M. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology 2018, 68, 1459–1475. [Google Scholar] [CrossRef] [Green Version]
- Han, L.L.; Yin, X.R.; Zhang, S.Q. miR-103 promotes the metastasis and EMT of hepatocellular carcinoma by directly inhibiting LATS2. Int. J. Oncol. 2018, 53, 2433–2444. [Google Scholar] [CrossRef] [Green Version]
- Xiao, D.; Gao, H.X. Mechanism of miR-107-targeting of regulator of G-protein signaling 4 in hepatocellular carcinoma. Oncol. Lett. 2019, 18, 5145–5154. [Google Scholar] [CrossRef] [Green Version]
- Su, S.G.; Yang, M.; Zhang, M.F.; Peng, Q.Z.; Li, M.Y.; Liu, L.P.; Bao, S.Y. miR-107-mediated decrease of HMGCS2 indicates poor outcomes and promotes cell migration in hepatocellular carcinoma. Int. J. Biochem. Cell Biol. 2017, 91 Pt A, 53–59. [Google Scholar] [CrossRef]
- McDonald, M.K.; Capasso, K.E.; Ajit, S.K. Purification and microRNA profiling of exosomes derived from blood and culture media. J. Vis. Exp. 2013, 76, e50294. [Google Scholar] [CrossRef] [Green Version]
- McDonald, J.S.; Milosevic, D.; Reddi, H.V.; Grebe, S.K.; Algeciras-Schimnich, A. Analysis of circulating microRNA: Preanalytical and analytical challenges. Clin. Chem. 2011, 57, 833–840. [Google Scholar] [CrossRef] [Green Version]
- Pimentel, F.; Bonilla, P.; Ravishankar, Y.G.; Contag, A.; Gopal, N.; LaCour, S.; Lee, T.; Niemz, A. Technology in MicroRNA Profiling: Circulating MicroRNAs as Noninvasive Cancer Biomarkers in Breast Cancer. J. Lab. Autom. 2015, 20, 574–588. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Rana, T.M. Therapeutic targeting of microRNAs: Current status and future challenges. Nat. Rev. Drug Discov. 2014, 13, 622–638. [Google Scholar] [CrossRef] [PubMed]
- Hoofnagle, J.H.; Peters, M.; Mullen, K.D.; Jones, D.B.; Rustgi, V.; Di Bisceglie, A.; Hallahan, C.; Park, Y.; Meschievitz, C.; Jones, E.A. Randomized, controlled trial of recombinant human alpha-interferon in patients with chronic hepatitis B. Gastroenterology 1988, 95, 1318–1325. [Google Scholar] [CrossRef]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.J. Therapeutic siRNA: State of the art. Signal Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef] [PubMed]
- Lennox, K.A.; Behlke, M.A. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011, 18, 1111–1120. [Google Scholar] [CrossRef] [Green Version]
- Mescalchin, A.; Restle, T. Oligomeric nucleic acids as antivirals. Molecules 2011, 16, 1271–1296. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Qi, P.; Ma, Z. Biology of MiR-17-92 Cluster and Its Progress in Lung Cancer. Int. J. Med. Sci. 2018, 15, 1443–1448. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Schutt, S.; Paz, K.; Zhang, M.; Flynn, R.P.; Bastian, D.; Sofi, M.H.; Nguyen, H.; Dai, M.; Liu, C.; et al. MicroRNA-17-92 is required for T-cell and B-cell pathogenicity in chronic graft-versus-host disease in mice. Blood 2018, 131, 1974–1986. [Google Scholar] [CrossRef]
- Kuo, G.; Wu, C.Y.; Yang, H.Y. MiR-17-92 cluster and immunity. J. Formos. Med. Assoc. 2019, 118 Pt 1, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Forterre, A.; Komuro, H.; Aminova, S.; Harada, M. A Comprehensive Review of Cancer MicroRNA Therapeutic Delivery Strategies. Cancers 2020, 12, 1852. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Xu, L.; Jiao, Y.; Luo, S.; Li, A.; Wu, K. The role of cancer-derived microRNAs in cancer immune escape. J. Hematol. Oncol. 2020, 13, 25. [Google Scholar] [CrossRef] [Green Version]
- Mittelbrunn, M.; Gutierrez-Vazquez, C.; Villarroya-Beltri, C.; Gonzalez, S.; Sanchez-Cabo, F.; Gonzalez, M.A.; Bernad, A.; Sanchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef]
- Jasirwan, C.O.M.; Fahira, A.; Siregar, L.; Loho, I. The alpha-fetoprotein serum is still reliable as a biomarker for the surveillance of hepatocellular carcinoma in Indonesia. BMC Gastroenterol. 2020, 20, 215. [Google Scholar] [CrossRef]
- Luo, P.; Wu, S.; Yu, Y.; Ming, X.; Li, S.; Zuo, X.; Tu, J. Current Status and Perspective Biomarkers in AFP Negative HCC: Towards Screening for and Diagnosing Hepatocellular Carcinoma at an Earlier Stage. Pathol. Oncol. Res. 2020, 26, 599–603. [Google Scholar] [CrossRef]
- Huang, A.; Yang, X.R.; Chung, W.Y.; Dennison, A.R.; Zhou, J. Targeted therapy for hepatocellular carcinoma. Signal Transduct. Target. Ther. 2020, 5, 146. [Google Scholar] [CrossRef]
- Jeng, K.S.; Chang, C.F.; Jeng, W.J.; Sheen, I.S.; Jeng, C.J. Heterogeneity of hepatocellular carcinoma contributes to cancer progression. Crit. Rev. Oncol. Hematol. 2015, 94, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.C.; Hsu, C.H.; Hsu, C.; Cheng, A.L. Tumor Heterogeneity in Hepatocellular Carcinoma: Facing the Challenges. Liver Cancer 2016, 5, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Awan, F.M.; Naz, A.; Obaid, A.; Ikram, A.; Ali, A.; Ahmad, J.; Naveed, A.K.; Janjua, H.A. MicroRNA pharmacogenomics based integrated model of miR-17-92 cluster in sorafenib resistant HCC cells reveals a strategy to forestall drug resistance. Sci. Rep. 2017, 7, 11448. [Google Scholar] [CrossRef] [Green Version]
- Ji, L.; Lin, Z.; Wan, Z.; Xia, S.; Jiang, S.; Cen, D.; Cai, L.; Xu, J.; Cai, X. miR-486-3p mediates hepatocellular carcinoma sorafenib resistance by targeting FGFR4 and EGFR. Cell Death Dis. 2020, 11, 250. [Google Scholar] [CrossRef] [Green Version]
- He, C.; Dong, X.; Zhai, B.; Jiang, X.; Dong, D.; Li, B.; Jiang, H.; Xu, S.; Sun, X. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget 2015, 6, 28867–28881. [Google Scholar] [CrossRef] [Green Version]
- Pratama, M.Y.; Pascut, D.; Massi, M.N.; Tiribelli, C. The role of microRNA in the resistance to treatment of hepatocellular carcinoma. Ann. Transl. Med. 2019, 7, 577. [Google Scholar] [CrossRef]
- Zhang, S.; Cheng, Z.; Wang, Y.; Han, T. The Risks of miRNA Therapeutics: In a Drug Target Perspective. Drug Des Devel. Ther. 2021, 15, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Binzel, D.W.; Shu, Y.; Li, H.; Sun, M.; Zhang, Q.; Shu, D.; Guo, B.; Guo, P. Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology. Mol. Ther. 2016, 24, 1267–1277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhary, V.; Jangra, S.; Yadav, N.R. Nanotechnology based approaches for detection and delivery of microRNA in healthcare and crop protection. J. Nanobiotechnol. 2018, 16, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, P.-S.; Liao, C.-J.; Huang, Y.-H.; Yeh, C.-T.; Chen, C.-Y.; Tang, H.-C.; Chang, C.-C.; Lin, K.-H. Functional and Clinical Significance of Dysregulated microRNAs in Liver Cancer. Cancers 2021, 13, 5361. https://doi.org/10.3390/cancers13215361
Huang P-S, Liao C-J, Huang Y-H, Yeh C-T, Chen C-Y, Tang H-C, Chang C-C, Lin K-H. Functional and Clinical Significance of Dysregulated microRNAs in Liver Cancer. Cancers. 2021; 13(21):5361. https://doi.org/10.3390/cancers13215361
Chicago/Turabian StyleHuang, Po-Shuan, Chia-Jung Liao, Ya-Hui Huang, Chau-Ting Yeh, Cheng-Yi Chen, Hui-Chi Tang, Cheng-Chih Chang, and Kwang-Huei Lin. 2021. "Functional and Clinical Significance of Dysregulated microRNAs in Liver Cancer" Cancers 13, no. 21: 5361. https://doi.org/10.3390/cancers13215361