ID1 and ID4 Are Biomarkers of Tumor Aggressiveness and Poor Outcome in Immunophenotypes of Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
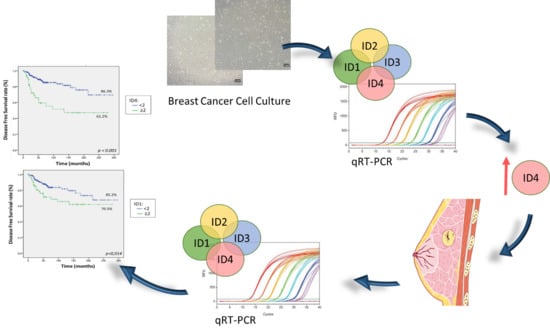
2.1. ID Genes Expression in BC Cell Lines
2.2. Patients’ and Tumor Characteristics
2.3. ID Genes Are Overexpressed in Most Analyzed Tumor Samples
2.4. ID1 and ID4 Overexpression Is Associated with Aggressive Clinicopathological Features
2.5. Patients Expressing High Levels of ID1 and ID4 Present Shorter OS and DFS
2.6. ID4 Is an Independent Marker of Poor Prognosis in Breast Cancer
2.7. Comparison with Available Databases
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Patients
4.3. Immunohistochemistry (IHC) and FISH
4.4. RNA Isolation and cDNA Synthesis
4.5. Quantitative Real-Time PCR (qRT-PCR)
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niell, B.L.; Freer, P.E.; Weinfurtner, R.J.; Arleo, E.K.; Drukteinis, J.S. Screening for Breast Cancer. Radiol. Clin. N. Am. 2017, 55, 1145–1162. [Google Scholar] [CrossRef] [PubMed]
- Coates, A.S.; Winer, E.P.; Goldhirsch, A.; Gelber, R.D.; Gnant, M.; Piccart-Gebhart, M.; Thurlimann, B.; Senn, H.J.; Panel, M. Tailoring therapies--improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann. Oncol. 2015, 26, 1533–1546. [Google Scholar] [CrossRef] [PubMed]
- SEER Cancer Statistics Review, 1975–2015. Available online: https://seer.cancer.gov/csr/1975_2015/ (accessed on 4 April 2019).
- Matsumoto, A.; Jinno, H.; Ando, T.; Fujii, T.; Nakamura, T.; Saito, J.; Takahashi, M.; Hayashida, T.; Kitagawa, Y. Biological markers of invasive breast cancer. Jpn. J. Clin. Oncol. 2016, 46, 99–105. [Google Scholar]
- American Cancer Society. Available online: https://www.cancer.org/cancer/breast-cancer/understanding-a-breast-cancer-diagnosis/breast-cancer-survival-rates.html (accessed on 5 April 2019).
- Stovgaard, E.S.; Nielsen, D.; Hogdall, E.; Balslev, E. Triple negative breast cancer - prognostic role of immune-related factors: A systematic review. Acta Oncol. 2018, 57, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Benezra, R.; Davis, R.L.; Lockshon, D.; Turner, D.L.; Weintraub, H. The protein Id: A negative regulator of helix-loop-helix DNA binding proteins. Cell 1990, 61, 49–59. [Google Scholar] [CrossRef]
- Ling, F.; Kang, B.; Sun, X.H. Id proteins: Small molecules, mighty regulators. Curr. Top. Dev. Biol. 2014, 110, 189–216. [Google Scholar]
- Riechmann, V.; van Cruchten, I.; Sablitzky, F. The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucleic Acids Res. 1994, 22, 749–755. [Google Scholar] [CrossRef]
- Roschger, C.; Cabrele, C. The Id-protein family in developmental and cancer-associated pathways. Cell Commun. Signal. 2017, 15, 7. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Yantiss, R.K.; Nam, H.S.; Chin, Y.; Zhou, X.K.; Scherl, E.J.; Bosworth, B.P.; Subbaramaiah, K.; Dannenberg, A.J.; Benezra, R. ID1 is a functional marker for intestinal stem and progenitor cells required for normal response to injury. Stem Cell Rep. 2014, 3, 716–724. [Google Scholar] [CrossRef] [Green Version]
- Colacino, J.A.; Azizi, E.; Brooks, M.D.; Harouaka, R.; Fouladdel, S.; McDermott, S.P.; Lee, M.; Hill, D.; Madden, J.; Boerner, J.; et al. Heterogeneity of Human Breast Stem and Progenitor Cells as Revealed by Transcriptional Profiling. Stem Cell Rep. 2018, 10, 1596–1609. [Google Scholar] [CrossRef] [Green Version]
- Nam, H.S.; Benezra, R. High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell. 2009, 5, 515–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaefer, B.M.; Koch, J.; Wirzbach, A.; Kramer, M.D. Expression of the helix-loop-helix protein ID1 in keratinocytes is upregulated by loss of cell-matrix contact. Exp. Cell Res. 2001, 266, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Lasorella, A.; Benezra, R.; Iavarone, A. The ID proteins: Master regulators of cancer stem cells and tumour aggressiveness. Nat. Rev. Cancer 2014, 14, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Amaral, L.H.P.; Bufalo, N.E.; Peres, K.C.; Barreto, I.S.; Campos, A.; Ward, L.S. ID Proteins May Reduce Aggressiveness of Thyroid Tumors. Endocr Pathol. 2019, 30, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Damdinsuren, B.; Nagano, H.; Kondo, M.; Yamamoto, H.; Hiraoka, N.; Yamamoto, T.; Marubashi, S.; Miyamoto, A.; Umeshita, K.; Dono, K.; et al. Expression of Id proteins in human hepatocellular carcinoma: Relevance to tumor dedifferentiation. Int. J. Oncol. 2005, 26, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Ding, R.; Han, S.; Lu, Y.; Guo, C.; Xie, H.; Zhang, N.; Song, Z.; Cai, L.; Liu, J.; Dou, K. Overexpressed Id-1 is associated with patient prognosis and HBx expression in hepatitis B virus-related hepatocellular carcinoma. Cancer Biol. Ther. 2010, 10, 299–307. [Google Scholar] [CrossRef] [Green Version]
- Ciarrocchi, A.; Piana, S.; Valcavi, R.; Gardini, G.; Casali, B. Inhibitor of DNA binding-1 induces mesenchymal features and promotes invasiveness in thyroid tumour cells. Eur. J. Cancer 2011, 47, 934–945. [Google Scholar] [CrossRef]
- Lin, C.Q.; Singh, J.; Murata, K.; Itahana, Y.; Parrinello, S.; Liang, S.H.; Gillett, C.E.; Campisi, J.; Desprez, P.Y. A role for Id-1 in the aggressive phenotype and steroid hormone response of human breast cancer cells. Cancer Res. 2000, 60, 1332–1340. [Google Scholar]
- Perk, J.; Gil-Bazo, I.; Chin, Y.; de Candia, P.; Chen, J.J.; Zhao, Y.; Chao, S.; Cheong, W.; Ke, Y.; Al-Ahmadie, H.; et al. Reassessment of id1 protein expression in human mammary, prostate, and bladder cancers using a monospecific rabbit monoclonal anti-id1 antibody. Cancer Res. 2006, 66, 10870–10877. [Google Scholar] [CrossRef] [Green Version]
- Schoppmann, S.F.; Schindl, M.; Bayer, G.; Aumayr, K.; Dienes, J.; Horvat, R.; Rudas, M.; Gnant, M.; Jakesz, R.; Birner, P. Overexpression of Id-1 is associated with poor clinical outcome in node negative breast cancer. Int. J. Cancer 2003, 104, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Wazir, U.; Jiang, W.G.; Sharma, A.K.; Newbold, R.F.; Mokbel, K. The mRNA expression of inhibitors of DNA binding-1 and -2 is associated with advanced tumour stage and adverse clinical outcome in human breast cancer. Anticancer Res. 2013, 33, 2179–2183. [Google Scholar] [PubMed]
- Zhou, X.L.; Zeng, D.E.; Ye, Y.H.; Sun, S.M.; Lu, X.F.; Liang, W.Q.; Chen, C.F.; Lin, H.Y. Prognostic values of the inhibitor of DNAbinding family members in breast cancer. Oncol. Rep. 2018, 40, 1897–1906. [Google Scholar]
- Li, K.; Yao, L.; Chen, L.; Cao, Z.G.; Yu, S.J.; Kuang, X.Y.; Hu, X.; Shao, Z.M. ID2 predicts poor prognosis in breast cancer, especially in triple-negative breast cancer, and inhibits E-cadherin expression. Onco. Targets Ther. 2014, 7, 1083–1094. [Google Scholar] [PubMed] [Green Version]
- Zhou, J.D.; Ma, J.C.; Zhang, T.J.; Li, X.X.; Zhang, W.; Wu, D.H.; Wen, X.M.; Xu, Z.J.; Lin, J.; Qian, J. High bone marrow ID2 expression predicts poor chemotherapy response and prognosis in acute myeloid leukemia. Oncotarget 2017, 8, 91979–91989. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, A.; Balwierz, W. The Role of Id2 Protein in Neuroblatoma in Children. Pathol. Oncol. Res. 2015, 21, 999–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stighall, M.; Manetopoulos, C.; Axelson, H.; Landberg, G. High ID2 protein expression correlates with a favourable prognosis in patients with primary breast cancer and reduces cellular invasiveness of breast cancer cells. Int. J. Cancer 2005, 115, 403–411. [Google Scholar] [CrossRef]
- Huang, L.; Cai, J.; Guo, H.; Gu, J.; Tong, Y.; Qiu, B.; Wang, C.; Li, M.; Xia, L.; Zhang, J.; et al. ID3 Promotes Stem Cell Features and Predicts Chemotherapeutic Response of Intrahepatic Cholangiocarcinoma. Hepatology 2019, 69, 1995–2012. [Google Scholar] [CrossRef]
- Sachindra, L.L.; Novak, D.; Wu, H.; Huser, L.; Granados, K.; Orouji, E.; Utikal, J. New role of ID3 in melanoma adaptive drug-resistance. Oncotarget 2017, 8, 110166–110175. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.S.; Aubee, J.; DiVito, K.A.; Zhou, H.; Zhang, W.; Chou, F.P.; Simbulan-Rosenthal, C.M.; Rosenthal, D.S. Id3 induces an Elk-1-caspase-8-dependent apoptotic pathway in squamous carcinoma cells. Cancer Med. 2015, 4, 914–924. [Google Scholar] [CrossRef]
- Chen, F.F.; Lv, X.; Zhao, Q.F.; Xu, Y.Z.; Song, S.S.; Yu, W.; Li, X.J. Inhibitor of DNA binding 3 reverses cisplatin resistance in human lung adenocarcinoma cells by regulating the PI3K/Akt pathway. Oncol. Lett. 2018, 16, 1634–1640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, D.; Morton, D.J.; Carey, J.; Havrda, M.C.; Chaudhary, J. Inhibitor of differentiation 4 (ID4): From development to cancer. Biochim. Biophys. Acta 2015, 1855, 92–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jen, Y.; Manova, K.; Benezra, R. Expression patterns of Id1, Id2, and Id3 are highly related but distinct from that of Id4 during mouse embryogenesis. Dev. Dyn. 1996, 207, 235–252. [Google Scholar] [CrossRef]
- Sharma, P.; Chinaranagari, S.; Chaudhary, J. Inhibitor of differentiation 4 (ID4) acts as an inhibitor of ID-1, -2 and -3 and promotes basic helix loop helix (bHLH) E47 DNA binding and transcriptional activity. Biochimie 2015, 112, 139–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhang, L.X.; Liu, X.Q.; Zhao, F.Y.; Ge, C.; Chen, T.Y.; Yao, M.; Li, J.J. Id4 promotes cell proliferation in hepatocellular carcinoma. Chin. J. Cancer 2017, 36, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, W.; Rushing, E.J.; Hartmann, D.P.; Azumi, N. Increased inhibitor of differentiation 4 (id4) expression in glioblastoma: A tissue microarray study. J. Cancer 2010, 1, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peretz, Y.; Wu, H.; Patel, S.; Bellacosa, A.; Katz, R.A. Inhibitor of DNA Binding 4 (ID4) is highly expressed in human melanoma tissues and may function to restrict normal differentiation of melanoma cells. PLoS ONE 2015, 10, e0116839. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Chinaranagari, S.; Patel, D.; Carey, J.; Chaudhary, J. Epigenetic inactivation of inhibitor of differentiation 4 (Id4) correlates with prostate cancer. Cancer Med. 2012, 1, 176–186. [Google Scholar] [CrossRef]
- Noetzel, E.; Veeck, J.; Niederacher, D.; Galm, O.; Horn, F.; Hartmann, A.; Knuchel, R.; Dahl, E. Promoter methylation-associated loss of ID4 expression is a marker of tumour recurrence in human breast cancer. BMC Cancer 2008, 8, 154. [Google Scholar] [CrossRef] [Green Version]
- Beger, C.; Pierce, L.N.; Kruger, M.; Marcusson, E.G.; Robbins, J.M.; Welcsh, P.; Welch, P.J.; Welte, K.; King, M.C.; Barber, J.R.; et al. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc. Natl. Acad. Sci. USA 2001, 98, 130–135. [Google Scholar] [CrossRef] [Green Version]
- Welcsh, P.L.; Lee, M.K.; Gonzalez-Hernandez, R.M.; Black, D.J.; Mahadevappa, M.; Swisher, E.M.; Warrington, J.A.; King, M.C. BRCA1 transcriptionally regulates genes involved in breast tumorigenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 7560–7565. [Google Scholar] [CrossRef] [Green Version]
- Cheang, M.C.; Chia, S.K.; Voduc, D.; Gao, D.; Leung, S.; Snider, J.; Watson, M.; Davies, S.; Bernard, P.S.; Parker, J.S.; et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J. Natl. Cancer Inst. 2009, 101, 736–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaplan-Meier Plotter. Available online: https://kmplot.com/analysis/ (accessed on 7 July 2019).
- Jang, K.S.; Han, H.X.; Paik, S.S.; Brown, P.H.; Kong, G. Id-1 overexpression in invasive ductal carcinoma cells is significantly associated with intratumoral microvessel density in ER-negative/node-positive breast cancer. Cancer Lett. 2006, 244, 203–210. [Google Scholar] [CrossRef] [PubMed]
- de Candia, P.; Akram, M.; Benezra, R.; Brogi, E. Id4 messenger RNA and estrogen receptor expression: Inverse correlation in human normal breast epithelium and carcinoma. Hum. Pathol. 2006, 37, 1032–1041. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.C.; Reis-Filho, J.S.; Russell, A.M.; Springall, R.J.; Ryder, K.; Steele, D.; Savage, K.; Gillett, C.E.; Schmitt, F.C.; Ashworth, A.; et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene 2007, 26, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- Agboola, A.O.J.; Banjo, A.A.F.; Anunobi, C.; Salami, B.; Deji-Agboola, M.; Musa, A.; Nolan, C.C.; Rakha, E.A.; Green Ellis, I.O.A.R. Helix-loop-helix protein inhibitor of differentiation 4 (ID4) expression is an indicator of poor survival in Nigerian breast cancer women. J. Afr. Cancer 2014, 6, 129–137. [Google Scholar] [CrossRef]
- Thike, A.A.; Tan, P.H.; Ikeda, M.; Iqbal, J. Increased ID4 expression, accompanied by mutant p53 accumulation and loss of BRCA1/2 proteins in triple-negative breast cancer, adversely affects survival. Histopathology 2016, 68, 702–712. [Google Scholar] [CrossRef]
- Donzelli, S.; Milano, E.; Pruszko, M.; Sacconi, A.; Masciarelli, S.; Iosue, I.; Melucci, E.; Gallo, E.; Terrenato, I.; Mottolese, M.; et al. Expression of ID4 protein in breast cancer cells induces reprogramming of tumour-associated macrophages. Breast Cancer Res. 2018, 20, 59. [Google Scholar] [CrossRef]
- Fontemaggi, G.; Dell’Orso, S.; Trisciuoglio, D.; Shay, T.; Melucci, E.; Fazi, F.; Terrenato, I.; Mottolese, M.; Muti, P.; Domany, E.; et al. The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nat. Struct. Mol. Biol. 2009, 16, 1086–1093. [Google Scholar] [CrossRef]
- Junankar, S.; Baker, L.A.; Roden, D.L.; Nair, R.; Elsworth, B.; Gallego-Ortega, D.; Lacaze, P.; Cazet, A.; Nikolic, I.; Teo, W.S.; et al. ID4 controls mammary stem cells and marks breast cancers with a stem cell-like phenotype. Nat. Commun. 2015, 6, 6548. [Google Scholar] [CrossRef]
- Baker, L.A.; Holliday, H.; Roden, D.; Krisp, C.; Wu, S.Z.; Junankar, S.; Serandour, A.A.; Mohammed, H.; Nair, R.; Sankaranarayanan, G.; et al. Proteogenomic analysis of Inhibitor of Differentiation 4 (ID4) in basal-like breast cancer. Breast Cancer Res. 2020, 22, 63. [Google Scholar] [CrossRef] [PubMed]
- Ruzinova, M.B.; Benezra, R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003, 13, 410–418. [Google Scholar] [CrossRef]
- Olmeda, D.; Moreno-Bueno, G.; Flores, J.M.; Fabra, A.; Portillo, F.; Cano, A. SNAI1 is required for tumor growth and lymph node metastasis of human breast carcinoma MDA-MB-231 cells. Cancer Res. 2007, 67, 11721–11731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wojnarowicz, P.M.; Lima, E.S.R.; Ohnaka, M.; Lee, S.B.; Chin, Y.; Kulukian, A.; Chang, S.H.; Desai, B.; Garcia Escolano, M.; Shah, R.; et al. A Small-Molecule Pan-Id Antagonist Inhibits Pathologic Ocular Neovascularization. Cell Rep. 2019, 29, 62–75. [Google Scholar] [CrossRef] [Green Version]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M. REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res. Treat. 2006, 100, 229–235. [Google Scholar] [CrossRef]
- Peiro, G.; Adrover, E.; Sanchez-Tejada, L.; Lerma, E.; Planelles, M.; Sanchez-Paya, J.; Aranda, F.I.; Giner, D.; Gutierrez-Avino, F.J. Increased insulin-like growth factor-1 receptor mRNA expression predicts poor survival in immunophenotypes of early breast carcinoma. Mod. Pathol. 2011, 24, 201–208. [Google Scholar] [CrossRef] [Green Version]
- Sanmartin, E.; Ortiz-Martinez, F.; Pomares-Navarro, E.; Garcia-Martinez, A.; Rodrigo-Banos, M.; Garcia-Escolano, M.; Andres, L.; Lerma, E.; Aranda, F.I.; Martinez-Peinado, P.; et al. CD44 induces FOXP3 expression and is related with favorable outcome in breast carcinoma. Virchows Arch. 2017, 470, 81–90. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.S.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 2018, 36, 2105–2122. [Google Scholar] [CrossRef] [Green Version]
- Maisonneuve, P.; Disalvatore, D.; Rotmensz, N.; Curigliano, G.; Colleoni, M.; Dellapasqua, S.; Pruneri, G.; Mastropasqua, M.G.; Luini, A.; Bassi, F.; et al. Proposed new clinicopathological surrogate definitions of luminal A and luminal B (HER2-negative) intrinsic breast cancer subtypes. Breast Cancer Res. 2014, 16, R65. [Google Scholar] [CrossRef] [Green Version]




| Patients (n = 297) | ID1 | ID4 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| FC < 2 (236) | FC ≥ 2 (61) | OR (95% CI) | p * | FC < 2 (248) | FC ≥ 2 (49) | OR (95% CI) | p * | ||
| % (n) | % (n) | % (n) | % (n) | ||||||
| Age | |||||||||
| ≥50 | 188 | 64.4% (152) | 59.0% (36) | 0.8 (0.5–1.4) | ns | 61.3% (152) | 73.5% (36) | 1.8 (0.9–3.5) | ns (0.071) |
| <50 | 109 | 35.6% (84) | 41.0% (25) | 1 | 38.7% (96) | 26.5% (13) | 1 | ||
| Size | |||||||||
| ≥20 | 162 | 51.3% (121) | 68.3% (41) | 2.1 (1.1–3.7) | 0.012 | 52.2% (129) | 67.3% (33) | 1.9 (1.0–3.6) | 0.036 |
| <20 | 134 | 48.7% (115) | 31.7% (19) | 1 | 47.8% (118) | 32.7% (16) | 1 | ||
| NA | 1 | ||||||||
| Histological Grade | |||||||||
| 1 | 30 | 11.9% (28) | 3.3% (2) | 1 | 11.7% (29) | 2.0% (1) | 1 | ||
| 2 | 89 | 33.1% (78) | 18.0% (11) | 2.0 (0.4–9.5) | ns | 32.3% (80) | 18.4% (9) | 3.3 (0.4–26.9) | ns |
| 3 | 178 | 55.1% (130) | 78.7% (48) | 5.2 (1.2–22.5) | 0.029 | 56.0% (139) | 79.6% (39) | 8.1 (1.1-61.6) | 0.042 |
| Necrosis | |||||||||
| Present | 100 | 29.9% (70) | 50.0% (30) | 2.3 (1.3–4.2) | 0.003 | 30.5% (75) | 52.1% (25) | 2.5 (1.3–4.6) | 0.004 |
| Absent | 194 | 70.1% (164) | 50.0% (30) | 1 | 69.5% (171) | 47.9% (23) | 1 | ||
| NA | 3 | ||||||||
| Vascular Invasion | |||||||||
| Present | 107 | 33.8% (79) | 45.9% (28) | 1.7 (0.9–3.0) | ns (0.055) | 33.3% (82) | 51.0% (25) | 2.1 (1.1–3.9) | 0.015 |
| Absent | 188 | 66.2% (155) | 54.1% (33) | 1 | 66.7% (164) | 49.0% (24) | 1 | ||
| NA | 2 | ||||||||
| Lymph Nodes Status | |||||||||
| Positive | 117 | 38.1% (90) | 44.3% (27) | 1.3 (0.7–2.3) | ns | 37.9% (94) | 46.9% (23) | 1.5 (0.8–2.7) | ns |
| Negative | 180 | 61.9% (146) | 55.7% (34) | 1 | 62.1% (154) | 53.1% (26) | 1 | ||
| Overall Survival | Disease-Free Survival | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | Univariate Analysis | Multivariate Analysis | |||||
| HR (95% CI) | p * | HR (95% CI) | p * | HR (95% CI) | p * | HR (95% CI) | p * | |
| Age | ||||||||
| ≥50 | 1.3 (0.7–2.3) | ns | 0.7 (0.4–1.1) | ns | ||||
| <50 | 1 | 1 | ||||||
| Size | ||||||||
| ≥20 | 2.8 (1.5–5.2) | 0.002 | 1.7 (0.8–3.5) | ns | 1.8 (1.0–3.2) | 0.034 | 1.3 (0.7–2.4) | ns |
| <20 | 1 | 1 | 1 | |||||
| Histological Grade | ||||||||
| 1 | 1 | 1 | 1 | |||||
| 2 | 4.2 (0.5–32.9) | ns | 2.5 (0.3–20.0) | ns | 1.3 (0.4–3.9) | 0.702 | ||
| 3 | 8.1 (1.1–58.8) | 0.040 | 2.6 (0.3–21.4) | ns | 2.1 (0.8–5.9) | 0.157 | ||
| Necrosis | ||||||||
| Present | 1.8 (1.0–3.2) | 0.042 | 1.4 (0.7–2.6) | ns | 1.8 (1.0–3.1) | 0.041 | 1.8 (1.0–3.1) | 0.050 |
| Absent | 1 | 1 | 1 | |||||
| Vascular Invasion | ||||||||
| Present | 2.6 (1.4–4.6) | 0.001 | 1.6 (0.8–3.1) | ns | 2.5 (1.4–4.2) | 0.001 | 1.9 (1.1–3.5) | 0.031 |
| Absent | 1 | 1 | 1 | |||||
| Lymph Nodes | ||||||||
| Positive | 2.6 (1.4–4.6) | 0.001 | 1.6 (0.8–3.1) | ns | 1.6 (0.9–2.7) | 0.107 | ||
| Negative | 1 | 1 | 1 | |||||
| HR Status | ||||||||
| Negative | 1.8 (1.0–3.3) | 0.039 | 1.3 (0.6–2.4) | ns | 1.6 (0.9–2.8) | ns (0.080) | ||
| Positive | 1 | 1 | 1 | |||||
| ID1 FC | ||||||||
| ≥2 | 1.3 (0.7–2.5) | ns | 2.0 (1.1–3.6) | 0.016 | 0.6 (0.3–1.5) | ns | ||
| <2 | 1 | 1 | ||||||
| ID2 FC | ||||||||
| ≥2 | 0.8 (0.4–1.7) | ns | 0.9 (0.4–1.7) | ns | ||||
| <2 | 1 | 1 | ||||||
| ID3 FC | ||||||||
| ≥2 | 1.0 (0.5–2.3) | ns | 0.9 (0.4–1.9) | ns | ||||
| <2 | 1 | 1 | ||||||
| ID4 FC | ||||||||
| ≥2 | 2.1 (1.2–3.9) | 0.016 | 1.6 (0.8–3.0) | ns (0.167) | 3.4 (1.9–6.0) | <0.001 | 4.0 (1.8–8.9) | 0.001 |
| <2 | 1 | 1 | 1 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia-Escolano, M.; Montoyo-Pujol, Y.G.; Ortiz-Martinez, F.; Ponce, J.J.; Delgado-Garcia, S.; Martin, T.A.; Ballester, H.; Aranda, F.I.; Castellon-Molla, E.; Sempere-Ortells, J.M.; et al. ID1 and ID4 Are Biomarkers of Tumor Aggressiveness and Poor Outcome in Immunophenotypes of Breast Cancer. Cancers 2021, 13, 492. https://doi.org/10.3390/cancers13030492
Garcia-Escolano M, Montoyo-Pujol YG, Ortiz-Martinez F, Ponce JJ, Delgado-Garcia S, Martin TA, Ballester H, Aranda FI, Castellon-Molla E, Sempere-Ortells JM, et al. ID1 and ID4 Are Biomarkers of Tumor Aggressiveness and Poor Outcome in Immunophenotypes of Breast Cancer. Cancers. 2021; 13(3):492. https://doi.org/10.3390/cancers13030492
Chicago/Turabian StyleGarcia-Escolano, Marta, Yoel G. Montoyo-Pujol, Fernando Ortiz-Martinez, Jose J. Ponce, Silvia Delgado-Garcia, Tina A. Martin, Hortensia Ballester, F. Ignacio Aranda, Elena Castellon-Molla, J. Miguel Sempere-Ortells, and et al. 2021. "ID1 and ID4 Are Biomarkers of Tumor Aggressiveness and Poor Outcome in Immunophenotypes of Breast Cancer" Cancers 13, no. 3: 492. https://doi.org/10.3390/cancers13030492