Comparison of the Tree-Based Machine Learning Algorithms to Cox Regression in Predicting the Survival of Oral and Pharyngeal Cancers: Analyses Based on SEER Database
Abstract
:Simple Summary
Abstract
1. Introduction
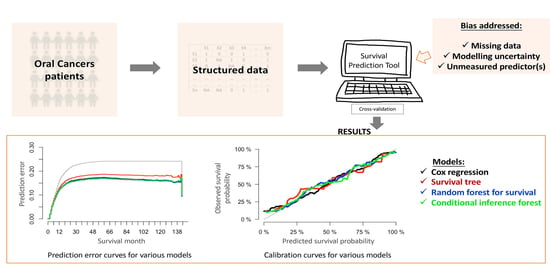
2. Results
2.1. Patient Selection
2.2. Characteristics of the Patients
2.3. Model Specification
2.4. Model Performance
2.5. Model Presentation and Development of an Online Survival Calculator
3. Discussion
4. Methods
4.1. Data Source and Study Population
4.2. Predictors and Outcome
4.3. Model Development
4.3.1. Cox Regression Model
4.3.2. Survival Tree Model
4.3.3. Random Forest Model
4.3.4. Random Forest Based on Conditional Inference Trees—Conditional Inference Forest (CF)
4.4. Missing Data
4.5. Model Validation and Performance Evaluation
4.6. Study Reporting and Software
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Du, M.; Nair, R.; Jamieson, L.; Liu, Z.; Bi, P. Incidence Trends of Lip, Oral Cavity, and Pharyngeal Cancers: Global Burden of Disease 1990–2017. J. Dent. Res. 2019, 99, 143–151. [Google Scholar] [CrossRef]
- Kioi, M. Recent advances in molecular-targeted therapy for oral cancer. Int. J. Oral Maxillofac. Surg. 2017, 46, 27. [Google Scholar] [CrossRef] [Green Version]
- Surveillance, Epidemiology, and End Results Program. Cancer Stat Facts: Oral Cavity and Pharynx Cancer. Available online: https://seer.cancer.gov/statfacts/html/oralcav.html (accessed on 30 November 2019).
- Patton, L.L. At the interface of medicine and dentistry: Shared decision-making using decision aids and clinical decision support tools. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2017, 123, 147–149. [Google Scholar] [CrossRef]
- Wolff, R.; Moons, K.G.; Riley, R.D.; Whiting, P.; Westwood, M.; Collins, G.S.; Reitsma, J.B.; Kleijnen, J.; Mallett, S. PROBAST: A Tool to Assess the Risk of Bias and Applicability of Prediction Model Studies. Ann. Intern. Med. 2019, 170, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (tripod): The tripod statement. Circulation 2015, 131, 211–219. [Google Scholar] [CrossRef] [Green Version]
- Du, M.; Haag, D.; Song, Y.; Lynch, J.; Mittinty, M. Examining Bias and Reporting in Oral Health Prediction Modeling Studies. J. Dent. Res. 2020, 99, 374–387. [Google Scholar] [CrossRef]
- Fakhry, C.; Zhang, Q.; Nguyen-Tân, P.F.; Rosenthal, D.; Weber, R.S.; Lambert, L.; Trotti, A.M.; Barrett, W.L.; Thorstad, W.L.; Jones, C.U.; et al. Development and Validation of Nomograms Predictive of Overall and Progression-Free Survival in Patients With Oropharyngeal Cancer. J. Clin. Oncol. 2017, 35, 4057–4065. [Google Scholar] [CrossRef]
- Chen, F.; Lin, L.; Yan, L.; Liu, F.; Qiu, Y.; Wang, J.; Hu, Z.; Wu, J.; Bao, X.; Lin, L.; et al. Nomograms and risk scores for predicting the risk of oral cancer in different sexes: A large-scale case-control study. J. Cancer 2018, 9, 2543–2548. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, H.; Wen, J.; Zhou, J.; Liu, Y.; Cheng, B.; Chen, X.; Wei, J. Nomograms forecasting long-term overall and cancer-specific survival of patients with oral squamous cell carcinoma. Cancer Med. 2018, 7, 943–952. [Google Scholar] [CrossRef]
- Breslow, N.E. Analysis of Survival Data under the Proportional Hazards Model. Int. Stat. Rev. 1975, 43, 45. [Google Scholar] [CrossRef]
- Chen, J.H.; Asch, S.M. Machine Learning and Prediction in Medicine—Beyond the Peak of Inflated Expectations. N. Engl. J. Med. 2017, 376, 2507–2509. [Google Scholar] [CrossRef] [Green Version]
- Ryo, M.; Rillig, M.C. Statistically reinforced machine learning for nonlinear patterns and variable interactions. Ecosphere 2017, 8, e01976. [Google Scholar] [CrossRef]
- Duda, R.O.; Hart, P.E.; Stork, D.G. Pattern Classification; John Wiley & Sons: New York, NY, USA, 2012; ISBN 0-471-05669-3. [Google Scholar]
- Kim, D.; Lee, S.; Kwon, S.; Nam, W.; Cha, I.; Kim, H.-J. Deep learning-based survival prediction of oral cancer patients. Sci. Rep. 2019, 9, 6994. [Google Scholar] [CrossRef] [Green Version]
- Tseng, W.-T.; Chiang, W.-F.; Liu, S.-Y.; Roan, J.; Lin, C.-N. The Application of Data Mining Techniques to Oral Cancer Prognosis. J. Med. Syst. 2015, 39, 59. [Google Scholar] [CrossRef]
- Harrell, F.E., Jr.; Califf, R.M.; Pryor, D.B.; Lee, K.L.; Rosati, R.A. Evaluating the yield of medical tests. JAMA 1982, 247, 2543–2546. [Google Scholar] [CrossRef]
- Mogensen, U.B.; Ishwaran, H.; Gerds, T.A. Evaluating random forests for survival analysis using prediction error curves. J. Stat. Softw. 2012, 50, 1–23. [Google Scholar] [CrossRef]
- Kamarudin, A.N.; Cox, T.; Kolamunage-Dona, R. Time-dependent ROC curve analysis in medical research: Current methods and applications. BMC Med. Res. Methodol. 2017, 17, 53. [Google Scholar] [CrossRef] [Green Version]
- Vittinghoff, E.; McCulloch, C.E. Relaxing the Rule of Ten Events per Variable in Logistic and Cox Regression. Am. J. Epidemiol. 2007, 165, 710–718. [Google Scholar] [CrossRef] [Green Version]
- Therneau, T.; Atkinson, B.; Ripley, B.; Ripley, M.B. Package ‘Rpart’. 2015. Available online: https://cran.r-project.org/web/packages/rpart/index.html (accessed on 23 September 2020).
- Ishwaran, H.; Kogalur, U.B.; Kogalur, M.U.B. Package ‘Randomforestsrc’. 2020. Available online: https://cran.r-project.org/web/packages/randomForestSRC/index.html (accessed on 23 September 2020).
- Breiman, L. Heuristics of instability and stabilization in model selection. Ann. Appl. Stat. 1996, 24, 2350–2383. [Google Scholar] [CrossRef]
- Ishwaran, H.; Kogalur, U.B.; Blackstone, E.H.; Lauer, M.S. Random survival forests. Ann. Appl. Stat. 2008, 2, 841–860. [Google Scholar] [CrossRef]
- Breiman, L. In Software for the masses, Wald Lectures. In Proceedings of the Meeting of the Institute of Mathematical Statistics, Banff, AB, Canada, 28–31 July 2002; pp. 255–294. [Google Scholar]
- Zhou, Y.; McArdle, J.J. Rationale and Applications of Survival Tree and Survival Ensemble Methods. Psychometrika 2014, 80, 811–833. [Google Scholar] [CrossRef]
- Nørregaard, C.; Grønhøj, C.; Jensen, D.; Friborg, J.; Andersen, E.; Von Buchwald, C. Cause-specific mortality in HPV+ and HPV− oropharyngeal cancer patients: Insights from a population-based cohort. Cancer Med. 2017, 7, 87–94. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Matsushita, Y.; Yamada, S.-I.; Yanamoto, S.; Shiraishi, T.; Asahina, I.; Umeda, M. Risk factors of distant metastasis in patients with squamous cell carcinoma of the oral cavity. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2016, 121, 474–480. [Google Scholar] [CrossRef]
- Snow, G.; Snow, M.G. Package ‘Obssens’. 2015. Available online: https://cran.r-project.org/web/packages/obsSens/index.html (accessed on 19 September 2020).
- Manual of Handlling Missing DATA in SEER Data. Available online: https://healthcaredelivery.cancer.gov/seer-cahps/researchers/missing-data-guidance.pdf (accessed on 15 September 2020).
- Segal, M.R. Regression Trees for Censored Data. Biometrics 1988, 44, 35. [Google Scholar] [CrossRef]
- Wright, M.N.; Dankowski, T.; Ziegler, A. Unbiased split variable selection for random survival forests using maximally selected rank statistics. Stat. Med. 2017, 36, 1272–1284. [Google Scholar] [CrossRef] [Green Version]
- Nasejje, J.; Mwambi, H.; Dheda, K.; Lesosky, M. A comparison of the conditional inference survival forest model to random survival forests based on a simulation study as well as on two applications with time-to-event data. BMC Med. Res. Methodol. 2017, 17, 115. [Google Scholar] [CrossRef]
- Allison, P. Handling Missing Data by Maximum Likelihood. Sas Global Forum Statistics and Data Analysis. 2012. Available online: http://www.statisticalhorizons.com/wp-content/uploads/MissingDataByML.pdf (accessed on 17 May 2020).
- Bartlett, J.W.; Seaman, S.R.; White, I.R.; Carpenter, J.R. Multiple imputation of covariates by fully conditional specification: Accommodating the substantive model. Stat. Methods Med. Res. 2014, 24, 462–487. [Google Scholar] [CrossRef]
- Pencina, M.J.; D’Agostino, R.B. Overall C as a measure of discrimination in survival analysis: Model specific population value and confidence interval estimation. Stat. Med. 2004, 23, 2109–2123. [Google Scholar] [CrossRef]
- Graf, E.; Schmoor, C.; Sauerbrei, W.; Schumacher, M. Assessment and comparison of prognostic classification schemes for survival data. Stat. Med. 1999, 18, 2529–2545. [Google Scholar] [CrossRef]
- Data and Codes Used in This Study. Available online: https://github.com/dumizai/SEER_OPCs_survival_prediction_code_data (accessed on 21 September 2020).




| Characteristics | 3-Year Cohort (Complete Cases, n = 11,888) | 5-Year Cohort (Complete Cases, n = 11,807) |
|---|---|---|
| Death Status | ||
| Alive | 7731 (65.0%) | 7092 (60.1%) |
| Dead | 4157 (35.0%) | 4715 (39.9%) |
| Survival months | ||
| Mean (SD) | 66.7 (44.1) | 66.8 (44.3) |
| Median [min, max] | 77.0 [2.00, 143] | 78.0 [2.00, 143] |
| Age(Years) | ||
| Mean (SD) | 59.0 (12.3) | 59.0 (12.3) |
| Median [min, max] | 58.0 [18.0, 103] | 58.0 [18.0, 103] |
| Sex | ||
| Female | 3114 (26.2%) | 3086 (26.1%) |
| Male | 8774 (73.8%) | 8721 (73.9%) |
| Race | ||
| American Indian/Alaska native | 55 (0.5%) | 54 (0.5%) |
| Asian or Pacific Islander | 762 (6.4%) | 750 (6.4%) |
| Black | 1051 (8.8%) | 1046 (8.9%) |
| White | 10,020 (84.3%) | 9957 (84.3%) |
| Marital Status | ||
| Divorced | 1532 (12.9%) | 1526 (12.9%) |
| Married (including common law) | 6997 (58.9%) | 6951 (58.9%) |
| Separated | 135 (1.1%) | 133 (1.1%) |
| Single (never married) | 2218 (18.7%) | 2198 (18.6%) |
| Widowed | 1006 (8.5%) | 999 (8.5%) |
| Characteristics | 3-Year Cohort (Complete Cases, n = 11,888) | 5-Year Cohort (Complete Cases, n = 11,807) |
|---|---|---|
| Differentiation Grade | ||
| Well differentiated; grade I | 1535 (12.9%) | 1521 (12.9%) |
| Moderately differentiated; grade II | 5667 (47.7%) | 5626 (47.6%) |
| Poorly differentiated; grade III | 4434 (37.3%) | 4410 (37.4%) |
| Undifferentiated; anaplastic; grade IV | 252 (2.1%) | 250 (2.1%) |
| T Category | ||
| T1 | 3956 (33.3%) | 3917 (33.2%) |
| T2 | 4204 (35.4%) | 4171 (35.3%) |
| T3 | 1562 (13.1%) | 1556 (13.2%) |
| T4 | 2132 (17.9%) | 2129 (18.0%) |
| TX | 34 (0.3%) | 34 (0.3%) |
| N Category | ||
| N0 | 4659 (39.2%) | 4615 (39.1%) |
| N1 | 2416 (20.3%) | 2404 (20.4%) |
| N2 | 4386 (36.9%) | 4362 (36.9%) |
| N3 | 391 (3.3%) | 390 (3.3%) |
| NX | 36 (0.3%) | 36 (0.3%) |
| M Category | ||
| M0 | 11,447 (96.3%) | 11367 (96.3%) |
| M1 | 348 (2.9%) | 347 (2.9%) |
| MX | 93 (0.8%) | 93 (0.8%) |
| Stage | ||
| I | 2183 (18.4%) | 2160 (18.3%) |
| II | 1540 (13.0%) | 1522 (12.9%) |
| III | 2356 (19.8%) | 2341 (19.8%) |
| IV | 5809 (48.9%) | 5784 (49.0%) |
| Lymph Nodes Removed | ||
| None | 6557 (55.2%) | 6515 (55.2%) |
| Yes | 5331 (44.8%) | 5292 (44.8%) |
| Tumor Size | ||
| 0~1 cm | 1420 (11.9%) | 1404 (11.9%) |
| 1~2 cm | 2806 (23.6%) | 2783 (23.6%) |
| 2~3 cm | 3120 (26.2%) | 3104 (26.3%) |
| 3~4 cm | 2097 (17.6%) | 2081 (17.6%) |
| 4~5 cm | 1369 (11.5%) | 1363 (11.5%) |
| 5~6 cm | 561 (4.7%) | 559 (4.7%) |
| 6~7 cm | 253 (2.1%) | 252 (2.1%) |
| 7~8 cm | 128 (1.1%) | 127 (1.1%) |
| 8~9 cm | 55 (0.5%) | 55 (0.5%) |
| 9~10 cm | 41 (0.3%) | 41 (0.3%) |
| >10 cm | 38 (0.3%) | 38 (0.3%) |
| Surgical Therapy | ||
| Surgery not performed | 4699 (39.5%) | 4673 (39.6%) |
| Surgery performed | 7189 (60.5%) | 7134 (60.4%) |
| Tumor Sites (ICD Code) | ||
| Lip (C00) | 540 (4.5%) | 536 (4.5%) |
| Base of tongue (C01) | 2192 (18.4%) | 2177 (18.4%) |
| Other parts of tongue (C02) | 2400 (20.2%) | 2378 (20.1%) |
| Gum (C03) | 412 (3.5%) | 410 (3.5%) |
| Floor of mouth (C04) | 786 (6.6%) | 783 (6.6%) |
| Palate (C05) | 318 (2.7%) | 314 (2.7%) |
| Other oral cavity (C06) | 645 (5.4%) | 639 (5.4%) |
| Parotid gland (C07) | 253 (2.1%) | 253 (2.1%) |
| Other salivary glands (C08) | 39 (0.3%) | 38 (0.3%) |
| Tonsil (C09) | 2858 (24.0%) | 2840 (24.1%) |
| Oropharynx (C10) | 363 (3.1%) | 362 (3.1%) |
| Nasopharynx (C11) | 408 (3.4%) | 404 (3.4%) |
| Pyriform sinus (C12) | 391 (3.3%) | 391 (3.3%) |
| Hypopharynx (C13) | 283 (2.4%) | 282 (2.4%) |
| Modelling Approaches | Development Dataset (80%) (Median (IQR)) | Test Dataset (20%) (Median (IQR)) |
|---|---|---|
| Three-Year Survival Cohort | ||
| Cox | 0.77 (0.77, 0.77) | 0.76 (0.76, 0.77) |
| Survival tree (ST) | 0.70 (0.70, 0.70) | 0.70 (0.69, 0.71) |
| Random forest for survival (RF) | 0.83 (0.83, 0.84) | 0.77 (0.76, 0.77) |
| Conditional inference forest (CF) | 0.83 (0.83, 0.86) | 0.76 (0.75, 0.76) |
| Five-Year Survival Cohort | ||
| Cox | 0.76 (0.76, 0.76) | 0.76 (0.76, 0.76) |
| Survival tree (ST) | 0.69 (0.69, 0.70) | 0.69 (0.68, 0.70) |
| Random forest for survival (RF) | 0.83 (0.83, 0.83) | 0.76 (0.76, 0.76) |
| Conditional inference forest (CF) | 0.85 (0.84, 0.86) | 0.75 (0.75, 0.76) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, M.; Haag, D.G.; Lynch, J.W.; Mittinty, M.N. Comparison of the Tree-Based Machine Learning Algorithms to Cox Regression in Predicting the Survival of Oral and Pharyngeal Cancers: Analyses Based on SEER Database. Cancers 2020, 12, 2802. https://doi.org/10.3390/cancers12102802
Du M, Haag DG, Lynch JW, Mittinty MN. Comparison of the Tree-Based Machine Learning Algorithms to Cox Regression in Predicting the Survival of Oral and Pharyngeal Cancers: Analyses Based on SEER Database. Cancers. 2020; 12(10):2802. https://doi.org/10.3390/cancers12102802
Chicago/Turabian StyleDu, Mi, Dandara G. Haag, John W. Lynch, and Murthy N. Mittinty. 2020. "Comparison of the Tree-Based Machine Learning Algorithms to Cox Regression in Predicting the Survival of Oral and Pharyngeal Cancers: Analyses Based on SEER Database" Cancers 12, no. 10: 2802. https://doi.org/10.3390/cancers12102802