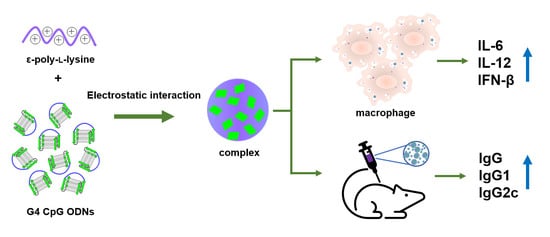
Non-Modified CpG Oligodeoxynucleotide Forming Guanine-Quadruplex Structure Complexes with ε-Poly-L-Lysine Induce Antibody Production as Vaccine Adjuvants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. G4 Structure Formation
2.3. Preparation of the ε-PLL/G4-CpG ODN Complexes
2.4. Particle Size, Surface Charge, and Morphological Analysis of the ε-PLL/G4-CpG ODN Complexes
2.5. Cell Culture
2.6. Cytotoxicity Assay
2.7. Stability of G4-CpG ODN Complexed with ε-PLL in Serum
2.8. Cellular Uptake
2.9. Immunostimulatory Stimulation Assay in RAW264 Cells
2.10. Immunization
2.11. Detection of OVA-Specific Antibodies via Enzyme-Linked Immunosorbent Assay (ELISA)
2.12. Statistical Analysis
3. Results
3.1. Preparation and Characterization of the ε-PLL/CpG ODN Complexes
3.2. Cytotoxicity of ε-PLL
3.3. Nuclease Resistance of G4-CpG ODNs in Complex
3.4. Cellular Uptake by the ε-PLL/G4-CpG ODN Complexes
3.5. Cytokine Induction by the ε-PLL/G4-CpG ODN Complexes at Various N/P Ratios
3.6. Adjuvant Effect of the ε-PLL/G4-CpG ODN Complexes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanagata, N. CpG oligodeoxynucleotide nanomedicines for the prophylaxis or treatment of cancers, infectious diseases, and allergies. Int. J. Nanomed. 2017, 12, 515–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rattanakiat, S.; Nishikawa, M.; Takakura, Y. Self-assembling CpG DNA nanoparticles for efficient antigen delivery and immunostimulation. Eur. J. Pharm. Sci. 2012, 47, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Hanagata, N.; Li, X.; Chen, M.-H.; Li, J.; Hattori, S. Double-stranded phosphodiester cytosine-guanine oligodeoxynucleotide complexed with calcium phosphate as a potent vaccine adjuvant for activating cellular and Th1-type humoral immunities. Int. J. Nanomed. 2018, 13, 43–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, W.; Sun, J.; Liu, F.; Yu, S.; Xu, Z.; Wang, F.; Liu, X. Enhanced Immunostimulatory Activity of a Cytosine-Phosphate-Guanosine Immunomodulator by the Assembly of Polymer DNA Wires and Spheres. ACS Appl. Mater. Interfaces 2020, 12, 17167–17176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, S.; Zhi, C.; Yamazaki, T.; Hanagata, N. Chitosan-coated boron nitride nanospheres enhance delivery of CpG oligodeoxynucleotides and induction of cytokines. Int. J. Nanomed. 2013, 8, 1783–1793. [Google Scholar] [CrossRef] [Green Version]
- Krieg, A.M.; Yi, A.-K.; Matson, S.; Waldschmidt, T.J.; Bishop, G.A.; Teasdale, R.; Koretzky, G.A.; Klinman, D.M. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 1995, 374, 546–549. [Google Scholar] [CrossRef]
- Latz, E.; Schoenemeyer, A.; Visintin, A.; Fitzgerald, K.A.; Monks, B.G.; Knetter, C.F.; Lien, E.; Nilsen, N.J.; Espevik, T.; Golenbock, D.T. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 2004, 5, 190–198. [Google Scholar] [CrossRef]
- Bauer, S.; Kirschning, C.J.; Häcker, H.; Redecke, V.; Hausmann, S.; Akira, S.; Wagner, H.; Lipford, G.B. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl. Acad. Sci. USA 2001, 98, 9237–9242. [Google Scholar] [CrossRef] [Green Version]
- Weiner, G.J.; Liu, H.-M.; Wooldridge, J.E.; Dahle, C.E.; Krieg, A.M. Immunostimulatory oligodeoxynucleotides containing the CpG motif are effective as immune adjuvants in tumor antigen immunization. Proc. Natl. Acad. Sci. USA 1997, 94, 10833–10837. [Google Scholar] [CrossRef] [Green Version]
- Manoharan, Y.; Ji, Q.; Yamazaki, T.; Shanmugavel; Chen, S.; Ganesan, S.; Hill, J.P.; Ariga, K.; Hanagata, N. Effect of molecular weight of polyethyleneimine on loading of CpG oligodeoxynucleotides onto flake-shell silica nanoparticles for enhanced TLR9-mediated induction of interferon-α. Int. J. Nanomed. 2012, 7, 3625–3635. [Google Scholar] [CrossRef]
- Hemmi, H.; Takeuchi, O.; Kawai, T.; Kaisho, T.; Sato, S.; Sanjo, H.; Matsumoto, M.; Hoshino, K.; Wagner, H.; Takeda, K.; et al. A Toll-like receptor recognizes bacterial DNA. Nature 2000, 408, 740–745. [Google Scholar] [CrossRef]
- Fu, J.; Cai, J.; Ling, G.; Li, A.; Zhao, J.; Guo, X.; Zhang, P. Cationic polymers for enhancing CpG oligodeoxynucleotides-mediated cancer immunotherapy. Eur. Polym. J. 2019, 113, 115–132. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell. Microbiol. 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krieg, A.M. Therapeutic potential of Toll-like receptor 9 activation. Nat. Rev. Drug Discov. 2006, 5, 471–484. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in Inflammation, Immunity, and Disease. Cold Spring Harb. Perspect. Biol. 2014, 6, 16295–16296. [Google Scholar] [CrossRef] [PubMed]
- Kieper, W.C.; Prlic, M.; Schmidt, C.S.; Mescher, M.F.; Jameson, S.C. Il-12 enhances CD8 T cell homeostatic expansion. J. Immunol. 2001, 166, 5515–5521. [Google Scholar] [CrossRef] [Green Version]
- Klinman, D.M. Immunotherapeutic uses of CpG oligodeoxynucleotides. Nat. Rev. Immunol. 2004, 4, 249–259. [Google Scholar] [CrossRef]
- Shirota, H.; Klinman, D.M. Recent progress concerning CpG DNA and its use as a vaccine adjuvant. Expert Rev. Vaccines 2014, 13, 299–312. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, X.-D. Nanodelivery systems for enhancing the immunostimulatory effect of CpG oligodeoxynucleotides. Mater. Sci. Eng. C 2017, 70, 935–946. [Google Scholar] [CrossRef]
- Wang, C.; Sun, W.; Wright, G.; Wang, A.Z.; Gu, Z. Inflammation-Triggered Cancer Immunotherapy by Programmed Delivery of CpG and Anti-PD1 Antibody. Adv. Mater. 2016, 28, 8912–8920. [Google Scholar] [CrossRef]
- Krieg, A.M. Toll-like receptor 9 (TLR9) agonists in the treatment of cancer. Oncogene 2008, 27, 161–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanagata, N. Structure-dependent immunostimulatory effect of CpG oligodeoxynucleotides and their delivery system. Int. J. Nanomed. 2012, 7, 2181–2195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshi, K.; Yamazaki, T.; Sugiyama, Y.; Tsukakoshi, K.; Tsugawa, W.; Sode, K.; Ikebukuro, K. G-Quadruplex Structure Improves the Immunostimulatory Effects of CpG Oligonucleotides. Nucleic Acid Ther. 2019, 29, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Tu, A.T.T.; Hoshi, K.; Ikebukuro, K.; Hanagata, N.; Yamazaki, T. Monomeric G-Quadruplex-Based CpG Oligodeoxynucleotides as Potent Toll-Like Receptor 9 Agonists. Biomacromolecules 2020, 21, 3644–3657. [Google Scholar] [CrossRef]
- Safitri, F.A.; Tu, A.T.T.; Hoshi, K.; Shobo, M.; Zhao, D.; Witarto, A.B.; Sumarsono, S.H.; Giri-Rachman, E.A.; Tsukakoshi, K.; Ikebukuro, K.; et al. Enhancement of the Immunostimulatory Effect of Phosphodiester CpG Oligodeoxynucleotides by an Antiparallel Guanine-Quadruplex Structural Scaffold. Biomolecules 2021, 11, 1617. [Google Scholar] [CrossRef]
- Tu, A.T.T.; Hoshi, K.; Shobo, M.; Yamazaki, T. G-quadruplex-based CpG oligodeoxynucleotide/DOTAP complex strongly stimulates immunity in CpG motif-specific and loop-length-dependent manners. Nanomed. Nanotechnol. Biol. Med. 2022, 40, 102508. [Google Scholar] [CrossRef]
- Dold, N.M.; Zeng, Q.; Zeng, X.; Jewell, C.M. A Poly (Beta-Amino Ester) Activates Macrophages Independent of NF-ΚB Signaling. Acta Biomater. 2018, 68, 168–177. [Google Scholar] [CrossRef]
- Perevyazko, I.Y.; Bauer, M.; Pavlov, G.M.; Hoeppener, S.; Schubert, S.; Fischer, D.; Schubert, U.S. Polyelectrolyte Complexes of DNA and Linear PEI: Formation, Composition and Properties. Langmuir 2012, 28, 16167–16176. [Google Scholar] [CrossRef]
- Cheng, T.; Miao, J.; Kai, D.; Zhang, H. Polyethylenimine-Mediated CpG Oligodeoxynucleotide Delivery Stimulates Bifurcated Cytokine Induction. ACS Biomater. Sci. Eng. 2018, 4, 1013–1018. [Google Scholar] [CrossRef]
- Ahn, H.H.; Lee, M.S.; Cho, M.H.; Na Shin, Y.; Lee, J.H.; Kim, K.S.; Kim, M.S.; Khang, G.; Hwang, K.C.; Lee, I.W.; et al. DNA/PEI nano-particles for gene delivery of rat bone marrow stem cells. Colloids Surfaces A Physicochem. Eng. Asp. 2008, 313–314, 116–120. [Google Scholar] [CrossRef]
- Pandey, A.P.; Sawant, K.K. Polyethylenimine: A Versatile, Multifunctional Non-Viral Vector for Nucleic Acid Delivery. Mater. Sci. Eng. C Mater. Biol. Appl. 2016, 68, 904–918. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-Y.; Heo, M.B.; Hwang, G.-S.; Jung, Y.; Choi, D.Y.; Park, Y.-M.; Lim, Y.T. Multivalent Polymer Nanocomplex Targeting Endosomal Receptor of Immune Cells for Enhanced Antitumor and Systemic Memory Response. Angew. Chem. Int. Ed. 2015, 54, 8139–8143. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Chen, X.; Lu, T.; Sun, J.; Tian, H.; Hu, J.; Wang, Y.; Zhang, P.; Jing, X. Poly(L-Lysine)-Graft-Chitosan Copolymers: Synthesis Characterization, and Gene Transfection Effect. Biomacromolecules 2007, 8, 1425–1435. [Google Scholar] [CrossRef] [PubMed]
- Green, J.J.; Langer, R.; Anderson, D.G. A Combinatorial Polymer Library Approach Yields Insight into Nonviral Gene Delivery. Acc. Chem. Res. 2008, 41, 749–759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.; Ryu, K.; Choi, Y.S.; Cho, Y.-Y.; Lee, J.Y.; Lee, H.S.; Kang, H.C. Effects of the Physicochemical, Colloidal, and Biological Characteristics of Different Polymer Structures between α-Poly(l-Lysine) and ε-Poly(l-Lysine) on Polymeric Gene Delivery. Biomacromolecules 2018, 19, 2483–2495. [Google Scholar] [CrossRef] [PubMed]
- Mandal, H.; Katiyar, S.S.; Swami, R.; Kushwah, V.; Katare, P.B.; Meka, A.K.; Banerjee, S.K.; Popat, A.; Jain, S. ε-Poly-l-Lysine/plasmid DNA nanoplexes for efficient gene delivery in vivo. Int. J. Pharm. 2018, 542, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Li, P.; Qi, X.; Sharif, A.R.M.; Poon, Y.F.; Cao, Y.; Chang, M.W.; Leong, S.S.J.; Chan-Park, M.B. A photopolymerized antimicrobial hydrogel coating derived from epsilon-poly-l-lysine. Biomaterials 2011, 32, 2704–2712. [Google Scholar] [CrossRef] [PubMed]
- Bo, T.; Han, P.-P.; Su, Q.-Z.; Fu, P.; Guo, F.-Z.; Zheng, Z.-X.; Tan, Z.-L.; Zhong, C.; Jia, S.-R. Antimicrobial ε-poly-l-lysine induced changes in cell membrane compositions and properties of Saccharomyces cerevisiae. Food Control. 2016, 61, 123–134. [Google Scholar] [CrossRef]
- Marzano, M.; Falanga, A.P.; Marasco, D.; Borbone, N.; D’Errico, S.; Piccialli, G.; Roviello, G.N.; Oliviero, G. Evaluation of an Analogue of the Marine ε-PLL Peptide as a Ligand of G-quadruplex DNA Structures. Mar. Drugs 2020, 18, 49. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, M.M.; Mori, Y.; Osada, K.; Murakami, H. Enhancement of Production of IgM and Interferon-β in Human Cell Lines by Poly-Lysine. Biosci. Biotechnol. Biochem. 1995, 59, 1842–1845. [Google Scholar] [CrossRef]
- Han, Y.; Duan, Q.; Li, Y.; Tian, J. In vitro and in vivo investigation of chitosan–polylysine polymeric nanoparticles for ovalbumin and CpG co-delivery. RSC Adv. 2017, 7, 39962–39969. [Google Scholar] [CrossRef] [Green Version]
- Gary, D.J.; Min, J.; Kim, Y.; Park, K.; Won, Y.-Y. The Effect of N/P Ratio on the In Vitro and In Vivo Interaction Properties of PEGylated Poly[2-(dimethylamino)ethyl methacrylate]-Based siRNA Complexes. Macromol. Biosci. 2013, 13, 1059–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachmann, M.F.; Jennings, G.T. Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010, 10, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, J.; Sequeira, R.A.; Vohra, A.; Devkar, R.V.; Maity, T.K.; Prasad, K. Ionic Liquid-Mediated Preparation of Noncytotoxic Hemocompatible Stable DNA–ε-Poly-l-lysine Polyplexes: A New Sustainable Approach for the Bulk Production of Potential Nonviral Vectors for Gene Delivery Applications. ACS Sustain. Chem. Eng. 2021, 9, 264–272. [Google Scholar] [CrossRef]
- Shirai, S.; Shibuya, M.; Kawai, A.; Tamiya, S.; Munakata, L.; Omata, D.; Suzuki, R.; Aoshi, T.; Yoshioka, Y. Lipid Nanoparticles Potentiate CpG-Oligodeoxynucleotide-Based Vaccine for Influenza Virus. Front. Immunol. 2019, 10, 3018. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Misato, K.; Aoshi, T.; Yamamoto, Y.; Kubota, Y.; Wu, X.; Kuroda, E.; Ishii, K.J.; Yamamoto, H.; Yoshioka, Y. Carbonate Apatite Nanoparticles Act as Potent Vaccine Adjuvant Delivery Vehicles by Enhancing Cytokine Production Induced by Encapsulated Cytosine-Phosphate-Guanine Oligodeoxynucleotides. Front. Immunol. 2018, 9, 783. [Google Scholar] [CrossRef]
- Markine-Goriaynoff, D.; Coutelier, J.-P. Increased Efficacy of the Immunoglobulin G2a Subclass in Antibody-Mediated Protection against Lactate Dehydrogenase-Elevating Virus-Induced Polioencephalomyelitis Revealed with Switch Mutants. J. Virol. 2002, 76, 432–435. [Google Scholar] [CrossRef] [Green Version]
- Beers, S.A.; Glennie, M.J.; White, A.L. Influence of immunoglobulin isotype on therapeutic antibody function. Blood 2016, 127, 1097–1101. [Google Scholar] [CrossRef] [PubMed]








| N/P Ratio | 1 | 2 | 3 | 4 | 5 | 10 | 20 |
|---|---|---|---|---|---|---|---|
| The concentration of ODNs in the complex solution | 12.5 μM (149 μg/mL) | ||||||
| The concentration of ε-PLL in the complex solution (μg/mL) | 60 | 120 | 180 | 240 | 300 | 600 | 1200 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, D.; Tu, A.T.T.; Shobo, M.; Le, N.B.T.; Yoshikawa, C.; Sugai, K.; Hakamata, Y.; Yamazaki, T. Non-Modified CpG Oligodeoxynucleotide Forming Guanine-Quadruplex Structure Complexes with ε-Poly-L-Lysine Induce Antibody Production as Vaccine Adjuvants. Biomolecules 2022, 12, 1868. https://doi.org/10.3390/biom12121868
Zhao D, Tu ATT, Shobo M, Le NBT, Yoshikawa C, Sugai K, Hakamata Y, Yamazaki T. Non-Modified CpG Oligodeoxynucleotide Forming Guanine-Quadruplex Structure Complexes with ε-Poly-L-Lysine Induce Antibody Production as Vaccine Adjuvants. Biomolecules. 2022; 12(12):1868. https://doi.org/10.3390/biom12121868
Chicago/Turabian StyleZhao, Dandan, Anh Thi Tram Tu, Miwako Shobo, Nguyen Bui Thao Le, Chiaki Yoshikawa, Kazuhisa Sugai, Yoji Hakamata, and Tomohiko Yamazaki. 2022. "Non-Modified CpG Oligodeoxynucleotide Forming Guanine-Quadruplex Structure Complexes with ε-Poly-L-Lysine Induce Antibody Production as Vaccine Adjuvants" Biomolecules 12, no. 12: 1868. https://doi.org/10.3390/biom12121868