Chemoinformatic Screening for the Selection of Potential Senolytic Compounds from Natural Products
Abstract
:1. Introduction
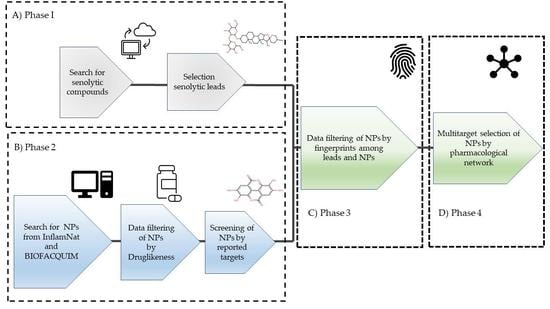
2. Materials and Methods
2.1. Creation of a Senolytic Lead Dataset
2.1.1. Collection of Senolytic Compounds
2.1.2. Selection of Senolytic Leads from the Dataset Based on Hierarchical Analysis and Chemical Space Analysis
2.2. Selection of Drug-Likeness of NPs from the BIOFACQUIM Database and the InflamNat Dataset Based on the Quantitative Estimate of Drug-Likeness (QED)
2.2.1. Data Collection for NPs
2.2.2. Determination of the Drug-Likeness of NPs
2.3. The Selection of Putative Senolytic Compounds Is Based on a Comparison of the Fingerprints of the Drug-Like NPs and Senolytic Leads
Fingerprints of Drug-Like NPs and Senolytic Leads
2.4. Selection of the Best Putative Senolytic Molecules through Their Multitarget Capacity
Compound-Target Network Generation
3. Results
3.1. Creation of a Senolytic Lead Dataset
3.2. Selection of Drug-Like NPs Based on QED
3.3. Selection of Putative Senolytic Compounds
3.4. Network Analysis to Obtain the Best Putative Senolytic Molecules through Their Multitarget Capacity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guerrero, A.; Herranz, N.; Sun, B. Cardiac glycosides are broad-spectrum senolytics. Nature 2019, 1, 1074–1088. [Google Scholar] [CrossRef]
- Pungsrinont, T.; Sutter, M.F.; Ertingshausen, M.; Lakshmana, G.; Kokal, M.; Khan, A.S.; Baniahmad, A. Senolytic compounds control a distinct fate of androgen receptor agonist- and antagonist-induced cellular senescent LNCaP prostate cancer cells. Cell Biosci. 2020, 10, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousefzadeh, M.J.; Zhu, Y.; McGowan, S.J.; Angelini, L.; Fuhrmann-Stroissnigg, H.; Xu, M.; Ling, Y.Y.; Melos, K.I.; Pirtskhalava, T.; Inman, C.L.; et al. Fisetin is a senotherapeutic that extends health and lifespan. EBioMedicine 2018, 36, 18–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkland, J.L.; Tchkonia, T. Senolytic drugs: From discovery to translation. J. Intern. Med. 2020, 288, 518–536. [Google Scholar] [CrossRef]
- Kameda, M.; Mikawa, T.; Yokode, M.; Inagaki, N.; Kondoh, H. Senescence research from historical theory to future clinical application. Geriatr. Gerontol. Int. 2021, 21, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Trendelenburg, A.U.; Scheuren, A.C.; Potter, P.; Müller, R.; Bellantuono, I. Geroprotectors: A role in the treatment of frailty. Mech. Ageing Dev. 2019, 180, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Justice, J.N.; Nambiar, A.M.; Tchkonia, T.; LeBrasseur, N.K.; Pascual, R.; Hashmi, S.K.; Prata, L.; Masternak, M.M.; Kritchevsky, S.B.; Musi, N.; et al. Senolytics in idiopathic pulmonary fibrosis: Results from a first-in-human, open-label, pilot study. EBioMedicine 2019, 40, 554–563. [Google Scholar] [CrossRef] [Green Version]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Kingston, D.G. Modern natural products drug discovery and its relevance to biodiversity conservation. J. Nat. Prod. 2011, 74, 496–511. [Google Scholar] [CrossRef] [Green Version]
- Lewinska, A.; Adamczyk-Grochala, J.; Bloniarz, D.; Olszowka, J.; Kulpa-Greszta, M.; Litwinienko, G.; Tomaszewska, A.; Wnuk, M.; Pazik, R. AMPK-mediated senolytic and senostatic activity of quercetin surface functionalized Fe3O4 nanoparticles during oxidant-induced senescence in human fibroblasts. Redox Biol. 2020, 28, 101337. [Google Scholar] [CrossRef]
- Zhu, Y.; Tchkonia, T.; Fuhrmann-Stroissnigg, H.; Dai, H.M.; Ling, Y.Y.; Stout, M.B.; Pirtskhalava, T.; Giorgadze, N.; Johnson, K.O.; Giles, C.B.; et al. Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of antiapoptotic factors. Aging Cell 2016, 15, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Lin, J.; Zou, Y.; Zhang, X.-J.; Xiao, W.-L. Chemical Space and Biological Target Network of Anti-Inflammatory Natural Products. J. Chem. Inf. Model. 2019, 59, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Pilón-Jiménez, B.A.; Saldívar-González, F.I.; Díaz-Eufracio, B.I.; Medina-Franco, J.L. BIOFACQUIM: A Mexican Compound Database of Natural Products. Biomolecules 2019, 9, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [Green Version]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]
- Sander, T.; Freyss, J.; von Korff, M.; Rufener, C. DataWarrior: An open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015, 55, 460–473. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Gui, Y.; Chen, L.; Yuan, G.; Lu, H.Z.; Xu, X. Use of natural products as chemical library for drug discovery and network pharmacology. PLoS ONE 2013, 8, e62839. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Dutta, P.; Sharma, M.; Rajput, N.K.; Dodiya, B.; Georrge, J.J.; Kholia, T.; Bhardwaj, A.; Consortium, O. BioPhytMol: A drug discovery community resource on anti-mycobacterial phytomolecules and plant extracts. J. Cheminform. 2014, 6, 46. [Google Scholar] [CrossRef] [Green Version]
- Sterling, T.; Irwin, J.J. ZINC 15–Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Voicu, A.; Duteanu, N.; Voicu, M.; Vlad, D.; Dumitrascu, V. The rcdk and cluster R packages applied to drug candidate selection. J. Cheminform. 2020, 12, 3. [Google Scholar] [CrossRef] [Green Version]
- Malhat, M.G.; Mousa, H.M.; El-Sisi, A.B. Parallel ward clustering for chemical compounds using mapreduce. In Proceedings of the International Conference on Advanced Machine Learning Technologies and Applications (AMLTA 2014), Cairo, Egypt, 28–30 November 2014; pp. 258–267. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Gilson, M.K.; Liu, T.; Baitaluk, M.; Nicola, G.; Hwang, L.; Chong, J. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016, 44, D1045–D1053. [Google Scholar] [CrossRef]
- Zhao, M.; Chen, L.; Qu, H. CSGene: A literature-based database for cell senescence genes and its application to identify critical cell aging pathways and associated diseases. Cell Death Dis. 2016, 7, e2053. [Google Scholar] [CrossRef] [PubMed]
- Marthandan, S.; Baumgart, M.; Priebe, S.; Groth, M.; Schaer, J.; Kaether, C.; Guthke, R.; Cellerino, A.; Platzer, M.; Diekmann, S.; et al. Conserved Senescence Associated Genes and Pathways in Primary Human Fibroblasts Detected by RNA-Seq. PLoS ONE 2016, 11, e0154531. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Jin, S.; Abraham, V.; Huang, X.; Liu, B.; Mitten, M.J.; Nimmer, P.; Lin, X.; Smith, M.; Shen, Y.; et al. The Bcl-2/Bcl-X(L)/Bcl-w inhibitor, navitoclax, enhances the activity of chemotherapeutic agents in vitro and in vivo. Mol. Cancer Ther. 2011, 10, 2340–2349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allaman-Pillet, N.; Oberson, A.; Munier, F.; Schorderet, D.F. The Bcl-2/Bcl-XL inhibitor ABT-737 promotes death of retinoblastoma cancer cells. Ophthalmic Genet. 2013, 34, 1–13. [Google Scholar] [CrossRef]
- Nami, B.; Donmez, H.; Kocak, N. Tunicamycin-induced endoplasmic reticulum stress reduces in vitro subpopulation and invasion of CD44+/CD24- phenotype breast cancer stem cells. Exp. Toxicol. Pathol. 2016, 68, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Wong, C.K.C.; Lai, H.-C.; Wong, A.S.T. Ginsenoside-Rb1 targets chemotherapy-resistant ovarian cancer stem cells via simultaneous inhibition of Wnt/β-catenin signaling and epithelial-to-mesenchymal transition. Oncotarget 2017, 8, 25897–25914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, D.; Wu, Q.; Chen, X.Q.; Du, Y.K.; Chen, T.; Li, H.; Tang, D.L.; Li, Q.H.; Zhang, Y.; Lu, L.Q.; et al. Azithromycin treats diffuse panbronchiolitis by targeting T cells via inhibition of mTOR pathway. Biomed. Pharmacother. 2019, 110, 440–448. [Google Scholar] [CrossRef]
- Dai, Y.; Li, F.; Wu, L.; Wang, R.; Li, P.; Yan, S.; Xu, H.; Xia, M.; Bai, C. Roxithromycin treatment inhibits TGF-β1-induced activation of ERK and AKT and down-regulation of caveolin-1 in rat airway smooth muscle cells. Respir. Res. 2014, 15, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, F.W.; Fong, S.; Griffin, C.; Shoemaker, M.; Staub, R.; Zhang, Y.-L.; Cohen, I.; Shtivelman, E. Timosaponin AIII is preferentially cytotoxic to tumor cells through inhibition of mTOR and induction of ER stress. PLoS ONE 2009, 4, e7283. [Google Scholar] [CrossRef]
- Belz, G.G.; Breithaupt-Grögler, K.; Osowski, U. Treatment of congestive heart failure—Current status of use of digitoxin. Eur. J. Clin. Investig. 2001, 31 (Suppl. 2), 10–17. [Google Scholar] [CrossRef]
- Lamming, DW Inhibition of the Mechanistic Target of Rapamycin (mTOR)-Rapamycin and Beyond. Cold Spring Harb. Perspect. Med. 2016, 6, a025924. [CrossRef] [PubMed] [Green Version]
- Alcaraz, M.J.; Payá, M. Marine sponge metabolites for the control of inflammatory diseases. Curr. Opin. Investig. Drugs 2006, 7, 974–979. [Google Scholar]
- Johnson, J.J. Carnosol: A promising anti-cancer and anti-inflammatory agent. Cancer Lett. 2011, 305, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.Y.; Kim, Y.I.; Park, S.J.; Kim, I.K.; Choi, Y.K.; Kim, S.H. Safe, high-throughput screening of natural compounds of MERS-CoV entry inhibitors using a pseudovirus expressing MERS-CoV spike protein. Int. J. Antimicrob. Agents 2018, 52, 730–732. [Google Scholar] [CrossRef]
- Wang, L.; Hu, T.; Shen, J.; Zhang, L.; Chan, R.L.; Lu, L.; Li, M.; Cho, C.H.; Wu, W.K. Dihydrotanshinone I induced apoptosis and autophagy through caspase-dependent pathway in colon cancer. Phytomedicine 2015, 22, 1079–1087. [Google Scholar] [CrossRef]
- Wang, M.; Dai, H.; Li, X.; Li, Y.; Wang, L.; Xue, M. Structural elucidation of metabolites of tanshinone I and its analogue dihydrotanshinone I in rats by HPLC–ESI-MSn. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2010, 878, 915–924. [Google Scholar] [CrossRef]
- Alam, A.; Haldar, S.; Thulasiram, H.V.; Kumar, R.; Goyal, M.; Iqbal, M.S.; Pal, C.; Dey, S.; Bindu, S.; Sarkar, S.; et al. Novel anti-inflammatory activity of epoxyazadiradione against macrophage migration inhibitory factor: Inhibition of tautomerase and proinflammatory activities of macrophage migration inhibitory factor. J. Biol. Chem. 2012, 287, 24844–24861. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Fu, X.; Gui, T.; Wang, T.; Wang, Z.; Kullak-Ublick, G.A.; Gai, Z. Effects of Farnesiferol B on Ischemia-Reperfusion-Induced Renal Damage, Inflammation, and NF-κB Signaling. Int. J. Mol. Sci. 2019, 20, 6280. [Google Scholar] [CrossRef] [Green Version]
- Antonisamy, P.; Duraipandiyan, V.; Ignacimuthu, S. Anti-inflammatory, analgesic and antipyretic effects of friedelin isolated from Azima tetracantha Lam. in mouse and rat models. J. Pharm. Pharmacol. 2011, 63, 1070–1077. [Google Scholar] [CrossRef]
- Marchbank, D.H.; Kerr, R.G. Semisynthesis of fuscoside B analogues and eunicosides, and analysis of anti-inflammatory activity. Tetrahedron 2011, 67, 3053–3061. [Google Scholar] [CrossRef]
- Reina, E.; Puentes, C.; Rojas, J.; García, J.; Ramos, F.A.; Castellanos, L.; Aragón, M.; Ospina, L.F. Fuscoside E: A strong anti-inflammatory diterpene from Caribbean octocoral Eunicea fusca. Bioorg. Med. Chem. Lett. 2011, 21, 5888–5891. [Google Scholar] [CrossRef] [PubMed]
- Reihill, J.A.; Malcomson, B.; Bertelsen, A.; Cheung, S.; Czerwiec, A.; Barsden, R.; Elborn, J.S.; Dürkop, H.; Hirsch, B.; Ennis, M.; et al. Induction of the inflammatory regulator A20 by gibberellic acid in airway epithelial cells. Br. J. Pharmacol. 2016, 173, 778–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolan, S.K.; O’Keeffe, G.; Jones, G.W.; Doyle, S. Resistance is not futile: Gliotoxin biosynthesis, functionality and utility. Trends Microbiol. 2015, 23, 419–428. [Google Scholar] [CrossRef]
- Jayakumar, T.; Liu, C.-H.; Wu, G.-Y.; Lee, T.-Y.; Manubolu, M.; Hsieh, C.-Y.; Yang, C.-H.; Sheu, J.-R. Hinokitiol Inhibits Migration of A549 Lung Cancer Cells via Suppression of MMPs and Induction of Antioxidant Enzymes and Apoptosis. Int. J. Mol. Sci. 2018, 19, 939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unger, C.; Kiss, I.; Vasas, A.; Lajter, I.; Kramer, N.; Atanasov, A.G.; Nguyen, C.H.; Chatuphonprasert, W.; Brenner, S.; Krieger, S.; et al. The germacranolide sesquiterpene lactone neurolenin B of the medicinal plant Neurolaena lobata (L.) R.Br. ex Cass inhibits NPM/ALK-driven cell expansion and NF-κB-driven tumour intravasation. Phytomedicine 2015, 22, 862–874. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Yu, N.H.; Jeon, S.J.; Lee, H.W.; Bae, C.H.; Yeo, J.H.; Lee, H.B.; Kim, I.S.; Park, H.W.; Kim, J.C. Antibacterial activities of penicillic acid isolated from Aspergillus persii against various plant pathogenic bacteria. Lett. Appl. Microbiol. 2016, 62, 488–493. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.Y.; Song, H.C.; Li, J.H.; Tang, Y.S.; Sun, R.; Wang, L.; Zhou, Y.P.; Wang, L.M.; Shen, K.Z.; Wang, C.R.; et al. Ymf 1029A-E, preussomerin analogues from the fresh-water-derived fungus YMF 1.01029. J. Nat. Prod. 2008, 71, 952–956. [Google Scholar] [CrossRef]
- Isaka, M.; Chinthanom, P.; Rachtawee, P.; Srichomthong, K.; Srikitikulchai, P.; Kongsaeree, P.; Prabpai, S. Cytotoxic hydroanthraquinones from the mangrove-derived fungus Paradictyoarthrinium diffractum BCC 8704. J. Antibiot. 2015, 68, 334–338. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.-Y.; Sher, H.-F.; Chen, H.-W.; Liu, C.-C.; Chen, C.-H.; Lin, C.-S.; Yang, P.-C.; Tsay, H.-S.; Chen, J.J. Anticancer effects of tanshinone I in human non-small cell lung cancer. Mol. Cancer Ther. 2008, 7, 3527–3538. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Xu, H. Anti-Inflammatory and Immunomodulatory Mechanism of Tanshinone IIA for Atherosclerosis. Evid. Based Complement. Altern. Med. 2014, 2014, 267976. [Google Scholar] [CrossRef]
- Ziaei, S.; Halaby, R. Immunosuppressive, anti-inflammatory and anti-cancer properties of triptolide: A mini review. Avicenna J. Phytomed. 2016, 6, 149–164. [Google Scholar]
- Hussain, H.; Green, I.R.; Ali, I.; Khan, I.A.; Ali, Z.; Al-Sadi, A.M.; Ahmed, I. Ursolic acid derivatives for pharmaceutical use: A patent review (2012–2016). Expert Opin. Ther. Patents 2017, 27, 1061–1072. [Google Scholar] [CrossRef]
- Ononye, S.N.; VanHeyst, M.D.; Oblak, E.Z.; Zhou, W.; Ammar, M.; Anderson, A.C.; Wright, D.L. Tropolones as lead-like natural products: The development of potent and selective histone deacetylase inhibitors. ACS Med. Chem. Lett. 2013, 4, 757–761. [Google Scholar] [CrossRef] [Green Version]
- Tsuchida, K.; Chaki, H.; Takakura, T.; Yokotani, J.; Aikawa, Y.; Shiozawa, S.; Gouda, H.; Hirono, S. Design, Synthesis, and Biological Evaluation of New Cyclic Disulfide Decapeptides That Inhibit the Binding of AP-1 to DNA. J. Med. Chem. 2004, 47, 4239–4246. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. PubChem Bioassay Record for AID 1436, Homologous Recombination_Rad51_DNA Binding Assay. Available online: https://pubchem.ncbi.nlm.nih.gov/gene/RAD51/human (accessed on 11 February 2021).
- Inoue, Y.; Suzuki, R.; Murata, I.; Nomura, H.; Isshiki, Y.; Kanamoto, I. Evaluation of Antibacterial Activity Expression of the Hinokitiol/Cyclodextrin Complex against Bacteria. ACS Omega 2020, 5, 27180–27187. [Google Scholar] [CrossRef] [PubMed]
- Saniewski, M.; Horbowicz, M.; Kanlayanarat, S. The Biological Activities of Troponoids and Their Use in Agriculture a Review. J. Hort. Res. 2014, 22, 5–19. [Google Scholar] [CrossRef] [Green Version]
- Fallik, E.; Grinberg, S. Hinokitiol: A natural substance that controls postharvest diseases in eggplant and pepper fruits. Postharvest Biol. Technol. 1992, 2, 137–144. [Google Scholar] [CrossRef]
- Chen, S.; Chen, D.; Cai, R.; Cui, H.; Long, Y.; Lu, Y.; Li, C.; She, Z. Cytotoxic and antibacterial preussomerins from the mangrove endophytic fungus Lasiodiplodia theobromae ZJ-HQ1. J. Nat. Prod. 2016, 79, 2397–2402. [Google Scholar] [CrossRef]
- Weber, H.A.; Gloer, J.B. The preussomerins: Novel antifungal metabolites from the coprophilous fungus Preussia isomera Cain. J. Org. Chem. 1991, 56, 4355–4360. [Google Scholar] [CrossRef]
- Macias-Rubalcava, M.L.; Ruiz-Velasco Sobrino, M.E.; Melendez-Gonzalez, C.; Hernandez-Ortega, S. Naphthoquinone spiroketals and organic extracts from the endophytic fungus Edenia gomezpompae as potential herbicides. J. Agric. Food Chem. 2014, 62, 3553–3562. [Google Scholar] [CrossRef]
- Seephonkai, P.; Isaka, M.; Kittakoop, P.; Palittapongarnpim, P.; Kamchonwongpaisan, S.; Tanticharoen, M.; Thebtaranonth, Y. Evaluation of antimycobacterial, antiplasmodial and cytotoxic activities of preussomerins isolated from the lichenicolous fungus Microsphaeropsis sp. BCC 3050. Planta Med. 2002, 68, 45–48. [Google Scholar] [CrossRef]
- Prieto-Martínez, F.D.; Norinder, U.; Medina-Franco, J.L. Cheminformatics Explorations of Natural Products. Prog. Chem. Org. Nat. Prod. 2019, 110, 1–35. [Google Scholar] [CrossRef]
- Wei, H.; Harper, M.T. Comparison of putative BH3 mimetics AT-101, HA14-1, sabutoclax and TW-37 with ABT-737 in platelets. Platelets 2021, 32, 105–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemming, F.W. Glycosyl phosphopolyprenols. In New Comprehensive Biochemistry; Elsevier: San Diego, CA, USA, 1985; Volume 10, pp. 261–305. [Google Scholar]
- Mohanan, P.; Subramaniyam, S.; Mathiyalagan, R.; Yang, D.-C. Molecular signaling of ginsenosides Rb1, Rg1, and Rg3 and their mode of actions. J. Ginseng. Res. 2018, 42, 123–132. [Google Scholar] [CrossRef]
- Thibodeaux, C.; Liu, H.-w.; Thorson, J. Complementary routes to natural product glycodiversification: Pathway engineering and glycorandomization. In Comprehensive Glycoscience; Elsevier: San Diego, CA, USA, 2007; Volume 1, pp. 373–396. [Google Scholar]
- Markham, A.; Faulds, D. Roxithromycin. An update of its antimicrobial activity, pharmacokinetic properties and therapeutic use. Drugs 1994, 48, 297–326. [Google Scholar] [CrossRef] [PubMed]
- McMillin, G.A.; Johnson-Davis, K.L. Chapter 13-Issues of Interferences in Therapeutic Drug Monitoring. In Accurate Results in the Clinical Laboratory; Dasgupta, A., Sepulveda, J.L., Eds.; Elsevier: San Diego, CA, USA, 2013; Volume 1, pp. 195–211. [Google Scholar] [CrossRef]
- Bickerton, G.R.; Paolini, G.V.; Besnard, J.; Muresan, S.; Hopkins, A.L. Quantifying the chemical beauty of drugs. Nat. Chem. 2012, 4, 90–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Docherty, M.H.; Baird, D.P.; Hughes, J.; Ferenbach, D.A. Cellular Senescence and Senotherapies in the Kidney: Current Evidence and Future Directions. Front. Pharmacol. 2020, 11, 755. [Google Scholar] [CrossRef]






| Senolytics | Pubchem Compound ID(CID) | Pharmacological Activity | K-Means Coefficient | References |
|---|---|---|---|---|
| Navitoclax | 24978538 | Inhibitor of Bcl-2 and Bcl-xL | 0.44 | [11,27] |
| ABT 737 | 11228183 | Inhibitor of Bcl-2 and Bcl-xL | 0.58 | [28] |
| Tunicamycin | 56927848 | Disturbs the endoplasmic reticulum (ER) homeostasis and causes the accumulation of misfolded or unfolded proteins in the ER, inducing cell death | 0.34 | [29] |
| Ginsenoside Rb1 | 9898279 | Affects the Wnt/β-catenin signalling pathway by downregulating β-catenin/T-cell factor-dependent transcription and expression of its target genes ATP-binding cassette G2 and P-glycoprotein | 0.31 | [30] |
| Azithromycin | 447043 | Enhances autophagosome formation of T cells by suppressing S6RP phosphorylation, which is a downstream target of the mammalian target of rapamycin (mTOR) pathway | 0.32 | [31] |
| Roxithromycin | 6915744 | Inhibitor of TGF-β1-induced activation of ERK and AKT and down-regulation of caveolin-1 | 0.23 | [32] |
| Timosaponin A-III | 15953793 | The inductor of selective cytotoxic activity that involves inhibition of mTOR, induction of ER stress, and protective autophagy | 0.29 | [33] |
| Digoxin | 2724385 | Positive inotropic and negative chronotropic agent | 0.21 | [34] |
| Rapamycin | 5284616 | Inhibitor of mTOR complex 1 (mTORC1), which phosphorylates substrates including S6 kinase 1 (S6K1), eIF4E-binding protein 1 (4E-BP1), transcription factor EB (TFEB), unc-51-like autophagy-activating kinase 1 (Ulk1), and growth factor receptor-bound protein 10 (GRB-10) | −0.2 | [35] |
| Compound Resulting from Fingerprint Analysis | ZINC ID | Pharmacological Activity Reported | References |
|---|---|---|---|
| Cacospongionolide B | ZINC26966472 | Anti-inflammatory agent | [36] |
| Carnosol | ZINC3871891 | Antineoplastic agent | [37] |
| Dihydrotanshinone I | ZINC2585546 | Antiviral, anti-mutagenic, anti-cancer agent | [38,39,40] |
| Epoxyazadiradione | ZINC58576553 | Anti-inflammatory agent | [41] |
| Farnesiferol B | ZINC29134693 | Anti-oxidant agent | [42] |
| Friedelin | ZINC4097720 | Anti-inflammatory and antipyretic agent | [43] |
| Fuscoside B | ZINC72123265 | Anti-inflammatory agent | [44,45] |
| Gibberellic acid | ZINC3860467 | Anti-inflammatory agent | [46] |
| Gliotoxin | ZINC3875454 | Anti-inflammatory agent | [47] |
| Hinokitiol | ZINC95911093 | Anti-cancer agent | [48] |
| Neurolenin B | ZINC100090140 | Anti-inflammatory agent | [49] |
| Penicillic acid | ZINC3874657 | Antibiotic | [50] |
| Preussomerin C | ZINC34383300 | Cytotoxic and anti-nematodal agent | [51,52] |
| Tanshinone I | ZINC2558154 | Anti-oxidant and anti-inflammatory agent | [40,53] |
| Tanshinone IIA | ZINC1650576 | Anti-oxidant and anti-inflammatory agent | [54] |
| Triptolide | ZINC6483512 | Anti-cancer, anti-inflammation, anti-obesity, and anti-diabetic | [55] |
| Ursolic acid | ZINC31356858 | Anti-inflammatory and antihyperlipidemic agent | [56] |
| Senolytic Candidate | Structure | Source | Pharmacological Activity | Targets of Senolytic Compound Network | References |
|---|---|---|---|---|---|
| Hinokitiol ZINC95911093 |  | The roots of the Hinoki tree, Hiba arboruitae (Japanese cypress). | Anti-cancer agent | 7 | [48] |
| Preussomerin C ZINC34383300 |  | Endophytic fungus Lasiodiplodia theobromae ZJ-HQ1 | Cytotoxic and anti-nematodal agent | 10 | [51,52] |
| Tanshinone I ZINC2558154 |  | Salvia miltiorrhiza (Danshen or Tanshen in Chinese) | Anti-inflammatory, anti-coagulant, and anti-neoplasic agent | 6 | [40,53] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barrera-Vázquez, O.S.; Gómez-Verjan, J.C.; Magos-Guerrero, G.A. Chemoinformatic Screening for the Selection of Potential Senolytic Compounds from Natural Products. Biomolecules 2021, 11, 467. https://doi.org/10.3390/biom11030467
Barrera-Vázquez OS, Gómez-Verjan JC, Magos-Guerrero GA. Chemoinformatic Screening for the Selection of Potential Senolytic Compounds from Natural Products. Biomolecules. 2021; 11(3):467. https://doi.org/10.3390/biom11030467
Chicago/Turabian StyleBarrera-Vázquez, Oscar Salvador, Juan Carlos Gómez-Verjan, and Gil Alfonso Magos-Guerrero. 2021. "Chemoinformatic Screening for the Selection of Potential Senolytic Compounds from Natural Products" Biomolecules 11, no. 3: 467. https://doi.org/10.3390/biom11030467