Polymorphic Protective Dps–DNA Co-Crystals by Cryo Electron Tomography and Small Angle X-Ray Scattering
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of DNA Sample
2.2. Overexpression and Purification of Dps
2.3. Sample Preparation for Cryo-EM
2.4. Cryo-Electron Tomography
2.5. Tomographic Reconstruction
2.6. Solution Scattering Experiments and Data Analysis
3. Results
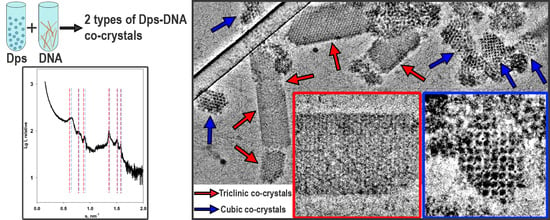
3.1. Cryo-Electron Tomography
3.2. Small-Angle X-Ray Scattering
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A






References
- Frenkiel-Krispin, D.; Ben-Avraham, I.; Englander, J.; Shimoni, E.; Wolf, S.G.; Minsky, A. Nucleoid restructuring in stationary-state bacteria. Mol. Microbiol. 2004, 51, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Reich, Z.; Wachtel, E.J.; Minsky, A. Liquid—Crystalline Mesophases of Plasmid DNA in Bacteria. Science 1994, 264, 1460–1463. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.G.; Frenkiel, D.; Arad, T.; Finkel, S.E.; Kolter, R.; Minsky, A. DNA protection by biocrystallization. Nature 1999, 400, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Almiron, M.; Link, A.J.; Furlong, D.; Kolter, R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992, 6, 2646–2654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nair, S.; Finkel, S.E. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 2004, 186, 4192–4198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Maier, R.J. Bacterial histone-like proteins: Roles in stress resistance. Curr. Genet. 2015, 61, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Azam, T.; Iwata, A.; Nishimura, A.; Ueda, S. Growth Phase-Dependent Variation in Protein Composition of the Escherichia coli Nucleoid. J. Bacteriol. 1999, 181, 6361–6370. [Google Scholar]
- Chiancone, E.; Ceci, P. The multifaceted capacity of Dps proteins to combat bacterial stress conditions: Detoxification of iron and hydrogen peroxide and DNA binding. Biochim. Biophys. Acta 2010, 1800, 798–805. [Google Scholar] [CrossRef]
- Dadinova, L.A.; Chesnokov, Y.M.; Kamyshinsky, R.A.; Orlov, I.A.; Petoukhov, M.V.; Mozhaev, A.A.; Soshinskaya, E.Y.; Lazarev, V.N.; Manuvera, V.A.; Orekhov, A.S.; et al. Protective Dps–DNA co-crystallization in stressed cells: An in vitro structural study by small-angle X-ray scattering and cryo-electron tomography. FEBS Lett. 2019, 593, 1360–1371. [Google Scholar] [CrossRef]
- Chowdhury, R.P.; Saraswathi, R.; Chatterji, D. Mycobacterial Stress Regulation: The Dps “Twin Sister” Defense Mechanism and Structure-Function Relationship. IUBMB Life 2010, 62, 67–77. [Google Scholar] [CrossRef]
- Pesek, J.; Buchler, R.; Albrecht, R.; Boland, W.; Zeth, K. Structure and Mechanism of Iron Translocation by a Dps Protein from Microbacterium arborescens. J. Biol. Chem. 2011, 286, 34872–34882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calhoun, L.N.; Kwon, Y.M. Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: A review. J. Appl. Microbiol. 2011, 110, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Pen, M.M.O.; Bullerjahn, G.S. The DpsA Protein of Synechococcus sp. Strain PCC7942 Is a DNA-binding Hemoprotein. J. Biol. Chem. 1995, 270, 22478–22482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takatsuka, M.; Osada-oka, M.; Satoh, E.F.; Kitadokoro, K.; Nishiuchi, Y.; Niki, M.; Inoue, M.; Iwai, K.; Arakawa, T.; Shimoji, Y.; et al. A Histone-Like Protein of Mycobacteria Possesses Ferritin Superfamily Protein-Like Activity and Protects against DNA Damage by Fenton Reaction. PLoS ONE 2011, 6, e20985. [Google Scholar] [CrossRef] [Green Version]
- Grant, R.A.; Filman, D.J.; Finkel, S.E.; Kolter, R.; Hogle, J.M. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat. Struct. Biol. 1998, 5, 294–303. [Google Scholar] [CrossRef]
- Kim, S.; Bhattacharyya, G.; Grove, A.; Lee, Y. Crystal Structure of Dps-1, a Functionally Distinct Dps Protein from Deinococcus radiodurans. J. Mol. Biol. 2006, 361, 105–114. [Google Scholar] [CrossRef]
- Andrews, S.C. The Ferritin-like superfamily: Evolution of the biological iron storeman from a rubrerythrin-like ancestor. Biochim. Biophys. Acta 2010, 1800, 691–705. [Google Scholar] [CrossRef]
- Facey, P.D.; Hitchings, M.D.; Saaverdra-Garcia, P.; Fernandez-Martinez, L.; Dyson, P.J.; Del Sol, R. Streptomyces coelicolor Dps-like proteins: Differential dual roles in response to stress during vegetative growth and in nucleoid condensation during reproductive cell division. Mol. Microbiol. 2009, 73, 1186–1202. [Google Scholar] [CrossRef]
- Nicodeme, M.; Perrin, C.; Hols, P.; Bracquart, P.; Gaillard, J. Identification of an Iron-Binding Protein of the Dps Family Expressed by Streptococcus thermophilus. Curr. Microbiol. 2004, 48, 51–56. [Google Scholar]
- Thieme, D.; Grass, G. The Dps protein of Escherichia coli is involved in copper homeostasis. Microbiol. Res. 2010, 165, 108–115. [Google Scholar] [CrossRef]
- Roy, S.; Saraswathi, R.; Chatterji, D.; Vijayan, M. Structural Studies on the Second Mycobacterium smegmatis Dps: Invariant and Variable Features of Structure, Assembly and Function. J. Mol. Biol. 2008, 375, 948–959. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, J.K.; Liu, X.S.; Tosha, T.; Diebold, A.; Theil, E.C.; Solomon, E.I. CD and MCD Spectroscopic Studies of the Two Dps Miniferritin Proteins from Bacillus anthracis: Role of O2 and H2O2 Substrates in Reactivity of the Diiron Catalytic Centers †. Biochemistry 2010, 49, 10516–10525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saraswathi, R.; Chowdhury, R.P.; Williams, S.M.; Ghatak, P.; Chatterji, D. The Mycobacterial MsDps2 Protein is a Nucleoid-Forming DNA Binding Protein Regulated by Sigma Factors s A and B. PLoS ONE 2009, 4, e8017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Facey, P.D.; Sevcikova, B.; Novakova, R.; Hitchings, M.D.; Crack, J.C.; Dyson, P.J.; Sol, R. Del The dpsA Gene of Streptomyces coelicolor: Induction of Expression from a Single Promoter in Response to Environmental Stress or during Development. PLoS ONE 2011, 6, e25593. [Google Scholar] [CrossRef]
- Chowdhury, R.P.; Gupta, S.; Chatterji, D. Identification and Characterization of the dps Promoter of Mycobacterium smegmatis: Promoter Recognition by Stress-Specific Extracytoplasmic Function Sigma Factors σH and σF. J. Bacteriol. 2007, 189, 8973–8981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceci, P.; Cellai, S.; Falvo, E.; Rivetti, C.; Rossi, G.L.; Chiancone, E. DNA condensation and self-aggregation of Escherichia coli Dps are coupled phenomena related to the properties of the N-terminus. Nucleic Acids Res. 2004, 32, 5935–5944. [Google Scholar] [CrossRef] [Green Version]
- Ceci, P.; Ilari, A.; Falvo, E.; Giangiacomo, L.; Chiancone, E. Reassessment of Protein Stability, DNA Binding, and Protection of Mycobacterium smegmatis Dps. J. Biol. Chem. 2005, 280, 34776–34785. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Chatterji, D. Bimodal Protection of DNA by Mycobacterium smegmatis DNA-binding Protein from Stationary Phase Cells. J. Biol. Chem. 2003, 278, 5235–5241. [Google Scholar] [CrossRef] [Green Version]
- Bozzi, M.; Mignogna, G.; Stefanini, S.; Barra, D.; Longhi, C.; Valenti, P.; Chiancone, E. A novel non-heme iron-binding ferritin related to the DNA-binding proteins of the Dps family in Listeria innocua. J. Biol. Chem. 1997, 272, 3259–3265. [Google Scholar] [CrossRef] [Green Version]
- Papinutto, E.; Dundon, W.G.; Pitulis, N.; Battistutta, R.; Montecucco, C.; Zanotti, G. Structure of two iron-binding proteins from Bacillus anthracis. J. Biol. Chem. 2002, 277, 15093–15098. [Google Scholar] [CrossRef] [Green Version]
- Zanotti, G.; Papinutto, E.; Dundon, W.G.; Battistutta, R.; Seveso, M.; Del Giudice, G.; Rappuoli, R.; Montecucco, C. Structure of the neutrophil-activating protein from Helicobacter pylori. J. Mol. Biol. 2002, 323, 125–130. [Google Scholar] [CrossRef]
- Frenkiel-Krispin, D.; Levin-Zaidman, S.; Shimoni, E.; Wolf, S.G.; Wachtel, E.J.; Arad, T.; Finkel, S.E.; Kolter, R.; Minsky, A. Regulated phase transitions of bacterial chromatin: A non-enzymatic pathway for generic DNA protection. EMBO J. 2001, 20, 1184–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, B.; Tibbelin, G.; Kajino, T.; Asami, O.; Ladenstein, R. The multi-layered structure of Dps with a novel di-nuclear ferroxidase center. J. Mol. Biol. 2003, 329, 467–477. [Google Scholar] [CrossRef]
- Zhao, G.; Ceci, P.; Ilari, A.; Giangiacomo, L.; Laue, T.M.; Chiancone, E.; Dennis Chasteen, N. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J. Biol. Chem. 2002, 277, 27689–27696. [Google Scholar] [CrossRef] [Green Version]
- Mozhaev, A.A.; Serova, O.V.; Orsa, A.N.; Boyko, A.A.; Goryashchenko, A.S.; Deyev, I.E.; Petrenko, A.G. The Hybrid Protein of the Alkaline Sensor IRR and the Fluorescent Protein GFP Retains the Functional Activity of the Receptor. Russ. J. Bioorganic Chem. 2019, 45, 179–182. [Google Scholar] [CrossRef]
- Marko, M.A.; Chipperfield, R.; Birnboim, H.C. A procedure for the large-scale isolation of highly purified plasmid DNA using alkaline extraction and binding to glass powder. Anal. Biochem. 1982, 121, 382–387. [Google Scholar] [CrossRef]
- Karas, V.O.; Westerlaken, I.; Meyer, A.S. Application of an in vitro DNA protection assay to visualize stress mediation properties of the Dps protein. J. Vis. Exp. 2013, 75, e50390. [Google Scholar] [CrossRef] [Green Version]
- Danev, R.; Buijsse, B.; Khoshouei, M.; Plitzko, J.M.; Baumeister, W. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc. Natl. Acad. Sci. USA 2014, 111, 15635–15640. [Google Scholar] [CrossRef] [Green Version]
- Kremer, J.R.; Mastronarde, D.N.; McIntosh, J.R. Computer Visualization of Three-Dimensional Image Data Using IMOD. J. Struct. Biol. 1996, 116, 71–76. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Dai, W.; Sun, S.Y.; Jonasch, D.; He, C.Y.; Schmid, M.F.; Chiu, W.; Ludtke, S.J. Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Nat. Methods 2017, 14, 983–985. [Google Scholar] [CrossRef] [Green Version]
- Tang, G.; Peng, L.; Baldwin, P.R.; Mann, D.S.; Jiang, W.; Rees, I.; Ludtke, S.J. EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 2007, 157, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheres, S.H.W. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012, 180, 519–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimanius, D.; Forsberg, B.O.; Scheres, S.H.W.; Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUS in RELION-2. eLife 2016, 5, e18722. [Google Scholar] [CrossRef] [PubMed]
- Bharat, T.A.M.; Scheres, S.H.W. Resolving macromolecular structures from electron cryo-Tomography data using subtomogram averaging in RELION. Nat. Protoc. 2016, 11, 2054–2065. [Google Scholar] [CrossRef] [Green Version]
- Rohou, A.; Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015, 192, 216–221. [Google Scholar] [CrossRef]
- Blanchet, C.E.; Spilotros, A.; Schwemmer, F.; Graewert, M.A.; Kikhney, A.; Jeffries, C.M.; Franke, D.; Mark, D.; Zengerle, R.; Cipriani, F.; et al. Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY). J. Appl. Crystallogr. 2015, 48, 431–443. [Google Scholar] [CrossRef]
- Jeffries, C.M.; Graewert, M.A.; Svergun, D.I.; Blanchet, C.E. Limiting radiation damage for high-brilliance biological solution scattering: Practical experience at the EMBL P12 beamline PETRAIII. J. Synchrotron Radiat. 2015, 22, 273–279. [Google Scholar] [CrossRef]
- Franke, D.; Petoukhov, M.V.; Konarev, P.V.; Panjkovich, A.; Tuukkanen, A.; Mertens, H.D.T.; Kikhney, A.G.; Hajizadeh, N.R.; Franklin, J.M.; Jeffries, C.M.; et al. ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017, 50, 1212–1225. [Google Scholar] [CrossRef] [Green Version]
- Feigin, L.A.; Svergun, D.I. Structure Analysis by Small-Angle X-ray and Neutron Scattering; Taylor, G.W., Ed.; Springer: Boston, MA, USA, 1987; ISBN 978-1-4757-6626-4. [Google Scholar]
- Porod, G. Small-Angle X-ray Scattering. In Small-Angle X-ray Scattering; Glatter, O., Kratky, O., Eds.; Academic Press: London, UK, 1982; pp. 17–51. [Google Scholar]
- Bilalov, A.; Olsson, U.; Lindman, B. DNA-lipid self-assembly: Phase behavior and phase structures of a DNA-surfactant complex mixed with lecithin and water. Soft Matter 2011, 7, 730–742. [Google Scholar] [CrossRef]
- Bilalov, A.; Olsson, U.; Lindman, B. A cubic DNA-lipid complex. Soft Matter 2009, 5, 3827–3830. [Google Scholar] [CrossRef]
- Loiko, N.G.; Danilova, Y.A.; Moiseenko, A.V.; Demkina, E.V.; Kovalenko, V.V.; Tereshkina, K.; El-Registan, G.I.; Sokolova, O.S.; Krupyanskii, Y.F. Condensation of nucleoid in Escherichia coli cell as a result of prolonged starvation. arXiV 2019, arXiv:1901.11322. [Google Scholar]




| Peak Position s, nm–1 (SAXS Data) | Miller Indices (hkl) | ||
|---|---|---|---|
| cubic crystal lattice system, a = 13.9 nm | |||
| 0.66 ± 0.02 | 0.64 | 9.81 | |
| 0.80 ± 0.01 | 0.78 | 8.05 | |
| 0.89 ± 0.04 | 0.90 | 6.98 | |
| 1.35 ± 0.02 | 1.36 | 4.62 | , |
| 1.50 ± 0.01 | 1.50 | 4.19 | |
| 1.57 ± 0.02 | 1.57 | 4.00 | |
| triclinic crystal lattice system, a = 9.1 nm; b = 9.5 nm; c = 10.4 nm; α = 75.2; β = 88.0; γ = 59.8 | |||
| 0.63 ± 0.02 | 0.63 | 9.97 | |
| 0.80 ± 0.01 | 0.80 | 7.85 | |
| 0.89 ± 0.04 | 0.87 | 7.22 | |
| 1.35 ± 0.02 | 1.35 | 4.65 | |
| 1.50 ± 0.01 | 1.50 | 4.19 | |
| 1.57 ± 0.02 | 1.57 | 4.00 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamyshinsky, R.; Chesnokov, Y.; Dadinova, L.; Mozhaev, A.; Orlov, I.; Petoukhov, M.; Orekhov, A.; Shtykova, E.; Vasiliev, A. Polymorphic Protective Dps–DNA Co-Crystals by Cryo Electron Tomography and Small Angle X-Ray Scattering. Biomolecules 2020, 10, 39. https://doi.org/10.3390/biom10010039
Kamyshinsky R, Chesnokov Y, Dadinova L, Mozhaev A, Orlov I, Petoukhov M, Orekhov A, Shtykova E, Vasiliev A. Polymorphic Protective Dps–DNA Co-Crystals by Cryo Electron Tomography and Small Angle X-Ray Scattering. Biomolecules. 2020; 10(1):39. https://doi.org/10.3390/biom10010039
Chicago/Turabian StyleKamyshinsky, Roman, Yury Chesnokov, Liubov Dadinova, Andrey Mozhaev, Ivan Orlov, Maxim Petoukhov, Anton Orekhov, Eleonora Shtykova, and Alexander Vasiliev. 2020. "Polymorphic Protective Dps–DNA Co-Crystals by Cryo Electron Tomography and Small Angle X-Ray Scattering" Biomolecules 10, no. 1: 39. https://doi.org/10.3390/biom10010039