The R Language as a Tool for Biometeorological Research
Abstract
:1. Introduction: The Biometeorology in the New Research Era
2. Methods and Tools in Biometeorology
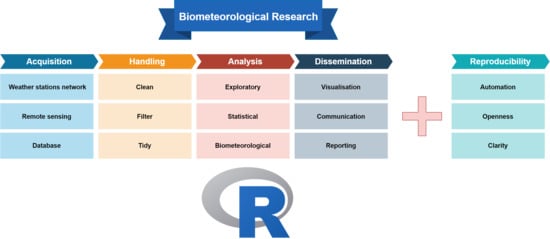
2.1. The Classic Research Workflow
2.1.1. Data Acquisition
2.1.2. Data Handling
2.1.3. Data Analysis
2.1.4. Results Dissemination
2.2. Reproducibility or Why We Use Code in Research
3. The R Biometeorological Research Workflow
3.1. R’s main Characteristics
3.2. Data Acquisition with R
3.3. Data Handling with R
3.4. Biometeorological Data Analysis with R
3.5. Results Dissemination with R
3.6. A Reproducible Research Example with the R Language
4. Conclusions
Supplementary Materials
Funding
Conflicts of Interest
References
- Tromp, S.W. Human biometeorology. Int. J. Biometeorol. 1963, 7, 145–158. [Google Scholar] [CrossRef]
- Tout, D.G. Biometeorology. Prog. Phys. Geogr. Earth Environ. 1987, 11, 473–486. [Google Scholar] [CrossRef]
- Flemming, G. The importance of air quality in human biometeorology. Int. J. Biometeorol. 1996, 39, 192–196. [Google Scholar] [CrossRef] [PubMed]
- McGregor, G.R. Human biometeorology. Prog. Phys. Geogr. Earth Environ. 2012, 36, 93–109. [Google Scholar] [CrossRef]
- Höppe, P. The physiological equivalent temperature—A universal index for the biometeorological assessment of the thermal environment. Int. J. Biometeorol. 1999, 43, 71–75. [Google Scholar] [CrossRef]
- Algeciras, J.A.; Consuegra, L.G.; Matzarakis, A. Spatial-temporal study on the effects of urban street configurations on human thermal comfort in the world heritage city of Camagüey-Cuba. Build. Environ. 2016, 101, 85–101. [Google Scholar] [CrossRef]
- Charalampopoulos, I.; Nouri, A.S. Investigating the behaviour of human thermal indices under divergent atmospheric conditions: A sensitivity analysis approach. Atmosphere 2019, 10, 580. [Google Scholar] [CrossRef] [Green Version]
- de Abreu-Harbich, L.V.; Labaki, L.C.; Matzarakis, A. Effect of tree planting design and tree species on human thermal comfort in the tropics. Landsc. Urban Plan. 2015, 138, 99–109. [Google Scholar] [CrossRef]
- Giannaros, T.M.; Kotroni, V.; Lagouvardos, K.; Matzarakis, A. Climatology and trends of the Euro-Mediterranean thermal bioclimate. Int. J. Climatol. 2018, 38, 3290–3308. [Google Scholar] [CrossRef]
- Kántor, N.; Chen, L.; Gál, C.V. Human-biometeorological significance of shading in urban public spaces—Summertime measurements in Pécs, Hungary. Landsc. Urban Plan. 2018, 170, 241–255. [Google Scholar] [CrossRef]
- Kaplan, S.; Peterson, C. Health and environment: A psychological analysis. Landsc. Urban Plan. 1993, 26, 17–23. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.-P.; Matzarakis, A. Tourism climate and thermal comfort in Sun Moon Lake, Taiwan. Int. J. Biometeorol. 2008, 52, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Matzarakis, A.; Mayer, H. The extreme heat wave in Athens in July 1987 from the point of view of human biometeorology. Atmos. Environ. Part B Urban Atmos. 1991, 25, 203–211. [Google Scholar] [CrossRef]
- Nastos, P.T.; Matzarakis, A. The effect of air temperature and human thermal indices on mortality in Athens, Greece. Theor. Appl. Climatol. 2012, 108, 591–599. [Google Scholar] [CrossRef]
- Bruse, M.; Fleer, H. Simulating surface–plant–air interactions inside urban environments with a three dimensional numerical model. Environ. Model. Softw. 1998, 13, 373–384. [Google Scholar] [CrossRef]
- Fröhlich, D.; Matzarakis, A. spatial estimation of thermal indices in Urban Areas—Basics of the SkyHelios Model. Atmosphere 2018, 9, 209. [Google Scholar] [CrossRef] [Green Version]
- Lindberg, F.; Grimmond, C.S.B.; Gabey, A.; Huang, B.; Kent, C.W.; Sun, T.; Theeuwes, N.E.; Järvi, L.; Ward, H.C.; Capel-Timms, I.; et al. Urban multi-scale environmental predictor (UMEP): An integrated tool for city-based climate services. Environ. Model. Softw. 2018, 99, 70–87. [Google Scholar] [CrossRef]
- Matzarakis, A.; Rutz, F.; Mayer, H. Modelling radiation fluxes in simple and complex environments—application of the RayMan model. Int. J. Biometeorol. 2007, 51, 323–334. [Google Scholar] [CrossRef]
- Gaughan, J.; Lacetera, N.; Valtorta, S.E.; Khalifa, H.H.; Hahn, L.; Mader, T. Response of domestic animals to climate challenges. In Biometeorology for Adaptation to Climate Variability and Change; Ebi, K.L., Burton, I., McGregor, G.R., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 131–170. ISBN 978-1-4020-8921-3. [Google Scholar]
- Hatfield, J.L.; Dold, C. Agroclimatology and wheat production: Coping with climate change. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Hondula, D.M.; Balling, R.C.; Andrade, R.; Scott Krayenhoff, E.; Middel, A.; Urban, A.; Georgescu, M.; Sailor, D.J. Biometeorology for cities. Int. J. Biometeorol. 2017, 61, 59–69. [Google Scholar] [CrossRef]
- Sofiev, M.; Bousquet, J.; Linkosalo, T.; Ranta, H.; Rantio-Lehtimaki, A.; Siljamo, P.; Valovirta, E.; Damialis, A. Pollen, Allergies and Adaptation. In Biometeorology for Adaptation to Climate Variability and Change; Ebi, K.L., Burton, I., McGregor, G.R., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 75–106. ISBN 978-1-4020-8921-3. [Google Scholar]
- Vasconcelos, J.; Freire, E.; Almendra, R.; Silva, G.L.; Santana, P. The impact of winter cold weather on acute myocardial infarctions in Portugal. Environ. Pollut. 2013, 183, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Quinn, A.; Tamerius, J.D.; Perzanowski, M.; Jacobson, J.S.; Goldstein, I.; Acosta, L.; Shaman, J. Predicting indoor heat exposure risk during extreme heat events. Sci. Total Environ. 2014, 490, 686–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Telfer, S.; Obradovich, N. Local weather is associated with rates of online searches for musculoskeletal pain symptoms. PLoS ONE 2017, 12, e0181266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charalampopoulos, I.; Nastos, P.T.; Didaskalou, E. Human thermal conditions and North Europeans’ web searching behavior (Google Trends) on mediterranean touristic destinations. Urban Sci. 2017, 1, 8. [Google Scholar] [CrossRef] [Green Version]
- Samson, D.R.; Crittenden, A.N.; Mabulla, I.A.; Mabulla, A.Z.P. The evolution of human sleep: Technological and cultural innovation associated with sleep-wake regulation among Hadza hunter-gatherers. J. Hum. Evol. 2017, 113, 91–102. [Google Scholar] [CrossRef]
- Charalampopoulos, I. A comparative sensitivity analysis of human thermal comfort indices with generalized additive models. Theor. Appl. Climatol. 2019. [Google Scholar] [CrossRef]
- Nouri, A.S.; Charalampopoulos, I.; Matzarakis, A. Beyond singular climatic variables—Identifying the dynamics of wholesome Thermo-Physiological factors for existing/future human thermal comfort during hot dry mediterranean summers. Int. J. Environ. Res. Public Health 2018, 15, 2362. [Google Scholar] [CrossRef] [Green Version]
- Chinazzo, G.; Wienold, J.; Andersen, M. Daylight affects human thermal perception. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Półrolniczak, M.; Tomczyk, A.M.; Kolendowicz, L. Thermal conditions in the city of Poznań (Poland) during selected heat waves. Atmosphere 2018, 9, 11. [Google Scholar] [CrossRef] [Green Version]
- Schweiker, M.; Wagner, A. Influences on the predictive performance of thermal sensation indices. Build. Res. Inf. 2017, 45, 745–758. [Google Scholar] [CrossRef]
- Quinn, A.; Kinney, P.; Shaman, J. Predictors of summertime heat index levels in New York City apartments. Indoor Air 2017, 27, 840–851. [Google Scholar] [CrossRef] [PubMed]
- Kolendowicz, L.; Półrolniczak, M.; Szyga-Pluta, K.; Bednorz, E. Human-biometeorological conditions in the southern Baltic coast based on the universal thermal climate index (UTCI). Theor. Appl. Climatol. 2018, 134, 363–379. [Google Scholar] [CrossRef] [Green Version]
- Just, M.G.; Nichols, L.M.; Dunn, R.R. Human indoor climate preferences approximate specific geographies. R. Soc. Open Sci. 2019, 6, 180695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salamone, F.; Bellazzi, A.; Belussi, L.; Damato, G.; Danza, L.; Dell’Aquila, F.; Ghellere, M.; Megale, V.; Meroni, I.; Vitaletti, W. Evaluation of the visual stimuli on personal thermal comfort perception in real and virtual environments using machine learning approaches. Sensors 2020, 20, 1627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, A.S.; Almeida, L.S.S.; Ghisi, E. Decision-making process for improving thermal and energy performance of residential buildings: A case study of constructive systems in Brazil. Energy Build. 2016, 128, 270–286. [Google Scholar] [CrossRef]
- Charalampopoulos, I.; Tsiros, I.; Chronopoulou-Sereli, A.; Matzarakis, A. A note on the evolution of the daily pattern of thermal comfort-related micrometeorological parameters in small urban sites in Athens. Int. J. Biometeorol. 2014, 59, 1223–1236. [Google Scholar] [CrossRef] [PubMed]
- Steiner, J.L.; Hatfield, J.L. Winds of change: A century of agroclimate research. Agron. J. 2008, 100, S-132–S-152. [Google Scholar] [CrossRef] [Green Version]
- Lees, J.M. Open and free: Software and scientific reproducibility. Seismol. Res. Lett. 2012, 83, 751–752. [Google Scholar] [CrossRef]
- Peng, R.D. Reproducible research in computational science. Science 2011, 334, 1226–1227. [Google Scholar] [CrossRef] [Green Version]
- Stodden, V. Reproducible research: Tools and strategies for scientific computing. Comput. Sci. Eng. 2012, 14, 11–12. [Google Scholar] [CrossRef] [Green Version]
- Lowndes, J.S.S.; Best, B.D.; Scarborough, C.; Afflerbach, J.C.; Frazier, M.R.; O’Hara, C.C.; Jiang, N.; Halpern, B.S. Our path to better science in less time using open data science tools. Nat. Ecol. Evol. 2017, 1, 0160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, R.D.; Matsui, E. The Art of Data Science; Leanpub: Victoria, BC, Canada, 2015; ISBN 978-1-365-06146-2. [Google Scholar]
- van den Burg, G.J.J.; Nazábal, A.; Sutton, C. Wrangling messy CSV files by detecting row and type patterns. Data Min. Knowl. Disc. 2019, 33, 1799–1820. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. Tidy Data. J. Stat. Softw. 2014, 59, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Kamber, M.; Pei, J. Data Mining Concepts and Techniques; Elsevier: Amsterdam, The Netherlands, 2011; ISBN 978-0-12-381479-1. [Google Scholar]
- Márquez, F.P.G.; Lev, B. Big Data Management; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; ISBN 978-3-319-45498-6. [Google Scholar]
- Leek, J. The Elements of Data Analytic Style; Leanpub: Victoria, BC, Canada, 2015. [Google Scholar]
- Sandve, G.K.; Nekrutenko, A.; Taylor, J.; Hovig, E. Ten simple rules for reproducible computational research. PLoS Comput. Biol. 2013, 9, e1003285. [Google Scholar] [CrossRef] [Green Version]
- Hothorn, T.; Everitt, B.S. A Handbook of Statistical Analyses Using R; CRC press: Boca Raton, FL, USA, 2014; ISBN 1-4822-0458-4. [Google Scholar]
- Weiss, N.A.; Weiss, C.A. Introductory Statistics; Pearson; Addison-Wesley: Boston, MA, USA, 2008; ISBN 0-321-39361-9. [Google Scholar]
- Munafò, M.R.; Nosek, B.A.; Bishop, D.V.M.; Button, K.S.; Chambers, C.D.; du Sert, N.P.; Simonsohn, U.; Wagenmakers, E.-J.; Ware, J.J.; Ioannidis, J.P.A. A manifesto for reproducible science. Nat. Hum. Behav. 2017, 1, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Powers, S.M.; Hampton, S.E. Open science, reproducibility, and transparency in ecology. Ecol. Appl. 2019, 29, e01822. [Google Scholar] [CrossRef] [Green Version]
- Eglen, S.J. A quick guide to teaching R Programming to computational biology students. PLoS Comput. Biol. 2009, 5, e1000482. [Google Scholar] [CrossRef] [Green Version]
- Ihaka, R.; Gentleman, R.R. A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- Grunsky, E.C.R. A data analysis and statistical programming environment—An emerging tool for the geosciences. Comput. Geosci. 2002, 28, 1219–1222. [Google Scholar] [CrossRef]
- Wickham, H.; Hester, J.; Francois, R. Readr: Read Rectangular Text Data. Available online: https://CRAN.R-project.org/package=readr (accessed on 25 April 2020).
- Peng, R.D. R Programming for Data Science; Leanpub: Victoria, BC, Canada, 2016; ISBN 978-1-365-05682-6. [Google Scholar]
- Dragulescu, A.; Arendt, C. Xlsx: Read, Write, Format Excel 2007 and Excel 97/2000/XP/2003 Files. Available online: https://CRAN.R-project.org/package=xlsx (accessed on 10 May 2020).
- R Core Team foreign. Read Data Stored by “Minitab”, “S”, “SAS”, “SPSS”, “Stata”, “Systat”, “Weka”, “dBase”. Available online: https://CRAN.R-project.org/package=foreign (accessed on 12 May 2020).
- Wickham, H.; Miller, E. Haven: Import and Export “SPSS”, “Stata” and “SAS” Files. Available online: https://CRAN.R-project.org/package=haven (accessed on 25 April 2020).
- Wickham, H. feather: R Bindings to the Feather “API”. Available online: https://CRAN.R-project.org/package=feather (accessed on 25 April 2020).
- Chamberlain, S. Rnoaa: “NOAA” Weather Data from R. Available online: https://CRAN.R-project.org/package=rnoaa (accessed on 10 May 2020).
- Sparks, A.H. Nasapower: NASA POWER API Client. Available online: https://CRAN.R-project.org/package=nasapower (accessed on 12 May 2020).
- Stevens, A. Copernicus. Available online: https://github.com/antoinestevens/copernicus (accessed on 4 June 2020).
- Mattiuzzi, M.; Detsch, F. MODIS: Acquisition and Processing of MODIS Products. Available online: https://CRAN.R-project.org/package=MODIS (accessed on 12 May 2020).
- Hart, E. RWBclimate: A package for accessing World Bank climate data. Available online: https://CRAN.R-project.org/package=rWBclimate (accessed on 12 May 2020).
- Kothe, S. Cmsaf: Tools for CM SAF NetCDF Data. Available online: https://CRAN.R-project.org/package=cmsaf (accessed on 12 May 2020).
- Mohammed, I. NASAaccess: Downloading and reformatting tool for NASA Earth observation data products. Available online https://github.com/nasa/NASAaccess: (accessed on 12 May 2020).
- Dowle, M.; Srinivasan, A. Data.table: Extension of ‘data.frame’. Available online: https://CRAN.R-project.org/package=data.table (accessed on 10 May 2020).
- Wickham, H.; François, R.; Henry, L.; Müller, K. Dplyr: A Grammar of Data Manipulation. Available online: https://CRAN.R-project.org/package=dplyr (accessed on 25 April 2020).
- Wickham, H. Reshape2: Flexibly Reshape Data: A Reboot of the Reshape Package. Available online: https://CRAN.R-project.org/package=reshape2 (accessed on 25 April 2020).
- Wickham, H. Reshaping data with the reshape package. J. Stat. Softw. 2007, 21, 1–20. [Google Scholar] [CrossRef]
- Spinu, V.; Grolemund, G.; Wickham, H. Lubridate: Make Dealing with Dates a Little Easier. Available online: https://CRAN.R-project.org/package=lubridate (accessed on 12 May 2020).
- Schweiker, M.; Mueller, S.; Kleber, M.; Kingma, B.; Shukuya, M. Comf: Functions for Thermal Comfort Research. Available online: https://CRAN.R-project.org/package=comf (accessed on 15 May 2020).
- Fanger, P.O. Thermal Comfort. Analysis and Applications in Environmental Engineering; McGraw-Hill Book Company: New York, NY, USA, 1970; ISBN 978-0-07-019915-6. [Google Scholar]
- Reig-Gracia, F.; Vicente-Serrano, S.M.; Dominguez-Castro, F.; Bedia-Jiménez, J. ClimInd: Climate Indices. Available online: https://CRAN.R-project.org/package=ClimInd (accessed on 10 May 2020).
- Crisci, A.; Morabito, M. RBiometeo: Biometeorological Functions in R. Available online: https://github.com/alfcrisci/rBiometeo (accessed on 15 May 2020).
- Czernecki, B.; Glogowski, A.; Nowosad, J. Climate: Interface to Download Meteorological (and Hydrological) Datasets. Available online: https://CRAN.R-project.org/package=climate (accessed on 17 May 2020).
- Kemp, M.U.; van Loon, E.E.; Shamoun-Baranes, J.; Bouten, W. RNCEP: Global weather and climate data at your fingertips. Methods Ecol. Evol. 2012, 3, 65–70. [Google Scholar] [CrossRef]
- Anderson, B.; Peng, R.; Ferreri, J. Weathermetrics: Functions to Convert Between Weather Metrics. Available online: https://CRAN.R-project.org/package=weathermetrics (accessed on 15 May 2020).
- Wood, S. Mgcv: Mixed GAM Computation Vehicle with Automatic Smoothness Estimation. Available online: https://CRAN.R-project.org/package=mgcv (accessed on 19 May 2020).
- Ho, D.; Imai, K.; King, G.; Stuart, E. MatchIt: Nonparametric Preprocessing for Parametric Causal Inference. Available online: https://CRAN.R-project.org/package=MatchIt (accessed on 17 May 2020).
- Sarkar, D. Lattice: Trellis Graphics for R. Available online: https://CRAN.R-project.org/package=lattice (accessed on 5 February 2020).
- Kabacoff, R. R in Action. Data Analysis and Graphics with R; Manning: New York, NY, USA, 2011. [Google Scholar]
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D. Ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. Available online: https://CRAN.R-project.org/package=ggplot2 (accessed on 5 February 2020).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Sievert, C.; Parmer, C.; Hocking, T.; Chamberlain, S.; Ram, K.; Corvellec, M.; Despouy, P. Plotly: Create Interactive Web Graphics via “plotly.js”. Available online: https://CRAN.R-project.org/package=plotly (accessed on 5 February 2020).
- Xie, Y.; Allaire, J.J.; Grolemund, G. R Markdown: The Definitive Guide; Chapman and Hall/CRC: New York, NY, USA, 2018; ISBN 978-1-138-35933-8. [Google Scholar]
- Gruber, J. Markdown. Available online: http://daringfireball.net/projects/markdown/ (accessed on 5 February 2020).
- Xie, Y. Blogdown: Create Blogs and Websites with R Markdown. Available online: https://CRAN.R-project.org/package=blogdown (accessed on 5 February 2020).
- Xie, Y. Bookdown: Authoring Books and Technical Documents with R Markdown. Available online: https://CRAN.R-project.org/package=bookdown (accessed on 5 February 2020).
- Chang, W.; Cheng, J.; Allaire, J.J.; Xie, Y.; McPherson, J. Shiny: Web Application Framework for R. Available online: https://CRAN.R-project.org/package=shiny (accessed on 5 February 2020).



| Package Name | Short Description | |
|---|---|---|
| Manual (local) input | readr | Fast and friendly way to read rectangular data (like csv, tsv, and fwf). |
| xlxs | Provides tools to read and write xlsx files and control the appearance of the spreadsheet files. | |
| foreign | Reading and writing data stored by some versions of ‘Epi Info’, ‘Minitab’, ‘S’, ‘SAS’, ‘SPSS’, ‘Stata’, ‘Systat’, ‘Weka’ and ‘dBase’ files. | |
| haven | Reading and writing data stored by of ‘SAS’, ‘SPSS’, ‘Stata’. | |
| feather | Read and write in an easy-to-use binary file format (feather) for storing data frames. | |
| Web database acquisition | rnoaa | Is an R wrapper to many NOAA APIs. Select and download each type of data |
| nasapower | Making quick and easy to automate downloading from NASA POWER global meteorology, surface solar energy and climatology data. | |
| MODIS | Downloading and processing functionalities for the Moderate Resolution Imaging Spectroradiometer (MODIS). | |
| Copernicus | Downloading data from the COPERNICUS data portal through the fast HTTP access. | |
| rWBclimate | Download model predictions from 15 different global circulation models in 20-year intervals from the World Bank. Access historical data and create maps at 2 different spatial scales. | |
| cmsaf | Provides satellite-based climate data records of essential climate variables of the energy budget and water cycle. The data records are generally distributed in NetCDF format. | |
| NASAaccess | Can generate gridded ASCII tables of climate (CIMP5) and weather data (GPM, TRMM, GLDAS) needed to drive various hydrological models (e.g., SWAT, VIC, RHESSys). |
| Package Name | Short Description |
|---|---|
| data.table | Integrated and quick data handling. |
| dplyr | A powerful package with a noticeably clear syntax to transform, summarise and do calculation into and between tabular data frames. |
| reshape2 | Is dedicated to the transformation processes between wide and long data frame format. |
| lubridate | Providing functions to deal with date and time formats and time spans. |
| Package Name | Short Description |
|---|---|
| comf | Calculates various common and less common thermal comfort indices, convert physical variables and evaluate the performance of thermal comfort indices. |
| ClimInd | Computes 138 standard climate indices at monthly, seasonal and annual resolution. |
| rBiometeo | Human thermal comfort and many biometeorological indices used in Institute of Biometeorology in Florence. Biometeorological indices used in Institute of Biometeorology in Florence |
| climate | Automized downloading of meteorological and hydrological data from publicly available repositories. |
| RNCEP | Contains functions to retrieve, organise and visualise weather data from the NCEP/NCAR Reanalysis and NCEP/DOE Reanalysis II datasets. |
| weathermetrics | Conversions between primary weather metrics. |
| Package Name | Short Description | |
|---|---|---|
| Visualisation | lattice | High-level data visualisation system, with an emphasis on multivariate data. |
| ggplot2 | A system for ‘declaratively’ creating graphics, based on “The Grammar of Graphics”. | |
| plotly | An all-in-one interactive graphics package (3D and animated). | |
| Communication | rmarkdown | Writing functional reports with embedded code and its results in .doc, .pdf, HTML format. |
| blogdown | Publishing web pages created with rmarkdown. | |
| bookdown | Facilitating writing ebooks, ready to print books, long articles and reports. | |
| shiny | Building interactive web applications focused on data sharing. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Charalampopoulos, I. The R Language as a Tool for Biometeorological Research. Atmosphere 2020, 11, 682. https://doi.org/10.3390/atmos11070682
Charalampopoulos I. The R Language as a Tool for Biometeorological Research. Atmosphere. 2020; 11(7):682. https://doi.org/10.3390/atmos11070682
Chicago/Turabian StyleCharalampopoulos, Ioannis. 2020. "The R Language as a Tool for Biometeorological Research" Atmosphere 11, no. 7: 682. https://doi.org/10.3390/atmos11070682