Multi-Task Topic Analysis Framework for Hallmarks of Cancer with Weak Supervision
Abstract
:1. Introduction
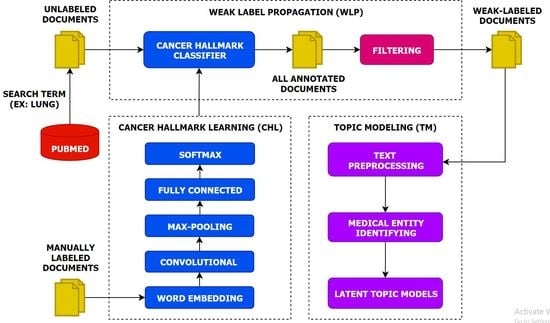
- We present a multi-task topic analysis (MTTA) framework to analyze cancer hallmark-specific topics in a multi-task manner by leveraging large amounts of unlabeled documents.
- We leverage a large number of unlabeled documents related to lung cancer according to the HoC. Experiments on the unlabeled documents have demonstrated that the MTTA framework is potentially valuable for the HoC analysis with an impressive superiority.
- We highlight the importance of the latent topic models on weak-labeled documents that are produced by the CNN model. The experimental results show that the CNN model can classify cancer hallmarks better than other approaches and conventional topic models can discover topics efficiently.
- We produce semantic representations for each hallmark category using the CNN model and capture this semantic information for each hallmark category using conventional topic models.
2. Related Work
2.1. Cancer Hallmarks Classification
2.2. Topic Modelling for Biomedical Text Mining
3. Materials and Methods
| Algorithm 1 Multi-task topic analysis framework |
| Input: A set of labeled documents; a set of unlabeled documents; |
| Output: A set of topics for each hallmark category; |
| begin |
|
| end |
3.1. Cancer Hallmarks Learning
| Algorithm 2 Cancer hallmark learning task |
| Input: A set of labeled documents; pre-trained word vectors; |
| Output: A model trained on ; |
| begin |
|
| end |
3.2. Weak Label Propagation
| Algorithm 3 Weak label propagation task |
| Input: A set of unlabeled documents; a model trained on ; |
| Output: A set of weak-labeled documents; |
| begin |
|
| end |
3.3. Topic Modeling
| Algorithm 4 Topic modeling task |
| Input: A set of weak-labeled documents; |
| Output: A set of topics for each hallmark category; |
| begin |
|
| end |
3.3.1. Text Preprocessing
- All numbers and special characters were removed.
- All uppercase characters were changed into lowercase.
- All non-ASCII character was removed.
- All stopword was removed.
- All classes in an abstract including background, objective, method, result, and conclusion were removed.
3.3.2. Medical Entity Identifier
3.3.3. Latent Topic Models
| Algorithm 5 Probabilistic latent semantic analysis |
| Input: A set of weak-labeled documents; a set of entities; |
| Output: A set of topics for each hallmark category; |
| begin |
|
| end |
| Algorithm 6 Latent Dirichlet allocation |
| Input: A set of weak-labeled documents; a set of entities; |
| Output: A set of topics for each hallmark category; |
| begin |
|
| end |
4. Experimental Setup
4.1. Dataset
4.1.1. Manually Labeled Documents
4.1.2. Unlabeled Documents
4.2. Deep Learning Model
- CNN [12]: a basic CNN model. It has three convolutional layers with a kernel width of 3, 4, and 5 with 100 output channels.
- RCNN [41]: a hybrid RNN and CNN model. The combined architecture CNN followed by Bidirectional Long Short Term Memory (BiLSTM).
- Gated Recurrent Unit (GRU) [42]: one of the basic RNN models. Only forward GRU with 256 hidden units.
- Bidirectional Gated Recurrent Unit (BiGRU) [43]: one of the basic RNN models. Forward and backward GRU with 256 hidden units.
- Long Short Term Memory (LSTM) [44]: one of the basic RNN models. Only forward LSTM with 256 hidden units.
- BiLSTM [45]: one of the basic RNN models. Forward and backward LSTM with 256 hidden units.
4.3. Topic Model
5. Experimental Result and Discussion
5.1. Cancer Hallmark Classification Result
5.2. Weak Label Propagation Result
5.3. Topic Modelling Result
5.4. Visualization
5.5. Optimal Number of Topics
6. Conclusions and Future Work
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mehmet Sitki Copur, M.D. State of Cancer Research around the Globe. Oncology 2019, 14, 33. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Gutschner, T.; Diederichs, S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012, 9, 703–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piao, Y.; Piao, M.; Ryu, K.H. Multiclass cancer classification using a feature subset-based ensemble from microRNA expression profiles. Comput. Biol. Med. 2017, 80, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Piao, M.; Piao, Y.; Li, M.; Ryu, K.H. A New direction of cancer classification: Positive effect of Low-ranking MicroRNAs. Osong Public Health Res. Perspect. 2014, 5, 279–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munkhdalai, T.; Li, M.; Batsuren, K.; Park, H.A.; Choi, N.H.; Ryu, K.H. Incorporating domain knowledge in chemical and biomedical named entity recognition with word representations. J. Chemin. 2015, 7, 9. [Google Scholar] [CrossRef] [Green Version]
- Munkhdalai, T.; Namsrai, O.E.; Ryu, K.H. Self-training in significance space of support vectors for imbalanced biomedical event data. BMC Bioinform. 2015, 16, 6. [Google Scholar] [CrossRef] [Green Version]
- Young, T.; Hazarika, D.; Poria, S.; Cambria, E. Recent trends in deep learning based natural language processing. IEEE Comput. Intell. Mag. 2018, 13, 55–75. [Google Scholar] [CrossRef]
- He, L.; Lee, K.; Lewis, M.; Zettlemoyer, L. Deep semantic role labeling: What works and what’s next. In Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics, Vancouver, BC, Canada, 30 July–4 August 2017; Volume 1, pp. 473–483. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; 2017; pp. 5998–6008. [Google Scholar]
- Kim, Y. Convolutional neural networks for sentence classification. arXiv 2014, arXiv:1408.5882. [Google Scholar]
- Mikolov, T.; Karafiát, M.; Burget, L.; Černocký, J.; Khudanpur, S. Recurrent neural network based language model. In Proceedings of the Eleventh Annual Conference of the International Speech Communication Association, Chiba, Japan, 26–30 September 2010. [Google Scholar]
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.S.; Dean, J. Distributed representations of words and phrases and their compositionality. In Advances in Neural Information Processing Systems; Curran Associates Inc.: Red Hook, NY, USA, 2013; pp. 3111–3119. [Google Scholar]
- Batbaatar, E.; Li, M.; Ryu, K.H. Semantic-emotion neural network for emotion recognition from text. IEEE Access 2019, 7, 111866–111878. [Google Scholar] [CrossRef]
- Blei, D.M.; Ng, A.Y.; Jordan, M.I. Latent dirichlet allocation. J. Mach. Learn. Res. 2003, 3, 993–1022. [Google Scholar]
- Hofmann, T. Unsupervised learning by probabilistic latent semantic analysis. Mach. Learn. 2001, 42, 177–196. [Google Scholar] [CrossRef]
- Baker, S.; Silins, I.; Guo, Y.; Ali, I.; Högberg, J.; Stenius, U.; Korhonen, A. Automatic semantic classification of scientific literature according to the hallmarks of cancer. Bioinformatics 2015, 32, 432–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, S.; Kiela, D.; Korhonen, A. Robust text classification for sparsely labelled data using multi-level embeddings. In Proceedings of the COLING 2016, the 26th International Conference on Computational Linguistics: Technical Papers, Osaka, Japan, 11–16 December 2016; pp. 2333–2343. [Google Scholar]
- Baker, S.; Korhonen, A.; Pyysalo, S. Cancer hallmark text classification using convolutional neural networks. In Proceedings of the Fifth Workshop on Building and Evaluating Resources for Biomedical Text Mining (BioTxtM 2016), Osaka, Japan, 11–16 December 2016; pp. 1–9. [Google Scholar]
- Baker, S.; Korhonen, A. Initializing Neural Networks for Hierarchical Multi-Label Text Classification; BioNLP: Vancouver, BC, Canada, 2017; pp. 307–315. [Google Scholar]
- Baker, S.; Ali, I.; Silins, I.; Pyysalo, S.; Guo, Y.; Högberg, J.; Stenius, U.; Korhonen, A. Cancer Hallmarks Analytics Tool (CHAT): A text mining approach to organize and evaluate scientific literature on cancer. Bioinformatics 2017, 33, 3973–3981. [Google Scholar] [CrossRef]
- Du, J.; Chen, Q.; Peng, Y.; Xiang, Y.; Tao, C.; Lu, Z. ML-Net: Multi-label classification of biomedical texts with deep neural networks. J. Am. Med. Inform. Assoc. 2019, 26, 1279–1285. [Google Scholar] [CrossRef] [Green Version]
- Pyysalo, S.; Baker, S.; Ali, I.; Haselwimmer, S.; Shah, T.; Young, A.; Guo, Y.; Högberg, J.; Stenius, U.; Narita, M. LION LBD: A literature-based discovery system for cancer biology. Bioinformatics 2018, 35, 1553–1561. [Google Scholar] [CrossRef]
- Peng, Y.; Yan, S.; Lu, Z. Transfer Learning in Biomedical Natural Language Processing: An Evaluation of BERT and ELMo on Ten Benchmarking Datasets. arXiv 2019, arXiv:1906.05474. [Google Scholar]
- Andrzejewski, D. Modeling Protein–Protein Interactions in Biomedical Abstracts with Latent Dirichlet Allocation; CS 838-Final Project; University of Wisconsin–Madison: Madison, WI, USA, 2006. [Google Scholar]
- Wang, H.; Huang, M.; Zhu, X. Extract interaction detection methods from the biological literature. BMC Bioinform. 2009, 10, 55. [Google Scholar] [CrossRef] [Green Version]
- Wang, V.; Xi, L.; Enayetallah, A.; Fauman, E.; Ziemek, D. GeneTopics-interpretation of gene sets via literature-driven topic models. BMC Syst. Biol. 2013, 7, 10. [Google Scholar] [CrossRef] [Green Version]
- Bisgin, H.; Liu, Z.; Fang, H.; Xu, X.; Tong, W. Mining FDA drug labels using an unsupervised learning technique-topic modeling. BMC Bioinform. 2011, 12, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bisgin, H.; Liu, Z.; Kelly, R.; Fang, H.; Xu, X.; Tong, W. Investigating drug repositioning opportunities in FDA drug labels through topic modeling. BMC Bioinform. 2012, 13, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Yin, X.; Li, Z.; Hu, X.; Huang, J.X. A LDA-based approach to promoting ranking diversity for genomics information retrieval. BMC Genomics 2012, 13, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hersh, W.R.; Cohen, A.M.; Roberts, P.M.; Rekapalli, H.K. TREC 2006 Genomics Track Overview; TREC: Gaithersburg, MD, USA, 2006. [Google Scholar]
- Song, M.; Kim, S.Y. Detecting the knowledge structure of bioinformatics by mining full-text collections. Scientometrics 2013, 96, 183–201. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, P.; Liu, T.; Xu, K. BioTopic: A topic-driven biological literature mining system. Int. J. Data Min. Bioinform. 2016, 14, 373–386. [Google Scholar] [CrossRef]
- Cui, M.; Liang, Y.; Li, Y.; Guan, R. Exploring Trends of Cancer Research Based on Topic Model. IWOST-1 2015, 1339, 7–18. [Google Scholar]
- Dunne, R.A.; Campbell, N.A. On the pairing of the softmax activation and cross-entropy penalty functions and the derivation of the softmax activation function. In Proceedings of the 8th Australian Conference on Neural Networks, Canberra, Australia, 10–12 April 1997; Volume 181, p. 185. [Google Scholar]
- Chiu, B.; Crichton, G.; Korhonen, A.; Pyysalo, S. How to train good word embeddings for biomedical NLP. In Proceedings of the 15th Workshop on Biomedical Natural Language Processing, Berlin, Germany, 12 August 2016; pp. 166–174. [Google Scholar]
- Aronson, A.R. Effective mapping of biomedical text to the UMLS Metathesaurus: The MetaMap program. In Proceedings of the AMIA Symposium. American Medical Informatics Association, Chicago, IL, USA, 14–18 November 2001; p. 17. [Google Scholar]
- Bodenreider, O. The unified medical language system (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 2004, 32, 267–270. [Google Scholar] [CrossRef] [Green Version]
- Chapman, B.; Chang, J. Biopython: Python tools for computational biology. ACM Sigbio Newsl. 2000, 20, 15–19. [Google Scholar] [CrossRef]
- Lai, S.; Xu, L.; Liu, K.; Zhao, J. Recurrent convolutional neural networks for text classification. In Proceedings of the Twenty-ninth AAAI Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015. [Google Scholar]
- Chung, J.; Gulcehre, C.; Cho, K.; Bengio, Y. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv 2014, arXiv:1412.3555. [Google Scholar]
- Luo, X.; Zhou, W.; Wang, W.; Zhu, Y.; Deng, J. Attention-based relation extraction with bidirectional gated recurrent unit and highway network in the analysis of geological data. IEEE Access 2017, 6, 5705–5715. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Liwicki, M.; Graves, A.; Fernàndez, S.; Bunke, H.; Schmidhuber, J. A novel approach to on-line handwriting recognition based on bidirectional long short-term memory networks. In Proceedings of the 9th International Conference on Document Analysis and Recognition, ICDAR 2007, Curitiba, Brazil, 23–26 September 2007. [Google Scholar]
- Řehůřek, R.; Sojka, P. Gensim—Statistical Semantics in Python. Statistical Semantics; Gensim; EuroScipy: Paris, France, 2011. [Google Scholar]
- Ketkar, N. Introduction to Pytorch; Apress: Berkeley, CA, USA, 2017. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Liu, T.C.; Jin, X.; Wang, Y.; Wang, K. Role of epidermal growth factor receptor in lung cancer and targeted therapies. Am. J. Cancer Res. 2017, 7, 187. [Google Scholar] [PubMed]
- Amin, A.R.; Karpowicz, P.A.; Carey, T.E.; Arbiser, J.; Nahta, R.; Chen, Z.G.; Dong, J.T.; Kucuk, O.; Khan, G.N.; Huang, G.S. Evasion of anti-growth signaling: A key step in tumorigenesis and potential target for treatment and prophylaxis by natural compounds. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2015; Volume 35, pp. 55–77. [Google Scholar]
- Liu, G.; Pei, F.; Yang, F.; Li, L.; Amin, A.D.; Liu, S.; Buchan, J.R.; Cho, W.C. Role of autophagy and apoptosis in non-small-cell lung cancer. Int. J. Mol. Sci. 2017, 18, 367. [Google Scholar] [CrossRef]
- Yaswen, P.; MacKenzie, K.L.; Keith, W.N.; Hentosh, P.; Rodier, F.; Zhu, J.; Firestone, G.L.; Matheu, A.; Carnero, A.; Bilsland, A. Therapeutic targeting of replicative immortality. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2015; Volume 35, pp. 104–128. [Google Scholar]
- Shimoyamada, H.; Yazawa, T.; Sato, H.; Okudela, K.; Ishii, J.; Sakaeda, M.; Kashiwagi, K.; Suzuki, T.; Mitsui, H.; Woo, T. Early growth response-1 induces and enhances vascular endothelial growth factor-A expression in lung cancer cells. Am. J. Pathol. 2010, 177, 70–83. [Google Scholar] [CrossRef]
- Martin, T.A.; Ye, L.; Sanders, A.J.; Lane, J.; Jiang, W.G. Cancer Invasion and Metastasis: Molecular and Cellular Perspective. Available online: https://www.ncbi.nlm.nih.gov/books/NBK164700/ (accessed onine 30 December 2019) (accessed on 30 December 2019).
- Ninomiya, H.; Nomura, K.; Satoh, Y.; Okumura, S.; Nakagawa, K.; Fujiwara, M.; Tsuchiya, E.; Ishikawa, Y. Genetic instability in lung cancer: Concurrent analysis of chromosomal, mini-and microsatellite instability and loss of heterozygosity. Br. J. Cancer 2006, 94, 1485. [Google Scholar] [CrossRef] [Green Version]
- Melkamu, T.; Qian, X.; Upadhyaya, P.; O’Sullivan, M.G.; Kassie, F. Lipopolysaccharide enhances mouse lung tumorigenesis: A model for inflammation-driven lung cancer. Vet. Pathol. 2013, 50, 895–902. [Google Scholar] [CrossRef]
- Harmey, J.H.; Bucana, C.D.; Lu, W.; Byrne, A.M.; McDonnell, S.; Lynch, C.; Bouchier-Hayes, D.; Dong, Z. Lipopolysaccharide-induced metastatic growth is associated with increased angiogenesis, vascular permeability and tumor cell invasion. Int. J. Cancer 2002, 101, 415–422. [Google Scholar] [CrossRef]
- Min, H.Y.; Lee, H.Y. Oncogene-driven metabolic alterations in cancer. Biomol. Amp Ther. 2018, 26, 45. [Google Scholar] [CrossRef] [Green Version]
- Gwin, J.L.; Klein-Szanto, A.J.; Zhang, S.Y.; Agarwal, P.; Rogatko, A.; Keller, S.M. Loss of blood group antigen A in non-small cell lung cancer. Ann. Surg. Oncol. 1994, 1, 423–427. [Google Scholar] [CrossRef]





















| Abbreviations | Definitions |
|---|---|
| Labeled documents | |
| Number of labeled documents | |
| Unlabeled documents | |
| Number of unlabeled documents | |
| Topics | |
| Number of topics | |
| Classification model | |
| Vocabulary for labeled documents | |
| Word vector | |
| Weak labeled documents | |
| Number of weak labeled documents | |
| Class probability for each class (positive and negative) | |
| Entities | |
| Number of entities | |
| Dirichlet prior (Topic distribution) | |
| Dirichlet prior (Word distribution) | |
| Multinomial distribution over Q topics | |
| Multinomial distribution over entities |
| Entity (CUI or AA) | |||||
|---|---|---|---|---|---|
| low-folate (LF) | 1 | 2 | 8 | 0 | 7 |
| metastasis (C4255448) | 3 | 4 | 5 | 4 | 9 |
| neoplasm metastasis (C0027627) | 3 | 2 | 6 | 3 | 6 |
| decreased folic acid (C0239623) | 9 | 4 | 2 | 1 | 0 |
| tryptophanase (C0041260) | 2 | 1 | 0 | 1 | 3 |
| Hallmark | Train | Development | Test | |||
|---|---|---|---|---|---|---|
| Y | N | Y | N | Y | N | |
| SPS | 328 | 975 | 43 | 140 | 91 | 275 |
| EGS | 172 | 1131 | 22 | 161 | 46 | 320 |
| RCD | 303 | 1000 | 42 | 141 | 84 | 282 |
| ERI | 81 | 1222 | 11 | 172 | 23 | 343 |
| IA | 99 | 1204 | 13 | 170 | 31 | 335 |
| AIM | 208 | 1095 | 29 | 154 | 54 | 312 |
| GIM | 227 | 1076 | 38 | 145 | 68 | 298 |
| TPI | 169 | 1134 | 24 | 159 | 47 | 319 |
| DCE | 74 | 1229 | 10 | 173 | 21 | 345 |
| AID | 77 | 1226 | 10 | 173 | 21 | 345 |
| Parameter | CNN | RNN |
|---|---|---|
| Learning rate | 0.001 | 0.001 |
| Batch size | 128 | 128 |
| Hidden dimension | 256 | - |
| Number of layers | 2 | - |
| Number of filters | - | 100 |
| Filter size | - | [3, 4, 5] |
| Early stopping patience | 20 | 20 |
| Dropout | 0.5 | 0.5 |
| Model | SPS | EGS | RCD | ERI | IA | AIM | GIM | TPI | DCE | AID | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Baker et al. [20] | Best | 70.00 | 71.50 | 86.90 | 91.50 | 85.70 | 82.60 | 81.70 | 84.20 | 88.30 | 75.80 |
| CNN | Base | 73.49 | 82.22 | 92.93 | 92.98 | 88.60 | 88.45 | 86.93 | 87.43 | 94.36 | 84.42 |
| Tuned | 74.66 | 80.15 | 92.12 | 93.18 | 89.10 | 90.15 | 87.62 | 87.93 | 95.83 | 84.31 | |
| RCNN | Base | 74.38 | 81.67 | 91.10 | 92.95 | 86.46 | 89.82 | 85.96 | 88.92 | 96.62 | 88.61 |
| Tuned | 75.13 | 79.77 | 91.32 | 92.82 | 87.41 | 89.57 | 86.05 | 88.70 | 95.52 | 88.55 | |
| GRU | Base | 74.76 | 79.14 | 85.95 | 91.44 | 88.12 | 87.78 | 82.30 | 86.48 | 90.48 | 83.74 |
| Tuned | 76.65 | 74.03 | 87.86 | 90.57 | 88.28 | 87.38 | 83.31 | 86.78 | 93.52 | 81.31 | |
| BiGRU | Base | 70.99 | 57.45 | 69.85 | 87.86 | 85.46 | 83.58 | 80.46 | 72.99 | 89.09 | 74.01 |
| Tuned | 71.16 | 58.73 | 70.61 | 85.87 | 85.39 | 82.69 | 81.05 | 71.03 | 90.40 | 70.37 | |
| LSTM | Base | 76.47 | 72.42 | 82.94 | 86.32 | 86.48 | 86.30 | 82.22 | 82.08 | 91.05 | 78.83 |
| Tuned | 76.62 | 69.62 | 82.84 | 87.22 | 87.10 | 84.84 | 82.60 | 82.26 | 90.43 | 78.88 | |
| BiLSTM | Base | 66.23 | 57.15 | 58.29 | 74.86 | 72.53 | 68.82 | 73.99 | 65.20 | 81.57 | 60.82 |
| Tuned | 65.33 | 56.75 | 56.36 | 73.44 | 72.61 | 69.73 | 73.00 | 64.00 | 80.90 | 62.40 | |
| Model | SPS | EGS | RCD | ERI | IA | AIM | GIM | TPI | DCE | AID | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CNN | Base | 81.99 | 92.31 | 95.08 | 98.13 | 96.68 | 94.06 | 91.68 | 94.96 | 98.64 | 96.92 |
| Tuned | 82.46 | 91.56 | 94.61 | 98.21 | 96.76 | 94.76 | 92.15 | 95.11 | 98.91 | 96.84 | |
| RCNN | Base | 82.15 | 91.92 | 93.94 | 98.13 | 95.97 | 94.96 | 91.29 | 95.31 | 99.10 | 97.27 |
| Tuned | 82.46 | 91.37 | 94.06 | 98.09 | 96.36 | 95.00 | 91.17 | 95.27 | 98.87 | 97.35 | |
| GRU | Base | 81.98 | 89.70 | 90.55 | 97.99 | 96.02 | 93.32 | 89.67 | 93.46 | 97.86 | 96.70 |
| Tuned | 82.99 | 89.64 | 91.68 | 97.83 | 95.93 | 93.13 | 90.25 | 93.57 | 98.52 | 96.62 | |
| BiGRU | Base | 78.98 | 82.55 | 78.71 | 97.11 | 95.25 | 91.37 | 88.49 | 88.27 | 97.77 | 94.62 |
| Tuned | 79.51 | 82.61 | 78.60 | 96.84 | 95.44 | 90.82 | 89.18 | 87.17 | 97.69 | 93.98 | |
| LSTM | Base | 82.97 | 87.94 | 88.19 | 96.95 | 95.58 | 92.61 | 89.48 | 91.13 | 98.05 | 95.88 |
| Tuned | 83.13 | 87.80 | 88.60 | 96.92 | 95.85 | 91.54 | 89.78 | 91.68 | 97.97 | 96.13 | |
| BiLSTM | Base | 76.24 | 82.52 | 71.57 | 94.34 | 92.78 | 84.78 | 85.11 | 84.97 | 96.32 | 92.17 |
| Tuned | 75.63 | 80.19 | 70.96 | 94.10 | 93.00 | 85.60 | 84.95 | 83.88 | 96.43 | 92.64 | |
| Hallmark | No. of Documents | No. of Entities | No. of Unique Entities |
|---|---|---|---|
| SPS | 4097 | 766,146 | 22,576 |
| EGS | 950 | 170,965 | 11,272 |
| RCD | 14,313 | 2,710,563 | 56,052 |
| ERI | 604 | 102,357 | 9531 |
| IA | 3009 | 582,378 | 22,362 |
| AIM | 48,953 | 7,698,235 | 80,444 |
| GIM | 7733 | 1,444,839 | 33,579 |
| TPI | 30,403 | 6,006,649 | 67,733 |
| DCE | 43 | 7701 | 2124 |
| AID | 13,098 | 2,094,029 | 40,055 |
| LDA | Patients; Epidermal Growth Factor Receptor Measurement; Epidermal Growth Factor Receptor; EGFR protein, human; Combined; Therapeutic procedure; Tryptophanase; Non-Small Cell Lung Carcinoma; EGFR gene; combination of objects. |
| PLSA | Epidermal Growth Factor Receptor Measurement; EGFR protein, human; Epidermal Growth Factor Receptor; EGFR gene; Non-Small Cell Lung Carcinoma; Patients; Tryptophanase; Therapeutic procedure; Non-Small Cell Lung Cancer Pathway; NCI CTEP SDC Non-Small Cell Lung Cancer Sub-Category Terminology. |
| LDA | Induce (action); Expression (foundation metadata concept); Expression procedure; Effect; Homo sapiens; Apoptosis; Tryptophanase; TP53 wt Allele; TP53 gene; Inhibition. |
| PLSA | Cell Count; Induce (action); Expression procedure; Expression (foundation metadata concept); Tryptophanase; Carcinoma of lung; Apoptosis; Malignant neoplasm of lung; Effect; TP53 gene. |
| LDA | Tryptophanase; Neoplasms; Treating; Therapeutic procedure; Patients; PSA Level Less than Two; 2+ Score, WHO; 2+ Score; therapeutic aspects; Administration procedure. |
| PLSA | Apoptosis; Induce (action); Tryptophanase; Cell Count; Expression procedure; Effect; Expression (foundation metadata concept); Therapeutic procedure; Treating; Increase. |
| LDA | Senility; Old age; Induce (action); Cellular Senescence; Tryptophanase; Fibroblasts; Cell Count; Homo sapiens; Associated with; Increase. |
| PLSA | Old age; Senility; Induce (action); Cellular Senescence; Tryptophanase; Cell Count; Expression procedure; Expression (foundation metadata concept); Fibroblasts; Homo sapiens. |
| LDA. | Vascular Endothelial Growth Factors; Recombinant Vascular Endothelial Growth Factor; Neoplasms; Tryptophanase; Angiogenic Process; Laboratory mice; Social group; Group Object; Tumor Mass; Population Group. |
| PLSA | Vascular Endothelial Growth Factors; Recombinant Vascular Endothelial Growth Factor; Tumor Angiogenesis; Angiogenic Process; Tryptophanase; Expression procedure; Expression (foundation metadata concept); Neoplasms; Patients; P prime. |
| LDA | Cell Count; Neoplasm Metastasis; Secondary Neoplasm; Tryptophanase; Metastatic to; Metastatic Neoplasm; metastatic qualifier; Metastatic Disease Clinical Trial Setting; Neoplasms; Metastasis. |
| PLSA | Patients; Neoplasm Metastasis; Secondary Neoplasm; Tryptophanase; Metastatic Neoplasm; Neoplasms; Metastasis; P Blood group antibodies; P prime; Expression procedure. |
| LDA | Mutation; Present; Tryptophanase; adduct; 1+ Score, WHO; 1+ Score; Greater than one; Carcinoma of lung; Mutation Abnormality; Malignant neoplasm of lung. |
| PLSA | Mutation; Patients; Tryptophanase; Mutation Abnormality; P Blood group antibodies; P prime; Induce (action); Exposure to; Present; EGFR protein, human. |
| LDA | Lipopolysaccharides; Tumor Necrosis Factor-alpha; Induce (action); TNF protein, human; Increase; Alpha tumor necrosis factor measurement; cytokine; Interleukin-1 beta; Protons; Hepatic Involvement. |
| PLSA | Population Group; Groups; Social group; User Group; Stage Grouping; Group Object; Tryptophanase; Induce (action); Increase; House mice. |
| LDA | Aerobic glycolysis; Inhibition; Tryptophanase; Glycolysis; Induce (action); Metabolic Process, Cellular; Glucose; Expression (foundation metadata concept); Increase; Mitochondria. |
| PLSA | Aerobic glycolysis; Glycolysis; Tryptophanase; Expression procedure; Expression (foundation metadata concept); Cell Count; Increase; production; Malignant Neoplasms; Induce (action). |
| LDA | Blood group antibody I; Iodides; Tryptophanase; Neoplasms; Tumor Mass; Specimen Source Codes—tumor; Neoplasm Metastasis; Induce (action); Vaccination; T-Lymphocyte. |
| PLSA | Tryptophanase; Patients; House mice; Laboratory mice; T-Lymphocyte; SNAP25 wt Allele; SNAP25 protein, human; HERPUD1 gene; HERPUD1 wt Allele; Negation. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batbaatar, E.; Pham, V.-H.; Ryu, K.H. Multi-Task Topic Analysis Framework for Hallmarks of Cancer with Weak Supervision. Appl. Sci. 2020, 10, 834. https://doi.org/10.3390/app10030834
Batbaatar E, Pham V-H, Ryu KH. Multi-Task Topic Analysis Framework for Hallmarks of Cancer with Weak Supervision. Applied Sciences. 2020; 10(3):834. https://doi.org/10.3390/app10030834
Chicago/Turabian StyleBatbaatar, Erdenebileg, Van-Huy Pham, and Keun Ho Ryu. 2020. "Multi-Task Topic Analysis Framework for Hallmarks of Cancer with Weak Supervision" Applied Sciences 10, no. 3: 834. https://doi.org/10.3390/app10030834