Excessive Substitution of Fish Meal with Fermented Soybean Meal Induces Oxidative Stress by Impairing Glutathione Metabolism in Largemouth Bass (Micropterus salmoides)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Diets
2.3. Animal Feeding Procedure and Sampling
2.4. Biochemical and Histological Analysis
2.5. Free Amino Acid Analyses in Serum and Muscle
2.6. RNA Extraction, Reverse Transcription, and Real-Time Polymerase Chain Reaction (qPCR)
2.7. Transcriptomic Analysis in the Liver
2.8. Untargeted Metabolomics in the Liver
2.9. Integrated Analysis of Transcriptomics and Metabolomics in the Liver
2.10. Statistical Analysis
3. Results
3.1. FSBM Substitution Reduced the Growth Performance of Juvenile Largemouth Bass
3.2. FSBM Substitution Inhibited Amino Acid Transporter Expression and Decreased Total Essential Amino Acid Contents in Muscle
3.3. FSBM Substitution Caused Liver Damage by Inducing Oxidative Stress
3.4. Transcriptome Analysis in the Liver Indicated That FSBM Substitution Affected Glycine, Serine and Threonine Metabolism and Glutathione Metabolism
3.5. FSBM Substitution Changed the Metabolome Profile in the Liver
3.6. Integrated Analysis of Transcriptomics and Metabolomics Data
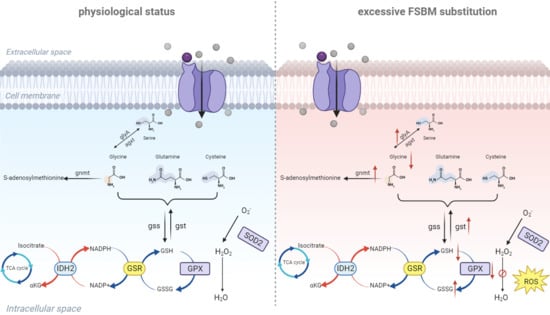
4. Discussion
5. Study Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, J.; Mai, K.; Ai, Q. Conventional Soybean Meal as Fishmeal Alternative in Diets of Japanese Seabass (Lateolabrax japonicus): Effects of Functional Additives on Growth, Immunity, Antioxidant Capacity and Disease Resistance. Antioxidants 2022, 11, 951. [Google Scholar] [CrossRef] [PubMed]
- Egounlety, M.; Aworh, O. Effect of soaking, dehulling, cooking and fermentation with Rhizopus oligosporus on the oligosaccharides, trypsin inhibitor, phytic acid and tannins of soybean (Glycine max Merr.), cowpea (Vigna unguiculata L. Walp) and groundbean (Macrotyloma geocarpa Harms). J. Food Eng. 2003, 56, 249–254. [Google Scholar]
- Muniyappan, M.; Shanmugam, S.; Park, J.H.; Han, K.; Kim, I.H. Effects of fermented soybean meal supplementation on the growth performance and apparent total tract digestibility by modulating the gut microbiome of weaned piglets. Sci. Rep. 2023, 13, 3691. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-H.; Chen, Y.-T. Lactobacillus spp. fermented soybean meal partially substitution to fish meal enhances innate immune responses and nutrient digestibility of white shrimp (Litopenaeus vannamei) fed diet with low fish meal. Aquaculture 2022, 548, 737634. [Google Scholar] [CrossRef]
- Zhang, Q.; Guo, M.; Li, F.; Qin, M.; Yang, Q.; Yu, H.; Xu, J.; Liu, Y.; Tong, T. Evaluation of Fermented Soybean Meal to Replace a Portion Fish Meal on Growth Performance, Antioxidant Capacity, Immunity, and mTOR Signaling Pathway of Coho Salmon (Oncorhynchus kisutch). Aquacult. Nutr. 2023, 2023, 2558173. [Google Scholar] [CrossRef] [PubMed]
- Novriadi, R.; Rhodes, M.; Powell, M.; Hanson, T.; Davis, D. Effects of soybean meal replacement with fermented soybean meal on growth, serum biochemistry and morphological condition of liver and distal intestine of Florida pompano Trachinotus carolinus. Aquacult. Nutr. 2018, 24, 1066–1075. [Google Scholar] [CrossRef]
- Weng, L.; Wang, Z.; Zhuang, W.; Yang, T.; Xu, X.; Liu, J.; Liu, J.; Xu, Z.; Chen, R.; Wang, Q. Effect of Replacing Fish Meal Using Fermented Soybean Meal on Growth Performance, Intestine Bacterial Diversity, and Key Gene Expression of Largemouth Bass (Micropterus salmoides). Fermentation 2023, 9, 520. [Google Scholar] [CrossRef]
- Zhang, W.; Tan, B.; Deng, J.; Dong, X.; Yang, Q.; Chi, S.; Liu, H.; Zhang, S.; Xie, S.; Zhang, H. Effects of high level of fermented soybean meal substitution for fish meal on the growth, enzyme activity, intestinal structure protein and immune-related gene expression and intestinal flora in juvenile pearl gentian grouper. Aquacult. Nutr. 2021, 27, 1433–1447. [Google Scholar] [CrossRef]
- Yamamoto, T.; Matsunari, H.; Sugita, T.; Furuita, H.; Masumoto, T.; Iwashita, Y.; Amano, S.; Suzuki, N. Optimization of the supplemental essential amino acids to a fish meal-free diet based on fermented soybean meal for rainbow trout Oncorhynchus mykiss. Fish. Sci. 2012, 78, 359–366. [Google Scholar] [CrossRef]
- Yuan, Y.-c.; Gong, S.-y.; Yang, H.-j.; Lin, Y.-c.; Yu, D.-h.; Luo, Z. Effects of supplementation of crystalline or coated lysine and/or methionine on growth performance and feed utilization of the Chinese sucker, Myxocyprinus asiaticus. Aquaculture 2011, 316, 31–36. [Google Scholar] [CrossRef]
- Wu, G. Functional Amino Acids in Nutrition and Health; Springer: Berlin/Heidelberg, Germany, 2013; Volume 45, pp. 407–411. [Google Scholar]
- Kelly, B.; Pearce, E.L. Amino assets: How amino acids support immunity. Cell Metab. 2020, 32, 154–175. [Google Scholar] [CrossRef] [PubMed]
- Mo, A.J.; Sun, J.X.; Wang, Y.H.; Yang, K.; Yang, H.S.; Yuan, Y.C. Apparent digestibility of protein, energy and amino acids in nine protein sources at two content levels for mandarin fish, Siniperca chuatsi. Aquaculture 2019, 499, 42–50. [Google Scholar] [CrossRef]
- Lin, C.; Han, G.; Ning, H.; Song, J.; Ran, N.; Yi, X.; Seow, Y.; Yin, H. Glycine enhances satellite cell proliferation, cell transplantation, and oligonucleotide efficacy in dystrophic muscle. Mol. Ther. 2020, 28, 1339–1358. [Google Scholar] [CrossRef] [PubMed]
- Rom, O.; Liu, Y.; Liu, Z.; Zhao, Y.; Wu, J.; Ghrayeb, A.; Villacorta, L.; Fan, Y.; Chang, L.; Wang, L. Glycine-based treatment ameliorates NAFLD by modulating fatty acid oxidation, glutathione synthesis, and the gut microbiome. Sci. Transl. Med. 2020, 12, eaaz2841. [Google Scholar] [CrossRef]
- Asantewaa, G.; Harris, I.S. Glutathione and its precursors in cancer. Curr. Opin. Biotechnol. 2021, 68, 292–299. [Google Scholar] [CrossRef]
- Kumar, P.; Liu, C.; Hsu, J.W.; Chacko, S.; Minard, C.; Jahoor, F.; Sekhar, R.V. Glycine and N-acetylcysteine (GlyNAC) supplementation in older adults improves glutathione deficiency, oxidative stress, mitochondrial dysfunction, inflammation, insulin resistance, endothelial dysfunction, genotoxicity, muscle strength, and cognition: Results of a pilot clinical trial. Clin. Transl. Med. 2021, 11, e372. [Google Scholar]
- Hoseini, S.M.; Majidiyan, N.; Mirghaed, A.T.; Hoseinifar, S.H.; Van Doan, H. Dietary glycine supplementation alleviates transportation-induced stress in common carp, Cyprinus carpio. Aquaculture 2022, 551, 737959. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, J.; Zhou, H.; Zhang, Y.; Huang, H.; Cao, Y.; Zhang, W.; Deng, J.; Tan, B. Feeding juvenile largemouth bass (Micropterus salmoides) with carboxymethyl cellulose with different viscous: Impacts on nutrient digestibility, growth, and hepatic and gut morphology. Front. Mar. 2022, 9, 1023872. [Google Scholar] [CrossRef]
- Yang, H.; Bian, Y.; Huang, L.; Lan, Q.; Ma, L.; Li, X.; Leng, X. Effects of replacing fish meal with fermented soybean meal on the growth performance, intestinal microbiota, morphology and disease resistance of largemouth bass (Micropterus salmoides). Aquacult. Rep. 2022, 22, 100954. [Google Scholar] [CrossRef]
- He, M.; Li, X.; Poolsawat, L.; Guo, Z.; Yao, W.; Zhang, C.; Leng, X. Effects of fish meal replaced by fermented soybean meal on growth performance, intestinal histology and microbiota of largemouth bass (Micropterus salmoides). Aquacult. Nutr. 2020, 26, 1058–1071. [Google Scholar] [CrossRef]
- Dai, C.; Ma, H.; He, R.; Huang, L.; Zhu, S.; Ding, Q.; Luo, L. Improvement of nutritional value and bioactivity of soybean meal by solid-state fermentation with Bacillus subtilis. LWT 2017, 86, 1–7. [Google Scholar] [CrossRef]
- Huang, D.; Wu, Y.; Lin, Y.; Chen, J.; Karrow, N.; Ren, X.; Wang, Y. Dietary protein and lipid requirements for juvenile largemouth bass, Micropterus salmoides. J. World Aquac. Soc. 2017, 48, 782–790. [Google Scholar] [CrossRef]
- Dairiki, J.K.; Dias, C.T.d.S.; Cyrino, J.E.P. Lysine requirements of largemouth bass, Micropterus salmoides: A comparison of methods of analysis of dose-response trials data. J. Appl. Aquacult. 2007, 19, 1–27. [Google Scholar] [CrossRef]
- Chen, N.; Ma, J.; Zhou, H.; Zhou, J.; Qiu, X.; Jin, L.; Lin, L. Assessment of dietary methionine requirement in largemouth bass, Micropterus salmoides. J. Fish. China 2010, 34, 1244–1253. [Google Scholar] [CrossRef]
- Li, X.; Zheng, S.; Ma, X.; Cheng, K.; Wu, G. Effects of dietary protein and lipid levels on the growth performance, feed utilization, and liver histology of largemouth bass (Micropterus salmoides). Amino Acids 2020, 52, 1043–1061. [Google Scholar] [CrossRef] [PubMed]
- AOAC. Official Methods of Analysis of the Association of Analytical Chemist, 15th ed.; AOAC International: Washington, DC, USA, 1995. [Google Scholar]
- Chen, Q.; Cui, K.; Zhao, Z.; Xu, X.; Liu, Y.; Shen, Y.; Chen, F.; Mai, K.; Ai, Q. LPS stimulation stabilizes HIF-1α by enhancing HIF-1α acetylation via the PARP1-SIRT1 and ACLY-Tip60 pathways in macrophages. FASEB J. 2022, 36, e22418. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sun, Y.; Yao, C.; Zhu, Y.; Wang, Y.; Zhang, Z. Metabolism response of fasting in Octopus sinensis paralarvae revealed by RNA-seq. Aquaculture 2022, 550, 737859. [Google Scholar] [CrossRef]
- Li, D.-L.; Zhu, R.; Yang, Z.-Y.; Li, L.; Qu, Z.-H.; Quan, Y.-N.; Wei, X.-F.; Shang, G.-J.; Wang, H.-T.; Wu, L.-F. Potential protective effects of sodium butyrate on glycinin-induced oxidative stress, inflammatory response and growth inhibition in Cyprinus carpio. Fish Physiol. Biochem. 2023. [Google Scholar] [CrossRef]
- Yang, H.; Qu, Y.; Li, J.; Liu, X.; Wu, R.; Wu, J. Improvement of the protein quality and degradation of allergens in soybean meal by combination fermentation and enzymatic hyd1rolysis. LWT 2020, 128, 109442. [Google Scholar] [CrossRef]
- da Silva Crozatti, T.T.; Miyoshi, J.H.; Tonin, A.P.P.; Tomazini, L.F.; Oliveira, M.A.S.; Maluf, J.U.; Meurer, E.C.; Matioli, G. Obtaining of bioactive di-and tripeptides from enzymatic hydrolysis of soybean meal and its protein isolate using Alcalase® and Neutrase®. Int. J. Food Sci. Technol. 2023, 58, 1586–1596. [Google Scholar] [CrossRef]
- Tibaldi, E.; Hakim, Y.; Uni, Z.; Tulli, F.; de Francesco, M.; Luzzana, U.; Harpaz, S. Effects of the partial substitution of dietary fish meal by differently processed soybean meals on growth performance, nutrient digestibility and activity of intestinal brush border enzymes in the European sea bass (Dicentrarchus labrax). Aquaculture 2006, 261, 182–193. [Google Scholar] [CrossRef]
- Dan, Z.; Zhang, W.; Zheng, J.; Gong, Y.; Cui, K.; Mai, K.; Ai, Q. Effects of fishmeal substitution by four fermented soybean meals on growth, antioxidant capacity and inflammation responses of turbot juveniles (Scophthalmus maximus L.). Aquaculture 2022, 560, 738414. [Google Scholar] [CrossRef]
- Cheng, A.C.; Yeh, S.P.; Hu, S.Y.; Lin, H.L.; Liu, C.H. Intestinal microbiota of white shrimp, Litopenaeus vannamei, fed diets containing Bacillus subtilis E20-fermented soybean meal (FSBM) or an antimicrobial peptide derived from B. subtilis E20-FSBM. Aquacult. Res. 2020, 51, 41–50. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, M.; Wang, L.; Lu, K.; Song, K.; Zhang, C. Bacillus subtilis LCBS1 supplementation and replacement of fish meal with fermented soybean meal in bullfrog (Lithobates catesbeianus) diets: Effects on growth performance, feed digestibility and gut health. Aquaculture 2021, 545, 737217. [Google Scholar] [CrossRef]
- Xu, C.; Liu, W.; Zhang, D.; Liu, J.; Zheng, X.; Zhang, C.; Yao, J.; Zhu, C.; Chi, C. Effects of partial fish meal replacement with two fermented soybean meals on the growth of and protein metabolism in the Chinese mitten crab (Eriocheir sinensis). Aquacult. Rep. 2020, 17, 100328. [Google Scholar] [CrossRef]
- He, M.; Yu, Y.; Li, X.; Poolsawat, L.; Yang, P.; Bian, Y.; Guo, Z.; Leng, X. An evaluation of replacing fish meal with fermented soybean meal in the diets of largemouth bass (Micropterus salmoides): Growth, nutrition utilization and intestinal histology. Aquacult. Res. 2020, 51, 4302–4314. [Google Scholar] [CrossRef]
- Prasad, P.D.; Wang, H.; Huang, W.; Kekuda, R.; Rajan, D.P.; Leibach, F.H.; Ganapathy, V. Human LAT1, a subunit of system L amino acid transporter: Molecular cloning and transport function. Biochem. Biophys. Res. Commun. 1999, 255, 283–288. [Google Scholar] [CrossRef]
- Torrents, D.; Estévez, R.; Pineda, M.; Fernández, E.; Lloberas, J.; Shi, Y.B.; Zorzano, A.; Palacín, M. Identification and characterization of a membrane protein (y+L amino acid transporter-1) that associates with 4F2hc to encode the amino acid transport activity y+L. A candidate gene for lysinuric protein intolerance. J. Biol. Chem. 1998, 273, 32437–32445. [Google Scholar] [CrossRef]
- Drummond, M.J.; Glynn, E.L.; Fry, C.S.; Timmerman, K.L.; Volpi, E.; Rasmussen, B.B. An increase in essential amino acid availability upregulates amino acid transporter expression in human skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E1011–E1018. [Google Scholar] [CrossRef]
- Wang, Z.; Tao, J.; Xie, R.; Zhang, Y.; Zhang, H.; Chen, N.; Li, S. Effects of Fish Meal Replacement with Composite Mixture of Soybean Protein Hydrolysates and Other Plant Proteins on Growth Performance, Antioxidant Capacity, and Target of Rapamycin Pathway in Largemouth Bass. N. Am. J. Aquacult. 2023, 85, 178–187. [Google Scholar] [CrossRef]
- Sheng, Z.; Xu, J.; Zhang, Y.; Wang, Z.; Chen, N.; Li, S. Dietary protein hydrolysate effects on growth, digestive enzymes activity, and expression of genes related to amino acid transport and metabolism of larval snakehead (Channa argus). Aquaculture 2023, 563, 738896. [Google Scholar] [CrossRef]
- Rubio-Aliaga, I.; Boll, M.; Daniel, H. Cloning and characterization of the gene encoding the mouse peptide transporter PEPT2. Biochem. Biophys. Res. Commun. 2000, 276, 734–741. [Google Scholar] [CrossRef]
- Ahmed, M.; Liang, H.; Chisomo Kasiya, H.; Ji, K.; Ge, X.; Ren, M.; Liu, B.; Zhu, X.; Sun, A. Complete replacement of fish meal by plant protein ingredients with dietary essential amino acids supplementation for juvenile blunt snout bream (Megalobrama amblycephala). Aquacult. Nutr. 2019, 25, 205–214. [Google Scholar] [CrossRef]
- Ding, Z.; Zhang, Y.; Ye, J.; Du, Z.; Kong, Y. An evaluation of replacing fish meal with fermented soybean meal in the diet of Macrobrachium nipponense: Growth, nonspecific immunity, and resistance to Aeromonas hydrophila. Fish Shellfish Immunol. 2015, 44, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Hu, L.; Dong, Y.; Wu, X.; Qin, Y.; Zheng, Y.; Shi, D.; Xue, M. Substitution of fish meal by fermented soybean meal affects the growth performance and flesh quality of Japanese seabass (Lateolabrax japonicus). Anim. Feed Sci. Technol. 2017, 229, 1–12. [Google Scholar] [CrossRef]
- Kaspar, J.W.; Niture, S.K.; Jaiswal, A.K. Nrf2: INrf2 (Keap1) signaling in oxidative stress. Free Radical Biol. Med. 2009, 47, 1304–1309. [Google Scholar] [CrossRef]
- Bell, E.L.; Guarente, L. The SirT3 divining rod points to oxidative stress. Mol. Cell 2011, 42, 561–568. [Google Scholar] [CrossRef]
- Wang, W.; Wu, Z.; Dai, Z.; Yang, Y.; Wang, J.; Wu, G. Glycine metabolism in animals and humans: Implications for nutrition and health. Amino Acids 2013, 45, 463–477. [Google Scholar] [CrossRef]
- Lu, S.C. Glutathione synthesis. Biochim. Biophys. Acta BBA-Gen. Subj. 2013, 1830, 3143–3153. [Google Scholar] [CrossRef]
- Adjoumani, J.-J.Y.; Wang, K.; Zhou, M.; Liu, W.; Zhang, D. Effect of dietary betaine on growth performance, antioxidant capacity and lipid metabolism in blunt snout bream fed a high-fat diet. Fish Physiol. Biochem. 2017, 43, 1733–1745. [Google Scholar] [CrossRef]
- Aljobaily, N.; Viereckl, M.J.; Hydock, D.S.; Aljobaily, H.; Wu, T.-Y.; Busekrus, R.; Jones, B.; Alberson, J.; Han, Y. Creatine alleviates doxorubicin-induced liver damage by inhibiting liver fibrosis, inflammation, oxidative stress, and cellular senescence. Nutrients 2020, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Zitka, O.; Skalickova, S.; Gumulec, J.; Masarik, M.; Adam, V.; Hubalek, J.; Trnkova, L.; Kruseova, J.; Eckschlager, T.; Kizek, R. Redox status expressed as GSH: GSSG ratio as a marker for oxidative stress in paediatric tumour patients. Oncol. Lett. 2012, 4, 1247–1253. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Jadhao, S.; Chandan, N.; Kumar, K.; Jha, A.; Bhushan, S.; Kumar, S.; Rana, R. Dietary choline, betaine and lecithin mitigates endosulfan-induced stress in Labeo rohita fingerlings. Fish Physiol. Biochem. 2012, 38, 989–1000. [Google Scholar] [CrossRef] [PubMed]






| Ingredients | Diets a | |
|---|---|---|
| CON | FSBM | |
| Fish meal | 62 | 31 |
| Fermented soybean meal b | 0 | 40.25 |
| Alpha-starch | 10 | 10 |
| Microcrystalline cellulose | 12.51 | 3.03 |
| Fish oil | 3 | 4 |
| Soybean oil | 4 | 3.23 |
| Soybean lecithin | 1 | 1 |
| Choline | 0.2 | 0.2 |
| Monocalcium phosphate | 1 | 1 |
| Sodium alginate | 1 | 1 |
| Vitamin C | 1 | 1 |
| Mineral mix c | 3 | 3 |
| Vitamin premix d | 0.05 | 0.05 |
| Mold inhibitor e | 0.2 | 0.2 |
| Ethoxyquin | 0.01 | 0.01 |
| L-lysine | 0 | 0.58 |
| L-methionine | 0 | 0.45 |
| Alanine | 1.03 | 0 |
| Total | 100 | 100 |
| Proximate analysis | ||
| Crude protein | 46.74 | 48.29 |
| Crude lipid | 11.9 | 11.67 |
| Amino Acid | Diets | |
|---|---|---|
| CON | FSBM | |
| Essential amino acids (EAAs) | ||
| Arginine | 2.98 | 2.89 |
| Histidine | 0.98 | 1.03 |
| Isoleucine | 2.04 | 1.92 |
| Lysine | 3.39 | 2.95 |
| Methionine | 1.26 | 1.1 |
| Phenylalanine | 1.90 | 2.03 |
| Threonine | 2.10 | 1.76 |
| Valine | 2.17 | 1.95 |
| Leucine | 3.58 | 3.23 |
| Non-essential amino acids (EAAs) | ||
| Alanine | 2.98 | 3.12 |
| Aspartate | 4.50 | 4.66 |
| Cysteine | 0.17 | 0.26 |
| Glycine | 3.06 | 2.37 |
| Glutamate | 6.52 | 7.76 |
| Proline | 2.38 | 2.38 |
| Serine | 2.22 | 2.09 |
| Tyrosine | 1.29 | 1.39 |
| Total | 43.53 | 42.88 |
| Gene Name | KEGG Annotation | Log2FC | Regulated | KEGG Pathway |
|---|---|---|---|---|
| cat | K03781 | −1.38 | down | Peroxisome |
| nqo1 | K00355 | 1.36 | up | Ubiquinone and other terpenoid quinones Biosynthesis |
| mpo | K10789 | −2.00 | down | Phagosome |
| gpx1b | K00432 | −1.68 | down | Glutathione metabolism |
| ptgs2b | K11987 | −2.15 | down | Arachidonic acid metabolism |
| trx | K03671 | −1.58 | down | NOD-like receptor signaling pathway |
| ptgs1 | K00509 | 1.67 | up | Arachidonic acid metabolism |
| keap1a | K10456 | 1.31 | up | Ubiquitin-mediated proteolysis |
| sod1 | K04565 | −1.49 | down | Peroxisome |
| gstt2 | K00799 | 1.32 | up | Glutathione metabolism |
| KEGG Annotation | Gene Name | Log2FC | Regulated |
|---|---|---|---|
| Glutathione metabolism | ggt5b | −1.59 | down |
| tm4sf4 | −1.53 | down | |
| gsta.1 | 1.41 | up | |
| gstt3 | 1.84 | up | |
| oplah | 2.01 | up | |
| gsto1 | −1.40 | down | |
| gpx3 | 2.01 | up | |
| odc1 | −2.23 | down | |
| idh2-like | −2.26 | down | |
| rrm1 | −1.08 | down | |
| gstt3-like | 1.95 | up | |
| gpx1b | −1.68 | down | |
| idh2 | −1.54 | down | |
| gstt2 | 1.32 | up |
| KEGG Annotation | Metabolite Name | Log2FC | Regulated |
|---|---|---|---|
| Glycine, serine and threonine metabolism | D-Glycerate 2-phosphate | 4.24 | Up |
| Methylglyoxal | −3.11 | Down | |
| Creatine | −0.89 | Down | |
| Betaine | −0.36 | Down | |
| 3-Phospho-D-glycerate | −0.61 | Down | |
| 5-Hydroxyectoine | −0.60 | Down | |
| L-2,4-Diaminobutanoate | −0.57 | Down | |
| Tetrahydrofolate Choline | 0.94 0.65 | Up Up |
| KEGG Annotation | Metabolite Name | Log2FC | Regulated |
|---|---|---|---|
| Glutathione metabolism | Ascorbic acid | −0.31 | Down |
| L-glutamate | −0.50 | Down | |
| Glutathione | −2.61 | Down | |
| Trypanothione | 1.23 | Up | |
| Glutathione, oxidized | 1.04 | Up | |
| Trypanothione disulfide | 4.26 | Up | |
| Glutathionylspermine | 0.36 | Up |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.; Wang, C.; Sun, Y.; Chen, Y.; Chen, S.; Han, T.; Wang, J. Excessive Substitution of Fish Meal with Fermented Soybean Meal Induces Oxidative Stress by Impairing Glutathione Metabolism in Largemouth Bass (Micropterus salmoides). Antioxidants 2023, 12, 2096. https://doi.org/10.3390/antiox12122096
Chen Q, Wang C, Sun Y, Chen Y, Chen S, Han T, Wang J. Excessive Substitution of Fish Meal with Fermented Soybean Meal Induces Oxidative Stress by Impairing Glutathione Metabolism in Largemouth Bass (Micropterus salmoides). Antioxidants. 2023; 12(12):2096. https://doi.org/10.3390/antiox12122096
Chicago/Turabian StyleChen, Qiang, Congcong Wang, Yulong Sun, Yan Chen, Songming Chen, Tao Han, and Jiteng Wang. 2023. "Excessive Substitution of Fish Meal with Fermented Soybean Meal Induces Oxidative Stress by Impairing Glutathione Metabolism in Largemouth Bass (Micropterus salmoides)" Antioxidants 12, no. 12: 2096. https://doi.org/10.3390/antiox12122096