Can Immobilized Artificial Membrane Chromatography Support the Characterization of Antimicrobial Peptide Origin Derivatives?
Abstract
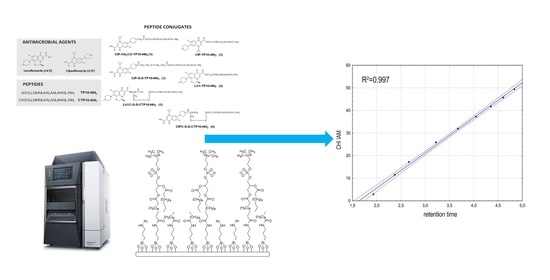
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Materials and Analytes
4.1.1. Short Cationic Lipopeptides
4.1.2. Citropin Analogs
4.1.3. Conjugates of Ciprofloxacin (CIP) and Levofloxacin (LVX) with a Cell-Penetrating Peptide
4.2. HPLC Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 2559. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hancock, R.E.; Chapple, D.S. Peptide antibiotics. Antimicrob. Agents Chemother. 1999, 43, 1317–1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colomb-Cotinat, M.; Lacoste, J.; Brun-Buisson, C.; Jarlier, V.; Coignard, B.; Vaux, S. Estimating the morbidity and mortality associated with infections due to multidrug-resistant bacteria (MDRB), France, 2012. Antimicrob. Resist. Infect. Control 2016, 5, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greber, K.E.; Zielińska, J.; Nierzwicki, Ł.; Ciura, K.; Kawczak, P.; Nowakowska, J.; Bączek, T.; Sawicki, W. Are the short cationic lipopeptides bacterial membrane disruptors? Structure-Activity Relationship and molecular dynamic evaluation. Biochim. Biophys. Acta Biomembr. 2019, 1861. [Google Scholar] [CrossRef]
- Khurshid, Z.; Naseem, M.; Sheikh, Z.; Najeeb, S.; Shahab, S.; Zafar, M.S. Oral antimicrobial peptides: Types and role in the oral cavity. Saudi Pharm. J. SPJ Off. Publ. Saudi Pharm. Soc. 2016, 24, 515–524. [Google Scholar] [CrossRef] [Green Version]
- Valko, K.; Ivanova-Berndt, G.; Beswick, P.; Kindey, M.; Ko, D. Application of biomimetic HPLC to estimate lipophilicity, protein and phospholipid binding of potential peptide therapeutics. ADMET DMPK 2018, 6, 162–175. [Google Scholar] [CrossRef] [Green Version]
- Vivès, E.; Schmidt, J.; Pèlegrin, A. Cell-penetrating and cell-targeting peptides in drug delivery. Biochim. Biophys. Acta Rev. Cancer 2008, 1786, 126–138. [Google Scholar] [CrossRef] [Green Version]
- Loureiro, D.R.P.; Soares, J.X.; Lopes, D.; Macedo, T.; Yordanova, D.; Jakobtorweihen, S.; Nunes, C.; Reis, S.; Pinto, M.M.M.; Afonso, C.M.M. Accessing lipophilicity of drugs with biomimetic models: A comparative study using liposomes and micelles. Eur. J. Pharm. Sci. 2018, 115, 369–380. [Google Scholar] [CrossRef]
- Grumetto, L.; Russo, G.; Barbato, F. Relationships between human intestinal absorption and polar interactions drug/phospholipids estimated by IAM-HPLC. Int. J. Pharm. 2015, 489, 186–194. [Google Scholar] [CrossRef]
- Russo, G.; Grumetto, L.; Barbato, F.; Vistoli, G.; Pedretti, A. Prediction and mechanism elucidation of analyte retention on phospholipid stationary phases (IAM-HPLC) by in silico calculated physico-chemical descriptors. Eur. J. Pharm. Sci. 2017, 99, 173–184. [Google Scholar] [CrossRef]
- Grumetto, L.; Russo, G.; Barbato, F. Immobilized Artificial Membrane HPLC Derived Parameters vs PAMPA-BBB Data in Estimating in Situ Measured Blood-Brain Barrier Permeation of Drugs. Mol. Pharm. 2016, 13, 2808–2816. [Google Scholar] [CrossRef] [PubMed]
- Valko, K.; Du, C.M.; Bevan, C.D.; Reynolds, D.P.; Abraham, M.H. Rapid-gradient HPLC method for measuring drug interactions with immobilized artificial membrane: Comparison with other lipophilicity measures. J. Pharm. Sci. 2000, 89, 1085–1096. [Google Scholar] [CrossRef]
- Valkó, K.L. Lipophilicity and biomimetic properties measured by HPLC to support drug discovery. J. Pharm. Biomed. Anal. 2016, 130, 35–54. [Google Scholar] [CrossRef] [PubMed]
- Valko, K.L. Chromatographic technique to support ADMET&DMPK in early drug discovery. ADMET DMPK 2018, 6, 71. [Google Scholar] [CrossRef] [Green Version]
- Teague, S.; Valko, K. How to identify and eliminate compounds with a risk of high clinical dose during the early phase of lead optimisation in drug discovery. Eur. J. Pharm. Sci. 2017, 110, 37–50. [Google Scholar] [CrossRef]
- Ciura, K.; Pastewska, M.; Ulenberg, S.; Kapica, H.; Kawczak, P.; Bączek, T. Chemometric analysis of bio-inspired micellar electrokinetic chromatographic systems – modelling of retention mechanism and prediction of biological properties using bile salts surfactants. Microchem. J. 2021, 167. [Google Scholar] [CrossRef]
- Ciura, K.; Fedorowicz, J.; Sączewski, J.; Kapica, H.; Pastewska, M.; Sawicki, W. Interaction between antifungal isoxazolo[3,4-b]pyridin 3(1h)-one derivatives and human serum proteins analyzed with biomimetic chromatography and qsar approach. Processes 2021, 9, 512. [Google Scholar] [CrossRef]
- Ciura, K.; Kapica, H.; Dziomba, S.; Kawczak, P.; Belka, M.; Bączek, T. Biopartitioning micellar electrokinetic chromatography – Concept study of cationic analytes. Microchem. J. 2020, 154, 104518. [Google Scholar] [CrossRef]
- Ciura, K.; Fedorowicz, J.; Kapica, H.; Adamkowska, A.; Sawicki, W.; Saczewski, J. Affinity of fluoroquinolone-safirinium dye hybrids to phospholipids estimated by IAM-HPLC. Processes 2020, 8, 1148. [Google Scholar] [CrossRef]
- Ciura, K.; Fedorowicz, J.; Žuvela, P.; Lovrić, M.; Kapica, H.; Baranowski, P.; Sawicki, W.; Wong, M.W.; Sączewski, J. Affinity of antifungal isoxazolo[3,4-b]pyridine-3(1H)-ones to phospholipids in immobilized artificial membrane (IAM) chromatography. Molecules 2020, 25, 4835. [Google Scholar] [CrossRef] [PubMed]
- Ciura, K.; Ulenberg, S.; Kapica, H.; Kawczak, P.; Belka, M.; Bączek, T. Drug affinity to human serum albumin prediction by retention of cetyltrimethylammonium bromide pseudostationary phase in micellar electrokinetic chromatography and chemically advanced template search descriptors. J. Pharm. Biomed. Anal. 2020, 188, 113423. [Google Scholar] [CrossRef] [PubMed]
- Ciura, K.; Ulenberg, S.; Kapica, H.; Kawczak, P.; Belka, M.; Bączek, T. Assessment of blood–brain barrier permeability using micellar electrokinetic chromatography and P_VSA-like descriptors. Microchem. J. 2020, 158, 105236. [Google Scholar] [CrossRef]
- Pastewska, M.; Bednarczyk-Cwynar, B.; Kovačević, S.; Buławska, N.; Ulenberg, S.; Georgiev, P.; Kapica, H.; Kawczak, P.; Bączek, T.; Sawicki, W.; et al. Multivariate assessment of anticancer oleanane triterpenoids lipophilicity. J. Chromatogr. A 2021, 1656, 462552. [Google Scholar] [CrossRef] [PubMed]
- Greber, K.E.; Ciura, K.; Belka, M.; Kawczak, P.; Nowakowska, J.; Bączek, T.; Sawicki, W. Characterization of antimicrobial and hemolytic properties of short synthetic cationic lipopeptides based on QSAR/QSTR approach. Amino Acids 2018, 50, 479–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ptaszyńska, N.; Gucwa, K.; Olkiewicz, K.; Heldt, M.; Serocki, M.; Stupak, A.; Martynow, D.; Dębowski, D.; Gitlin-Domagalska, A.; Lica, J.; et al. Conjugates of Ciprofloxacin and Levofloxacin with Cell-Penetrating Peptide Exhibit Antifungal Activity and Mammalian Cytotoxicity. Int. J. Mol. Sci. 2020, 21, 4696. [Google Scholar] [CrossRef]
- Marshall, A.J.H.; Piddock, L.J.V. Interaction of divalent cations, quinolones and bacteria. J. Antimicrob. Chemother. 1994, 34, 465–483. [Google Scholar] [CrossRef]
- Chapman, J.S.; Georgopapdakou, N.H. Routes of quinolone permeation in Escherichia coli. Antimicrob. Agents Chemother. 1988, 32, 438–442. [Google Scholar] [CrossRef] [Green Version]
- Fields, G.B.; Noble, R.L. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept. Protein Res. 1990, 35, 161–214. [Google Scholar] [CrossRef]
- Ptaszyńska, N.; Olkiewicz, K.; Okońska, J.; Gucwa, K.; Łęgowska, A.; Gitlin-Domagalska, A.; Dębowski, D.; Lica, J.; Heldt, M.; Milewski, S.; et al. Peptide conjugates of lactoferricin analogues and antimicrobials-Design, chemical synthesis, and evaluation of antimicrobial activity and mammalian cytotoxicity. Peptides 2019, 117, 170079. [Google Scholar] [CrossRef]


| Analyte | TR1 | TR2 | TR3 | Mean TR | CHIIAM |
|---|---|---|---|---|---|
| (4–16) Citropin | 3.987 | 3.979 | 3.985 | 3.984 | 38.24 |
| (8–16) Citropin | 4.482 | 4.496 | 4.506 | 4.495 | 45.95 |
| (1–7) Citropin | 4.240 | 4.244 | 4.244 | 4.243 | 42.15 |
| (4–14) Citropin | 3.305 | 3.289 | 3.293 | 3.295 | 27.85 |
| (1–7)–(10–16) Citropin | 3.888 | 3.901 | 3.934 | 3.908 | 37.09 |
| (1–5)–(12–16) Citropin | 3.349 | 3.348 | 3.351 | 3.349 | 28.66 |
| Levofloxacin | 3.087 | 3.089 | 3.101 | 3.092 | 24.88 |
| Ciprofloxacin | 2.886 | 2.921 | 2.899 | 2.902 | 19.70 |
| Double Fatty Acid Chain Lipopeptides | Tetradecanoic Fatty Acid Lipopeptides | Hexadecanoic Fatty Acid Lipopeptides |
|---|---|---|
| (C8)2-KKKK-NH2 | C14-K-NH2 | C16-K-NH2 |
| (C10)2-KKKK-NH2 | C14-KG-NH2 | C16-KGK-NH2 |
| (C12)2-KKKK-NH2 | C14-KGK-NH2 | C16-KGKG-NH2 |
| (C14)2-KKKK-NH2 | C14-KGKG-NH2 | C16-KK-NH2 |
| (C16)2-KKKK-NH2 | C14-KKK-NH2 | C16-KKKK-NH2 |
| C14-KKKK-NH2 | C16-KKY-NH2 | |
| C16-KKS-NH2 | ||
| C16-KKD-NH2 |
| Antimicrobial Peptides | Amino Acid Sequences |
|---|---|
| Citropin 1.1 | GLFDVIKKVASVIGGL-NH2 |
| (4–16) Citropin | DVIKKVASVIGGL-NH2 |
| (8–16) Citropin | KVASVIGGL-NH2 |
| (1–7) Citropin | GLFDVIK-NH2 |
| (4–14) Citropin | DVIKKVASVIG-NH2 |
| (1–7)–(10–16) Citropin | GLFDVIKASVIGGL-NH2 |
| (1–5)–(12–16) Citropin | GLFDVVIGGL-NH2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciura, K.; Ptaszyńska, N.; Kapica, H.; Pastewska, M.; Łęgowska, A.; Rolka, K.; Kamysz, W.; Sawicki, W.; Greber, K.E. Can Immobilized Artificial Membrane Chromatography Support the Characterization of Antimicrobial Peptide Origin Derivatives? Antibiotics 2021, 10, 1237. https://doi.org/10.3390/antibiotics10101237
Ciura K, Ptaszyńska N, Kapica H, Pastewska M, Łęgowska A, Rolka K, Kamysz W, Sawicki W, Greber KE. Can Immobilized Artificial Membrane Chromatography Support the Characterization of Antimicrobial Peptide Origin Derivatives? Antibiotics. 2021; 10(10):1237. https://doi.org/10.3390/antibiotics10101237
Chicago/Turabian StyleCiura, Krzesimir, Natalia Ptaszyńska, Hanna Kapica, Monika Pastewska, Anna Łęgowska, Krzysztof Rolka, Wojciech Kamysz, Wiesław Sawicki, and Katarzyna E. Greber. 2021. "Can Immobilized Artificial Membrane Chromatography Support the Characterization of Antimicrobial Peptide Origin Derivatives?" Antibiotics 10, no. 10: 1237. https://doi.org/10.3390/antibiotics10101237