1. Introduction
The lip is a unique tissue anatomically and histologically in humans for a transition zone, which is referred to as vermilion and is located between keratinized skin and non-keratinized labial oral mucosa [
1,
2]. The skin is covered by a thin, stratified squamous keratinized epithelium involved in hair follicles. The skin transitions to the vermillion zone, forming the red portion of the lip, and continues to the moist oral mucosa. The stratified epithelium continues to become thicker across the lip (from vermillion to oral mucosa) [
3]. There are no adnexal structures (hair follicles and sweat glands) in the vermilion dermis [
1]. The numerous, densely-packed dermal papillae of the lamina propria underlying the vermillion epithelium allow blood vessels close access to the surface, conferring a red color to vermillion. Minor salivary glands are located in the underlying tissue of the oral mucosa. Orbicularis oris muscle presents at the central core of the lip [
1,
4]. In addition to the distinct epithelial morphology of the human lip (skin/vermillion/oral mucosa), the histological characteristics are closely pertinent to its barrier function because the lip is located at the boundary of dry and moist conditions caused by severe environmental damages [
5]. Therefore, it is useful to produce an efficient cellular model to investigate the correlation of the characteristics of the lip epithelium with the barrier function under cellular and molecular bases [
6,
7]. Additionally, the model can be used to evaluate a route via the vermilion that can be another drug delivery system due to those unique tissue properties of the lip [
4,
8].
Our final goal is to develop a specific human lip/vermilion epithelium in vitro model to assess the efficacy and safety of various pharmaceutical products and consumer drugs for topical application to the vermilion. Due to the vermilion epithelium’s unique features, compared with skin and oral mucosa, currently available in vitro skin and oral mucosa models have limitations in use when accurately evaluating and confirming the effects of testing products on the lip/vermilion epithelium [
9]. Thus, there is a need to explore and detect single potential markers specific to the vermilion epithelium that differentiate from adjacent skin and oral mucosa. However, procuring a fresh human lip tissue sample that includes all epithelia that continuously originate from the skin, vermilion, and oral mucosa is particularly challenging due to aesthetic issues. Lack of sufficient tissue samples for research use can be attributable for the limitations in biological study of the lip.
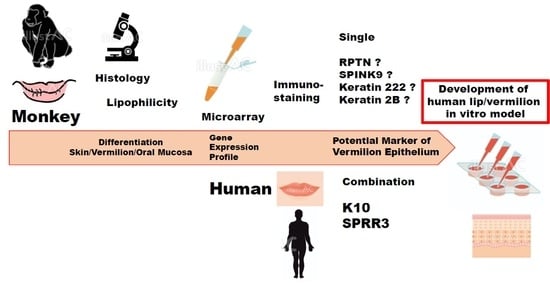
According to a prior histological study, possibly for the first time, this study showed that a Japanese macaque presented with vermilion epithelium in the lip distinct from the adjacent skin and oral mucosa, which is very similar to the characteristics of the human lip. This has allowed us to obtain a sufficient amount of the lip tissue to thoroughly investigate the vermilion epithelium. Therefore, in the present study, we aimed to perform microarray analysis for in vivo tissues as well as primary keratinocytes in a culture derived from three distinct epithelia in the lip of a Japanese macaque, and examined the gene expression profile that distinguished vermilion epithelium from adjacent skin and oral mucosa epithelia. Since gene clustering analysis determines differentially expressed genes specific to the vermilion epithelium, results should contribute to detecting a single potential marker specific to the human vermilion epithelium. Furthermore, herein, we have used immunohistochemistry to test specific protein expression detected by the gene clustering analysis and their expression patterns in human and monkey lip in vivo to support the gene expression profile of the Japanese macaque.
2. Materials and Methods
2.1. Preparation for Lip Tissue of a Japanese Macaque (Skin, Vermilion and Oral Mucosa)
The procurement of the entire lip tissue of a Japanese macaque (male, 7-year and 9-month old) complied with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. The Niigata University Institutional Animal Care and Use Committee approved the study’s experimental procedures (SA00008, 31 March 2017).
The total harvested lip tissue was dissected through the orbicularis oris muscle tissue using scissors, which developed an entire section of lip tissue that consisted of the skin, lip vermilion, and labial oral mucosa. The muscle tissue was trimmed off beneath the dermis/submucosal connective tissue. The tissue was cut into three different sections of the lip, such as skin, vermilion, and oral mucosa using a scalpel. The skin was macroscopically cut off from the vermilion by differentiating the tissues that had hair from those without hair. To differentiate the vermilion from the oral mucosa, 0.1% solution of Sudan black B (Merck, Darmstadt, Germany), dissolved in 70% ethanol (Wako chemical, Osaka, Japan) followed by filtration with a 0.2 μm membrane filter (Merck, Darmstadt, Germany), was applied to the rest of the (hairless) tissue surface using a cotton swab on which the vermilion was stained in black. This resulted in three different lip tissue sections. Each tissue was dissected into several pieces, which were approximately 5 mm2 in size, for the below-mentioned examinations.
2.2. RNA Sampling from Lip Tissue of a Japanese Macaque (Skin, Vermilion, and Oral Mucosa)
After removing the adipose tissue to the maximum extent, 1500 PU/mL of dispase II (#383-02281, Godo-shusei, Tokyo, Japan) that was dissolved in Hanks’ balanced salt solution (Wako Chemical, Osaka, Japan) was intra-cutaneously injected using a 1 mL syringe with a 30-gauge needle to develop blisters over the entire surface of the piece of tissue piece. This helped detach the epithelial layer from the underlying connective tissue. The pieces of tissue, which were approximately 5–10 mm2, were incubated with dispase II at 37 °C for 60 m (oral mucosa) or 30 m (skin and vermilion). Finally, the pieces of tissue were washed with cold defined phosphate buffered saline (D-PBS; Wako Chemical, Osaka, Japan), and forceps were used to separate the epithelial layer from the underlying tissue. The detached epithelial layer was washed with cold D-PBS and weighed.
For microarray analysis, the epithelial layer was homogenized in an appropriate volume of QIAzol (Qiagen, Valencia, CA, USA), 100 mg of the tissue per 1 mL in a 1.5 mL tube (Eppendorf, Tokyo, Japan) using a homogenizer (Thermo Fisher Scientific, Waltham, MA, USA), and the total RNA was extracted using an RNeasy mini kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s instruction.
2.3. Culturing Primary Lip Keratinocytes (Skin, Vermilion, and Labial Oral Mucosa)
Primary keratinocyte cultures from three sections of lip tissue were established using the explant culture technique. Small explants (dermal side up) were placed in a 60 mm Petri dish (Corning, Corning, NY, USA). They were incubated in a moist atmosphere at 37 °C and 5% CO2 with complete EpiLife that contained EpiLife-defined growth supplements (Thermo Fisher Scientific, Waltham, MA, USA), 0.06 mM Ca2+, gentamicin (5.0 μg/mL; Thermo Fisher Scientific, Waltham, MA, USA), and amphotericin B (0.375 μg/mL; Thermo Fisher Scientific, Waltham, MA, USA), which is a serum-free culture medium. The culture medium was refreshed with complete EpiLife medium every other day. When cell outgrowth reached 80% confluence, the p0 keratinocytes were detached with 0.025% trypsin and ethylenediaminetetraacetic acid (Thermo Fisher Scientific, Waltham, MA, USA), neutralized with defined trypsin inhibitor (Thermo Fisher Scientific, Waltham, MA, USA), and replated as p1 cells into another tissue-culture Petri dish (1.0 × 104 cells/cm2) with complete EpiLife medium. Passage 1 (p1) keratinocytes obtained from the skin, vermilion, and oral mucosa were fed with the same culture medium every other day. After a confluence of approximately 80% was reached, the total RNA was extracted from the cells using a RNeasy mini kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s instructions.
2.4. Microarray Analysis
The microarray data were deposited into the NCBI Gene Expression Omnibus Database (GSE 172126). The labeled aRNA was fragmented and hybridized to an Affymetrix GeneChip Cynomolgus + Rhesus Gene 1.0 ST array (Cat# 901941), which covers 5319 and 37,293 genes of Cynomolgus and Rhesus monkeys, respectively. The signals detected for each gene were normalized using the Robust Multi-array Average algorithm.
Microarray data were analyzed using a microarray data analysis tool (Filgen, Nagoya, Japan). To exclude data with low reliability, following normalization, the probe sets were excluded, which showed expression values below the average expression value of 2318 negative control probes. These were putative intronic-based probe sets from 100 adult putative housekeeping genes. Then, six pairwise comparisons were performed with keratinocytes in the vermilion, oral mucosa, and epidermis in vivo and in vitro. The expression values for vermilion and epidermal keratinocytes were always used as test and control samples, respectively. After the ratio of expression value had been calculated between the test and control sample, it was converted to log2 ratio, which was referred to as logFC. Then, the up-regulated and down-regulated genes were determined by the test sample’s expression values with a logFC of |≥1| compared with the control, respectively. This resulted in 2059 genes, which were filtered by applying them to at least one or more up-regulated or down-regulated genes. To generate a heat map, the logFC values of filtered genes were clustered using R software’s Euclidean distance and Ward’s method (
http://www.r-project.org/ (accessed on 29 November 2021)).
2.5. Histological and Immunohistochemical Examination of Lip Tissue from a Japanese Monkey and Human
The lip tissue of a Japanese macaque not used for cell culture was fixed with 4% paraformaldehyde in 100 mM D-PBS and embedded in paraffin for the histological examination. The paraffin-embedded samples were deparaffinized, rehydrated, cut into 5-μM thick sections, and stained with hematoxylin and eosin (H & E) for histological examination. In addition, the sections were prepared for immunohistochemical examination of keratin 10 (K10) and small proline-rich protein 3 (SPRR3). The sections were deparaffinized in xylene and rehydrated in ethanol. Endogenous peroxidase was blocked with 0.3% hydrogen peroxide in methanol for 30 m. Antigen retrieval was achieved by autoclaving with 10 mM citric sodium buffer (pH 6.0) at 100 °C for 20 m. After incubating with 5% BSA in phosphate-buffered saline (PBS) for 30 m, sections were incubated with either a mouse monoclonal antibody against K10 (ab9026) (Abcam, Cambridge, UK) at 1:200 or a mouse monoclonal antibody against SPRR3 (ab58233) (Abcam, Cambridge, UK) at of 1:100, at 4 °C overnight. After washing with TBS, the sections were reacted with EnVision FLEX Plus (Dako, Carpinteria, CA, USA) at room temperature for 1 h. The immunoreactions were then visualized with 3,3’diaminobenzidine (Dojindo Co. Ltd., Kumamoto, Japan), and the sections were counterstained with hematoxylin.
For the histological examination of the human lip (female, 53-year-old), frozen whole lip tissue was purchased and imported from Science Care (Phoenix, AZ, USA). The donor died of acute myeloblastic leukemia. The median part of the lower lip was excised and embedded in SCEM (SECTION-LAB, Hiroshima, Japan), cut into 6 μm sections, and fixed with methanol at −30 °C for 10 m. The sections were washed with D-PBS three times and stained with H & E. For immunohistochemical examination, the sections were incubated in a blocking buffer (1% new-born calf serum in D-PBS) for 60 m at room temperature and incubated with primary antibodies against K10, SPRR3, and blocking buffer overnight at 4 °C. Then, the samples were washed three times with D-PBS and incubated with secondary antibodies for 60 m at room temperature. After washing, the nuclei were stained with DAPI (1:700, D1306, Thermo Fisher Scientific, Waltham, MA, USA) for 1 h and mounted. The primary antibodies that were used and their dilution rates were: guinea pig anti-K10 (GP-K10) (1:200, Progen, Heidelberg, Germany) and rabbit anti-SPRR3 (HPA044467) (1:1000, Atlas Antibodies, Bromma, Sweden). Secondary antibodies (Alexa 488 (A-11070) and 647 (A-21450, Thermo Fisher Scientific, Waltham, MA, USA) were used at 1:1000 dilution.
4. Discussion
The vermilion epithelium’s histological characteristics are distinct from the adjacent epidermis and oral mucosa in human lips; however, our findings demonstrated that those in the monkey lip were similar to humans, despite the fact that vermilion is known to be inherent in
Homo sapiens [
1,
10]. These findings could help us understand the vermilion epithelium’s unique differentiation and keratinization. Furthermore, it could lead to the discovery of specific epithelial markers for vermilion and might facilitate the development of a human lip/vermilion model as a tool for quality assurance of therapeutic products of lips [
8].
Nonetheless, it is unclear why the Japanese macaque has vermilion similar to histological features of the human lip. A recent hypothetical study revealed that non-aggressive friction occurring in the lip may decrease the density of hair follicles in human skin, suggesting the relationship between pronunciation and the histological appearance of the vermillion [
11]. Although Chimpanzees do not speak, the lip movement associated with facial expressions and social functions, which is not as much as that in human, can produce mild mechanical stimuli to the vermillion. This could be due to the similarities of vermilion of human and monkey [
12]. According to a previous report, the porcine snout has an area with absence of adnexal structures like the human lips, which is a distinct characteristic relative to other porcine and human skin [
4]. Altogether, long-term mild friction to the perioral area may play a role, in part, in vermillion histology development.
The key issue of this study was to macroscopically determine the border between vermilion and skin/oral mucosa, which can avoid major contamination of keratinocytes derived from other different epithelial tissues. To achieve it, we devised a method to especially distinguish vermilion from the oral mucosa within monkey lip. Vermilion surface lipids originate from blood vessel penetration, not sebum secreted from sebaceous glands, which demonstrates the lipophilicity of vermilion [
13]. Sudan black B is used to stain various lipids such as phospholipids, sterols, and neutral triglycerides. Therefore, because Sudan black dye rendered the border between hairless vermilion and oral mucosa discriminable, it was successful to separate the skin, vermilion, and oral mucosa within the lip epithelium. Additionally, this technique facilitated the development of monkey primary keratinocytes in culture, possibly for the first time, despite the use of the explant culture technique, which often causes fibroblast contamination. This could be due to the primary keratinocyte culture system of serum-free and low Ca
++ (0.06 mM) conditions, identical to the human clinical application [
14]. Conversely, because this culture condition maintains cultured keratinocytes in a proliferative and undifferentiated state [
15], there were little morphological differences among the cultured keratinocytes harvested from the skin, vermilion, and oral mucosa. Further studies are required to develop an in vitro lip model that exhibits the in vivo lip keratinocytes’ profile.
Our clustering analysis demonstrated differences in the vermilion between the skin and oral mucosa within the lip of a Japanese macaque, although the statistical power was limited. First, the gene expression profile of vermilion keratinocyte in vivo was relatively similar to that of any keratinocytes in vitro. This suggests a more proliferative potential of vermilion epithelium, which is important to develop an in vitro model. Second, in vivo, the profile of skin epithelium was much different from that of the vermilion. This could be attributed to the presence of hair and terminal differentiation in the epidermis. Third, more importantly, the expression levels of genes clustered into C11 was highly up-regulated in vermilion keratinocytes in vivo. Therefore, RPTN, SPINK9, keratin222, and keratin2B were expected to be single potential markers specific to vermilion. However, our results failed to indicate that the microarray data were consistent with the immunohistochemical findings, although the increase in mRNA levels did not necessarily reflect the same in the protein level. Nonetheless, all RPTN, SPINK9, keratin222, and keratin2B have an important and unique role in desquamation process and epithelial differentiation in squamous epithelium. Therefore, these proteins associated with genes in the C11 cluster require further studies to explore the specific functions of vermilion as well as a potential marker.
Instead, we focused on K10 and SPRR3 categorized into C6 and C13, respectively, because the proteins associated with these clustered genes could be used as combination markers to distinguish vermilion epithelium from the epidermis and oral mucosa. Additionally, previous studies on developing lip in vitro models reported their immunoreaction independently [
16,
17]. Considering that gene expression levels of K10 and SPRR3 agreed with the associated protein expression and localization in human lips, we concluded that K10 and SPRR3 can be used as specific double combination markers for vermilion epithelium in human, although their expression pattern was a little different from that in the monkey lip.
Vermilion’s phenotypic features between the adjacent epidermis and oral mucosa in the lip are not well-known, despite its distinct characteristics. The expression levels of terminal differentiation markers in stratified squamous epithelia, such as loricrin, involucrin, and SPRRs, are regulated and triggered by various intrinsic factors and internal and external stimuli, such as the underlying connective tissue and moist conditions [
18,
19,
20]. Also, the functional assay to measure transepithelial water loss suggested incomplete cornified layer formation of the lip surface [
6]. Moreover, attention should be paid to lipid metabolism and cholesterol sulphate in vermilion and crosstalk through soluble factors from the underlying tissue [
21,
22]. Further investigation is required to determine the three distinct epithelia within the lip to facilitate the development of a human in vitro model to understand unique lip biology.
We recognize that the very small sample size is a limitation of this study, although harvesting the entire lip tissue from humans and monkeys is very challenging due to cosmetic and ethical issues. Additionally, because our immunohistochemical analysis to verify the protein expression specific to vermillion epithelium did not provide any significant results, this incompletion to demonstrate a single marker was another weak point of this study. It may be required to produce higher quality antibodies for immunostaining or to perform in situ hybridization clustered in C11 to overcome this state.