High Rate of Non-Human Feeding by Aedes aegypti Reduces Zika Virus Transmission in South Texas
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Mosquito Collection
2.2. Blood Meal Analysis
2.3. Molecular Verification of Mosquito Species
2.4. Quantitative Synthesis of Published Literature
2.5. Mosquito Relative Abundance
2.6. Vertebrate Surveys
2.7. Human Density Estimation
2.8. Host Selection Indices
2.9. Tamaulipas Human Disease Data
2.10. Mathematical Modeling
| Feeding Patterns on Vertebrates (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Citation | Location | Method a | Site b | Human | Mix/Human | Dog | Cat | Other Mammal | Avian | Unknown | Total |
| [45] | Nigeria | Ab | In/Out | 7 (44%) | 1 (6%) | 8 (50%) | 16 | ||||
| [46] | Tanzania | Ab | In | 45 (100%) | 45 | ||||||
| [47] | Kenya—coast | Ab | In/Out | 165 (94%) | 1 (0.5%) | 1 (0.5%) | 9 (5%) | 176 | |||
| [48] | South Africa | Ab | Out | 3 (75%) | 1 (25%) | 4 | |||||
| [49] | India, Poona | Ab | In | 17 (81%) | 4 (19%) | 21 | |||||
| [50] | India | Ab | In | 49 (96%) | 2 (4%) | 51 | |||||
| [49] | Malaya | Ab | In | 109 (99%) | 1 (1%) | 110 | |||||
| [51] | Hawaii | Ab | Out | 339 (54%) | 117 (19%) | 21 (3%) | 71 (11%) | 3 (0.5%) | 80 (13%) | 631 | |
| [52] | Thailand | Ab | In/Out | 789 (88%) | 66 (7.4%) | 2 (2.2%) | 4 (0.5%) | 8 (1%) | 9 (1%) | 896 | |
| [53] | Puerto Rico | Ab | In | 1483 (95%) | 31 (2%) | 47 (3%) | 1561 | ||||
| [54] | Thailand—single host | Ab | In/Out | 658 (99%) | 1 | 4 (0.6%) | 1 | 664 | |||
| [54] | Thailand—mixed | Ab | In/Out | 86 (98%) | 88 | ||||||
| [55] | E. Australia | DNA | Out | 131 (75%) | 7 (4%) | 23 (13%) | 2 (1%) | 1 (0.5%) | 10 (6%) | 174 | |
| [56] | Thailand | DNA | N/A | 766 (86.1%) | 32 (3.6%) * | 18 (2%) | 39 (4.4%) | 35 (3.9%) | 890 | ||
| [57] | Puerto Rico-P | DNA | Out | 101 (76.2%) | 27 (20.8%) | 3 (2.3%) | 1 (0.8%) | 132 | |||
| [57] | Puerto Rico-R | DNA | Out | 210 (78.9%) | 1 (0.4%) | 49 (18.4%) | 3 (1.1%) | 3 (1.1%) | 266 | ||
| [58] | India | Gel precip | In/Out | 129 (87.8%) | 11 (7.5%) | 1 (0.7%) | 6 (4%) | 147 | |||
| [59] | India | Gel precip | Out | 54 (96.4%) | 2 (3.6%) | 56 | |||||
| [60] | Mexico | DNA | In/Out | 223 (98%) | 5 (2%) | 228 | |||||
| [61] | Florida—IR | DNA | Out | 111 (90.2%) | 11 (8.9%) | 1 (0.8%) | 123 | ||||
| [61] | Florida—M | DNA | Out | 8 (61.5%) | 5 (38.5%) | 13 | |||||
| [62] | Grenada | DNA | Out | 22 (70%) | 2 (6%) | 1 (3%) | 6 (18%) | 1 (3%) | 32 | ||
3. Results
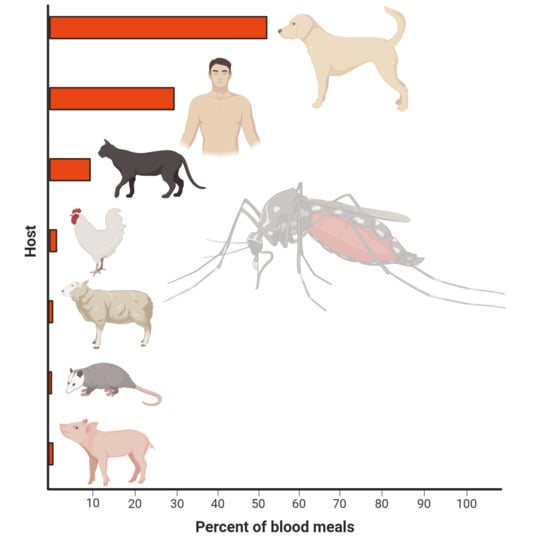
3.1. Blood Meal Analysis
3.2. Mosquito Relative Abundance
3.3. Vertebrate Surveys and Population Density
3.4. Host Selection
3.5. Mathematical Modeling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Huang, Y.-J.S.; Higgs, S.; Vanlandingham, D.L. Emergence and re-emergence of mosquito-borne arboviruses. Curr. Opin. Virol. 2019, 34, 104–109. [Google Scholar] [CrossRef]
- Gould, E.; Pettersson, J.; Higgs, S.; Charrel, R.; de Lamballerie, X. Emerging arboviruses: Why today? One Health 2017, 4, 1–13. [Google Scholar] [CrossRef]
- Espinal, M.A.; Andrus, J.K.; Jauregui, B.; Waterman, S.H.; Morens, D.M.; Santos, J.I.; Horstick, O.; Francis, L.A.; Olson, D. Emerging and reemerging Aedes-transmitted arbovirus infections in the region of the Americas: Implications for health policy. Am. J. Public Health 2019, 109, 387–392. [Google Scholar] [CrossRef]
- Macpherson, C.; Noël, T.; Fields, P.; Jungkind, D.; Yearwood, K.; Simmons, M.; Widjaja, S.; Mitchell, G.; Noel, D.; Bidaisee, S. Clinical and serological insights from the Asian lineage chikungunya outbreak in Grenada, 2014: An observational study. Am. J. Trop. Med. Hyg. 2016, 95, 890–893. [Google Scholar] [CrossRef] [Green Version]
- PAHO. Chikungunya. Available online: https://www.paho.org/hq/index.php?option=com_topics&view=article&id=343&Itemid=40931&lang=en (accessed on 12 July 2019).
- Faria, N.R.; da Silva Azevedo, R.d.S.; Kraemer, M.U.; Souza, R.; Cunha, M.S.; Hill, S.C.; Thézé, J.; Bonsall, M.B.; Bowden, T.A.; Rissanen, I. Zika virus in the Americas: Early epidemiological and genetic findings. Science 2016, 352, 345–349. [Google Scholar] [CrossRef] [Green Version]
- Brady, O.J.; Osgood-Zimmerman, A.; Kassebaum, N.J.; Ray, S.E.; de Araújo, V.E.; da Nóbrega, A.A.; Frutuoso, L.C.; Lecca, R.C.; Stevens, A.; de Oliveira, B.Z. The association between Zika virus infection and microcephaly in Brazil 2015–2017: An observational analysis of over 4 million births. PLoS Med. 2019, 16, e1002755. [Google Scholar] [CrossRef]
- Hahn, M.B.; Eisen, R.J.; Eisen, L.; Boegler, K.A.; Moore, C.G.; McAllister, J.; Savage, H.M.; Mutebi, J.-P. Reported distribution of Aedes (stegomyia) aegypti and Aedes (stegomyia) albopictus in the United States, 1995–2016 (diptera: Culicidae). J. Med. Entomol. 2016, 53, 1169–1175. [Google Scholar] [CrossRef] [Green Version]
- Thomas, D.L.; Santiago, G.A.; Abeyta, R.; Hinojosa, S.; Torres-Velasquez, B.; Adam, J.K.; Evert, N.; Caraballo, E.; Hunsperger, E.; Muñoz-Jordán, J.L. Reemergence of dengue in southern Texas, 2013. Emerg. Infect. Dis. 2016, 22, 1002. [Google Scholar] [CrossRef] [Green Version]
- Rey, J.R. Dengue in Florida (USA). Insects 2014, 5, 991–1000. [Google Scholar] [CrossRef] [Green Version]
- Secretaría de Salud, M. Dirección General de Epidemiología. 2019. Available online: https://www.gob.mx/salud/acciones-y-programas/historico-boletin-epidemiologico (accessed on 21 February 2020).
- Centers for Disease Control and Prevention. Dengue hemorrhagic fever—US-Mexico border, 2005. MMWR. Morbidity and Mortality Weekly Report. 2007. Available online: https://www.ncbi.nlm.nih.gov/pubmed/17687243 (accessed on 6 July 2019).
- Ramos, M.M.; Mohammed, H.; Zielinski-Gutierrez, E.; Hayden, M.H.; Lopez, J.L.R.; Fournier, M.; Trujillo, A.R.; Burton, R.; Brunkard, J.M.; Anaya-Lopez, L. Epidemic dengue and dengue hemorrhagic fever at the Texas–Mexico border: Results of a household-based seroepidemiologic survey, December 2005. Am. J. Trop. Med. Hyg. 2008, 78, 364–369. [Google Scholar] [CrossRef]
- Martin, E.; Medeiros, M.C.; Carbajal, E.; Valdez, E.; Juarez, J.G.; Gracia-Luna, S.; Salazar, A.; Qualls, W.A.; Hinojosa, S.; Borucki, M.K. Surveillance of Aedes aegypti indoors and outdoors using Autocidal Gravid Ovitraps in South Texas during local transmission of Zika virus, 2016 to 2018. Acta Trop. 2019, 192, 129–137. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Ladner, J.T.; Kraemer, M.U.; Dudas, G.; Tan, A.L.; Gangavarapu, K.; Wiley, M.R.; White, S.; Thézé, J.; Magnani, D.M. Genomic epidemiology reveals multiple introductions of Zika virus into the United States. Nature 2017, 546, 401. [Google Scholar] [CrossRef]
- Adams, L.E.; Martin, S.W.; Lindsey, N.P.; Lehman, J.A.; Rivera, A.; Kolsin, J.; Landry, K.; Staples, J.E.; Sharp, T.M.; Paz-Bailey, G. Epidemiology of dengue, chikungunya, and Zika virus disease in the US states and territories, 2017. Am. J. Trop. Med. Hyg. 2019, 101, 884–890. [Google Scholar] [CrossRef]
- Walker, K.R.; Williamson, D.; Carrière, Y.; Reyes-Castro, P.A.; Haenchen, S.; Hayden, M.H.; Jeffrey Gutierrez, E.; Ernst, K.C. Socioeconomic and human behavioral factors associated with Aedes aegypti (Diptera: Culicidae) immature habitat in Tucson, AZ. J. Med. Entomol. 2018, 55, 955–963. [Google Scholar] [CrossRef] [Green Version]
- Reiter, P.; Lathrop, S.; Bunning, M.; Biggerstaff, B.; Singer, D.; Tiwari, T.; Baber, L.; Amador, M.; Thirion, J.; Hayes, J. Texas lifestyle limits transmission of dengue virus. Emerg. Infect. Dis. 2003, 9, 86. [Google Scholar] [CrossRef]
- Fox, M. Illustrated key to common mosquitoes of Louisiana. In Mosquito Control Training Manual; Louisiana Mosquito Control Association: Baton Rouge, LA, USA, 2007; pp. 86–150. [Google Scholar]
- Sella, M. The antimalaria campaign at Fiumicino (Rome), with epidemiological and biological notes. Int. J. Public Health 1920, 1, 316–346. [Google Scholar]
- Graham, C.B.; Black Iv, W.C.; Boegler, K.A.; Montenieri, J.A.; Holmes, J.L.; Gage, K.L.; Eisen, R.J. Combining real-time polymerase chain reaction using SYBR Green I detection and sequencing to identify vertebrate bloodmeals in fleas. J. Med. Entomol. 2012, 49, 1442–1452. [Google Scholar] [CrossRef] [Green Version]
- Hamer, S.A.; Weghorst, A.C.; Auckland, L.D.; Roark, E.B.; Strey, O.F.; Teel, P.D.; Hamer, G.L. Comparison of DNA and carbon and nitrogen stable isotope-based techniques for identification of prior vertebrate hosts of ticks. J. Med. Entomol. 2015, 52, 1043–1049. [Google Scholar] [CrossRef]
- Greenstone, M.H.; Weber, D.C.; Coudron, T.A.; Payton, M.E.; Hu, J.S. Removing external DNA contamination from arthropod predators destined for molecular gut-content analysis. Mol. Ecol. Resour. 2012, 12, 464–469. [Google Scholar] [CrossRef]
- Hughes, G.L.; Dodson, B.L.; Johnson, R.M.; Murdock, C.C.; Tsujimoto, H.; Suzuki, Y.; Patt, A.A.; Cui, L.; Nossa, C.W.; Barry, R.M.; et al. Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc. Natl. Acad. Sci. USA 2014, 111, 12498–12503. [Google Scholar] [CrossRef] [Green Version]
- Hamer, G.L.; Kitron, U.D.; Goldberg, T.L.; Brawn, J.D.; Loss, S.R.; Ruiz, M.O.; Hayes, D.B.; Walker, E.D. Host selection by Culex pipiens mosquitoes and West Nile virus amplification. Am. J. Trop. Med. Hyg. 2009, 80, 268–278. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Triana, L.M.; Brugman, V.A.; Prosser, S.W.J.; Weland, C.; Nikolova, N.; Thorne, L.; De Marco, M.F.; Fooks, A.R.; Johnson, N. Molecular approaches for blood meal analysis and species identification of mosquitoes (Insecta: Diptera: Culicidae) in rural locations in southern England, United Kingdom. Zootaxa 2017, 4250, 67–76. [Google Scholar] [CrossRef]
- Medeiros, M.C.; Ricklefs, R.E.; Brawn, J.D.; Hamer, G.L. Plasmodium prevalence across avian host species is positively associated with exposure to mosquito vectors. Parasitology 2015, 142, 1612–1620. [Google Scholar] [CrossRef] [Green Version]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- McClelland, G.; Weitz, B. Serological identification of the natural hosts of Aedes aegypti (L.) and some other mosquitoes (Diptera, Culicidae) caught resting in vegetation in Kenya and Uganda. Ann. Trop. Med. Parasitol. 1963, 57, 214–224. [Google Scholar] [CrossRef]
- Teesdale, C. Studies on the bionomics of Aedes aegypti (L.) in its natural habitats in a coastal region of Kenya. Bull. Entomol. Res. 1955, 46, 711–742. [Google Scholar] [CrossRef]
- Chepkorir, E.; Venter, M.; Lutomiah, J.; Mulwa, F.; Arum, S.; Tchouassi, D.; Sang, R. The occurrence, diversity and blood feeding patterns of potential vectors of dengue and yellow fever in Kacheliba, West Pokot County, Kenya. Acta Trop. 2018, 186, 50–57. [Google Scholar] [CrossRef] [Green Version]
- Feria-Arroyo, T.P.; Aguilar, C.; Oraby, T. A tale of two cities: Aedes Mosquito surveillance across the Texas-Mexico Border. Subtrop. Agric. Environ. 2020, 71, 12. [Google Scholar]
- Brown, L.D.; Cai, T.T.; DasGupta, A. Interval estimation for a binomial proportion. Stat. Sci. 2001, 16, 101–117. [Google Scholar]
- Komar, N.; Panella, N.A.; Golnar, A.J.; Hamer, G.L. Forage ratio analysis of the southern house mosquito in College Station, Texas. Vector Borne Zoonotic Dis. 2018, 18, 485–490. [Google Scholar] [CrossRef]
- Garcia-Rejon, J.E.; Blitvich, B.J.; Farfan-Ale, J.A.; Lorono-Pino, M.A.; Chi Chim, W.A.; Flores-Flores, L.F.; Rosado-Paredes, E.; Baak-Baak, C.; Perez-Mutul, J.; Suarez-Solis, V.; et al. Host-feeding preference of the mosquito, Culex quinquefasciatus, in Yucatan State, Mexico. J. Insect. Sci. 2010, 10, 32. [Google Scholar] [CrossRef] [Green Version]
- Garrett-Jones, C. The human blood index of malaria vectors in relation to epidemiological assessment. Bull. World Health Organ. 1964, 30, 241–261. [Google Scholar]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.J.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008, 14, 1232. [Google Scholar] [CrossRef]
- Perkins, T.A.; Siraj, A.S.; Ruktanonchai, C.W.; Kraemer, M.U.; Tatem, A.J. Model-based projections of Zika virus infections in childbearing women in the Americas. Nat. Microbiol. 2016, 1, 16126. [Google Scholar] [CrossRef] [Green Version]
- Ndeffo-Mbah, M.L.; Pandey, A. Global risk and elimination of yellow fever epidemics. J. Inf. Dis. 2019. [Google Scholar] [CrossRef]
- Zhang, Q.; Sun, K.; Chinazzi, M.; y Piontti, A.P.; Dean, N.E.; Rojas, D.P.; Merler, S.; Mistry, D.; Poletti, P.; Rossi, L. Spread of Zika virus in the Americas. Proc. Natl. Acad. Sci. USA 2017, 114, E4334–E4343. [Google Scholar] [CrossRef] [Green Version]
- Reiner, R.C., Jr.; Stoddard, S.T.; Scott, T.W. Socially structured human movement shapes dengue transmission despite the diffusive effect of mosquito dispersal. Epidemics 2014, 6, 30–36. [Google Scholar] [CrossRef] [Green Version]
- Padmanabha, H.; Correa, F.; Rubio, C.; Baeza, A.; Osorio, S.; Mendez, J.; Jones, J.H.; Diuk-Wasser, M.A. Human social behavior and demography drive patterns of fine-scale Dengue transmission in endemic areas of Colombia. PLoS ONE 2015, 10, e0144451. [Google Scholar] [CrossRef] [Green Version]
- Cori, A.; Ferguson, N.M.; Fraser, C.; Cauchemez, S. A new framework and software to estimate time-varying reproduction numbers during epidemics. Am. J. Epidemiol. 2013, 178, 1505–1512. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, N.M.; Cucunubá, Z.M.; Dorigatti, I.; Nedjati-Gilani, G.L.; Donnelly, C.A.; Basáñez, M.-G.; Nouvellet, P.; Lessler, J. Countering the Zika epidemic in Latin America. Science 2016, 353, 353–354. [Google Scholar] [CrossRef] [Green Version]
- Davis, G.E.; Philip, C.B. The Identification of the Blood-meal in West African Mosquitoes by means of the Precipitin Test. A preliminary Report. Am. J. Hyg. 1931, 14, 130–141. [Google Scholar] [CrossRef]
- Lumsden, W. An epidemic of virus disease in Southern Province, Tanganyika territory, in 1952–1953 II. General description and epidemiology. Trans. R. Soc. Trop. Med. Hyg. 1955, 49, 33–57. [Google Scholar] [CrossRef]
- Heisch, R.; Nelson, G.; Furlong, M. Studies in filariasis in East Africa. 1. Filariasis on the Island of Pate, Kenya. Trans. R. Soc. Trop. Med. Hyg. 1959, 53, 41–53. [Google Scholar] [CrossRef]
- Paterson, H.; Bronsden, P.; Levitt, J.; Worth, C. Some Culicine Mosquitoes (Diptera, Culicidae) at Ndumu, Republic of South. Africa. A Study of Their Host Preferences and Host Range. Med. Proc. 1964, 10, 188–192. [Google Scholar]
- Macdonald, W. Host feeding preferences. Bull. World Health Organ. 1967, 36, 597. [Google Scholar]
- Krishnamurthy, B.; Kalra, N.; Joshi, G.; Singh, N. Reconnaissance Survey of Aedes Mosquitoes in Delhi. Bull. Indian Soc. Malar. Commun. Dis. 1965, 2, 56–67. [Google Scholar]
- Tempelis, C.; Hayes, R.; Hess, A.; Reeves, W. Blood-feeding habits of four species of mosquito found in Hawaii. Am. J. Trop. Med. Hyg. 1970, 19, 335–341. [Google Scholar] [CrossRef]
- Scott, T.W.; Chow, E.; Strickman, D.; Kittayapong, P.; Wirtz, R.A.; Lorenz, L.H.; Edman, J.D. Blood-feeding patterns of Aedes aegypti (Diptera: Culicidae) collected in a rural Thai village. J. Med. Entomol. 1993, 30, 922–927. [Google Scholar] [CrossRef]
- Scott, T.W.; Amerasinghe, P.H.; Morrison, A.C.; Lorenz, L.H.; Clark, G.G.; Strickman, D.; Kittayapong, P.; Edman, J.D. Longitudinal studies of Aedes aegypti (Diptera: Culicidae) in Thailand and Puerto Rico: Blood feeding frequency. J. Med. Entomol. 2000, 37, 89–101. [Google Scholar] [CrossRef]
- Ponlawat, A.; Harrington, L.C. Blood feeding patterns of Aedes aegypti and Aedes albopictus in Thailand. J. Med. Entomol. 2005, 42, 844–849. [Google Scholar] [CrossRef] [Green Version]
- Jansen, C.C.; Webb, C.E.; Graham, G.C.; Craig, S.B.; Zborowski, P.; Ritchie, S.A.; Russell, R.C.; Van den Hurk, A.F. Blood sources of mosquitoes collected from urban and peri-urban environments in eastern Australia with species-specific molecular analysis of avian blood meals. Am. J. Trop. Med. Hyg. 2009, 81, 849–857. [Google Scholar] [CrossRef] [Green Version]
- Siriyasatien, P.; Pengsakul, T.; Kittichai, V.; Phumee, A.; Kaewsaitiam, S.; Thavara, U.; Tawatsin, A.; Asavadachanukorn, P.; Mulla, M.S. Identification of blood meal of field caught Aedes aegypti (L.) by multiplex PCR. Southeast Asian J. Trop. Med. Public Health 2010, 41, 43. [Google Scholar]
- Barrera, R.; Bingham, A.M.; Hassan, H.K.; Amador, M.; Mackay, A.J.; Unnasch, T.R. Vertebrate hosts of Aedes aegypti and Aedes mediovittatus (Diptera: Culicidae) in rural Puerto Rico. J. Med. Entomol. 2012, 49, 917–921. [Google Scholar] [CrossRef] [Green Version]
- Sivan, A.; Shriram, A.; Sunish, I.; Vidhya, P. Host-feeding pattern of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) in heterogeneous landscapes of South Andaman, Andaman and Nicobar Islands, India. Parasitol. Res. 2015, 114, 3539–3546. [Google Scholar] [CrossRef]
- Wilson, J.J.; Sevarkodiyone, S. Host preference of blood feeding mosquitoes in rural areas of southern Tamil Nadu, India. Acad. J. Entomol. 2015, 8, 80–83. [Google Scholar]
- Baak-Baak, C.M.; Cigarroa-Toledo, N.; Cruz-Escalona, G.A.; Machain-Williams, C.; Rubi-Castellanos, R.; Torres-Chable, O.M.; Torres-Zapata, R.; Garcia-Rejon, J.E. Human blood as the only source of Aedes aegypti in churches from Merida, Yucatan, Mexico. J. Vector Borne Dis. 2018, 55, 58. [Google Scholar]
- Stenn, T.; Peck, K.J.; Rocha Pereira, G.; Burkett-Cadena, N.D. Vertebrate Hosts of Aedes aegypti, Aedes albopictus, and Culex quinquefasciatus (Diptera: Culicidae) as Potential Vectors of Zika Virus in Florida. J. Med. Entomol. 2018, 56, 10–17. [Google Scholar] [CrossRef]
- Fitzpatrick, D.M.; Hattaway, L.M.; Hsueh, A.N.; Ramos-Niño, M.E.; Cheetham, S.M. PCR-Based Bloodmeal Analysis of Aedes aegypti and Culex quinquefasciatus (Diptera: Culicidae) in St. George Parish, Grenada. J. Med. Entomol. 2019, 56, 1170–1175. [Google Scholar] [CrossRef] [Green Version]
- Chaves, L.F.; Harrington, L.C.; Keogh, C.L.; Nguyen, A.M.; Kitron, U.D. Blood feeding patterns of mosquitoes: Random or structured? Front. Zool. 2010, 7, 3. [Google Scholar] [CrossRef] [Green Version]
- Hess, A.; Hayes, R.O. Relative potentials of domestic animals for zooprophylaxis against mosquito vectors of encephalitis. Am. J. Trop. Med. Hyg. 1970, 19, 327–334. [Google Scholar] [CrossRef]
- Hamer, G.L.; Chaves, L.F.; Anderson, T.K.; Kitron, U.D.; Brawn, J.D.; Ruiz, M.O.; Loss, S.R.; Walker, E.D.; Goldberg, T.L. Fine-scale variation in vector host use and force of infection drive localized patterns of West Nile virus transmission. PLoS ONE 2011, 6, e23767. [Google Scholar] [CrossRef] [Green Version]
- Koolhof, I.; Carver, S. Epidemic host community contribution to mosquito-borne disease transmission: Ross River virus. Epidemiol. Infect. 2017, 145, 656–666. [Google Scholar] [CrossRef] [Green Version]
- Hackett, L.; Missiroli, A. The Natural Disappearance of Malaria in Certain Regions of Europe. Am. J. Hyg. 1931, 13, 57–78. [Google Scholar] [CrossRef]
- Fantini, B. Anophelism without malaria: An ecological and epidemiological puzzle. Parassitologia 1994, 36, 83–106. [Google Scholar]
- Russell, P.F.; West, L.S.; Manwell, R.D.; Macdonald, G. Practical malariology. In Practical Malariology; Oxford University Press: London, UK, 1963. [Google Scholar]
- Mahande, A.; Mosha, F.; Mahande, J.; Kweka, E. Feeding and resting behaviour of malaria vector, Anopheles arabiensis with reference to zooprophylaxis. Malar. J. 2007, 6, 100. [Google Scholar] [CrossRef] [Green Version]
- Iwashita, H.; Dida, G.O.; Sonye, G.O.; Sunahara, T.; Futami, K.; Njenga, S.M.; Chaves, L.F.; Minakawa, N. Push by a net, pull by a cow: Can zooprophylaxis enhance the impact of insecticide treated bed nets on malaria control? Parasit. Vectors 2014, 7, 52. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, W.-P.; Suzuki, M.; Thiem, V.D.; White, R.G.; Tsuzuki, A.; Yoshida, L.-M.; Yanai, H.; Haque, U.; Tho, L.H.; Anh, D.D. Population density, water supply, and the risk of dengue fever in Vietnam: Cohort study and spatial analysis. PLoS Med. 2011, 8, e1001082. [Google Scholar] [CrossRef]
- Fritz, M.; Walker, E.; Miller, J.; Severson, D.; Dworkin, I. Divergent host preferences of above-and below-ground Culex pipiens mosquitoes and their hybrid offspring. Med. Vet. Entomol. 2015, 29, 115–123. [Google Scholar] [CrossRef]
- Main, B.J.; Lee, Y.; Ferguson, H.M.; Kreppel, K.S.; Kihonda, A.; Govella, N.J.; Collier, T.C.; Cornel, A.J.; Eskin, E.; Kang, E.Y. The genetic basis of host preference and resting behavior in the major African malaria vector, Anopheles arabiensis. PLoS Genet. 2016, 12, e1006303. [Google Scholar] [CrossRef]
- Kotsakiozi, P.; Evans, B.R.; Gloria-Soria, A.; Kamgang, B.; Mayanja, M.; Lutwama, J.; Le Goff, G.; Ayala, D.; Paupy, C.; Badolo, A. Population structure of a vector of human diseases: Aedes aegypti in its ancestral range, Africa. Ecol. Evolut. 2018, 8, 7835–7848. [Google Scholar] [CrossRef] [Green Version]
- Crawford, J.E.; Alves, J.M.; Palmer, W.J.; Day, J.P.; Sylla, M.; Ramasamy, R.; Surendran, S.N.; Black, W.C.; Pain, A.; Jiggins, F.M. Population genomics reveals that an anthropophilic population of Aedes aegypti mosquitoes in West Africa recently gave rise to American and Asian populations of this major disease vector. BMC Biol. 2017, 15, 16. [Google Scholar] [CrossRef] [Green Version]
- McBride, C.S.; Baier, F.; Omondi, A.B.; Spitzer, S.A.; Lutomiah, J.; Sang, R.; Ignell, R.; Vosshall, L.B. Evolution of mosquito preference for humans linked to an odorant receptor. Nature 2014, 515, 222–227. [Google Scholar] [CrossRef]
- Paris, V.; Cottingham, E.; Ross, P.A.; Axford, J.K.; Hoffmann, A.A. Effects of alternative blood sources on Wolbachia infected Aedes aegypti females within and across generations. Insects 2018, 9, 140. [Google Scholar] [CrossRef] [Green Version]
- Hodo, C.L.; Rodriguez, J.Y.; Curtis-Robles, R.; Zecca, I.B.; Snowden, K.F.; Cummings, K.J.; Hamer, S.A. Repeated cross-sectional study of Trypanosoma cruzi in shelter dogs in Texas, in the context of Dirofilaria immitis and tick-borne pathogen prevalence. J. Vet. Intern. Med. 2019, 33, 158–166. [Google Scholar] [CrossRef] [Green Version]
- Tiawsirisup, S.; Nithiuthai, S. Vector competence of Aedes aegypti (L.) and Culex quinquefasciatus (Say) for Dirofilaria immitis (Leidy). Southeast Asian J. Trop. Med. Public Health 2006, 37, 110. [Google Scholar]
- Serrão, M.L.; Labarthe, N.; Lourenço-de-Oliveira, R. Vectorial competence of Aedes aegypti (Linnaeus 1762) Rio de Janeiro strain, to Dirofilaria immitis (Leidy 1856). Mem. Inst. Oswaldo Cruz 2001, 96, 593–598. [Google Scholar] [CrossRef] [Green Version]
- Ledesma, N.A.; Kaufman, P.E.; Xue, R.-D.; Leyen, C.; Macapagal, M.J.; Winokur, O.C.; Harrington, L.C. Entomological and sociobehavioral components of heartworm (Dirofilaria immitis) infection in two Florida communities with a high or low prevalence of dogs with heartworm infection. J. Am. Vet. Med. Assoc. 2019, 254, 93–103. [Google Scholar] [CrossRef]
- Thoisy, B.D.; Lacoste, V.; Germain, A.; Muñoz-Jordán, J.; Colón, C.; Mauffrey, J.F.; Delaval, M.; Catzeflis, F.; Kazanji, M.; Matheus, S.; et al. Dengue infection in neotropical forest mammals. Vector Borne Zoonotic Dis. 2009, 9, 157–170. [Google Scholar] [CrossRef] [Green Version]
- Pauvolid-Corrêa, A.; Gonçalves Dias, H.; Marina Siqueira Maia, L.; Porfírio, G.; Oliveira Morgado, T.; Sabino-Santos, G.; Helena Santa Rita, P.; Teixeira Gomes Barreto, W.; Carvalho de Macedo, G.; Marinho Torres, J. Zika Virus Surveillance at the Human–Animal Interface in West-Central Brazil, 2017–2018. Viruses 2019, 11, 1164. [Google Scholar] [CrossRef] [Green Version]
- Farajollahi, A.; Fonseca, D.M.; Kramer, L.D.; Kilpatrick, A.M. “Bird biting” mosquitoes and human disease: A review of the role of Culex pipiens complex mosquitoes in epidemiology. Inf. Genet. Evolut. 2011, 11, 1577–1585. [Google Scholar] [CrossRef] [Green Version]
- Kent, R.J.; Reiche, A.S.G.; Morales-Betoulle, M.E.; Komar, N. Comparison of engorged Culex quinquefasciatus collection and blood-feeding pattern among four mosquito collection methods in Puerto Barrios, Guatemala, 2007. J. Am. Mosq. Control. Assoc. 2010, 26, 332–337. [Google Scholar] [CrossRef]
- Janssen, N.; Fernandez-Salas, I.; Diaz Gonzalez, E.E.; Gaytan-Burns, A.; Medina-de la Garza, C.E.; Sanchez-Casas, R.M.; Borstler, J.; Cadar, D.; Schmidt-Chanasit, J.; Jost, H. Mammalophilic feeding behaviour of Culex quinquefasciatus mosquitoes collected in the cities of Chetumal and Cancun, Yucatan Peninsula, Mexico. Trop. Med. Int. Health 2015, 20, 1488–1491. [Google Scholar] [CrossRef] [Green Version]
- Molaei, G.; Andreadis, T.G.; Armstrong, P.M.; Bueno, R., Jr.; Dennett, J.A.; Real, S.V.; Sargent, C.; Bala, A.; Randle, Y.; Guzman, H.; et al. Host feeding pattern of Culex quinquefasciatus (Diptera: Culicidae) and its role in transmission of West Nile virus in Harris County, Texas. Am. J. Trop. Med. Hyg. 2007, 77, 73–81. [Google Scholar] [CrossRef] [Green Version]
- McCall, J.W.; Genchi, C.; Kramer, L.H.; Guerrero, J.; Venco, L. Heartworm disease in animals and humans. Adv. Parasitol. 2008, 66, 193–285. [Google Scholar]


| Host | Count (%) | Forage Ratio (95% CI) |
|---|---|---|
| Dog | 93 (50%) | 1.61 (1.43–1.84) |
| Human | 57 * (31%) | 0.81 (0.73–0.91) |
| Cat | 22 (12%) | 0.91 (0.73–1.13) |
| Chicken | 6 (3%) | 0.19 (0.16–0.24) |
| Sheep | 3 (1.6%) | 2.69 ** (1.01–8.06) |
| Opossum | 2 (1%) | 1.19 (0.51–2.69) |
| Pig | 3 (1.6%) | 2.69 (1.01–8.06) |
| Total | 186 |
| Host | Count (%) | Forage Ratio (95% CI) |
|---|---|---|
| Chicken | 82 (67%) | 3.92 (3.33–4.87) |
| Dog | 27 (22%) | 0.71 (0.63–0.81) |
| House sparrow | 6 (5%) | - |
| Western kingbird | 1 (0.8%) | - |
| Human | 1 (0.8%) | 0.02 (0.02–0.02) |
| Cat | 1 (0.8%) | 0.06 (0.05–0.08) |
| Pig | 1 (0.8%) | 1.36 (0.51–4.07) |
| Plain chachalaca | 1 (0.8%) | - |
| Curvebilled thrasher | 1 (0.8%) | - |
| Northern mockingbird | 1 (0.8%) | - |
| Rock dove | 1 (0.8%) | - |
| Total | 123 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olson, M.F.; Ndeffo-Mbah, M.L.; Juarez, J.G.; Garcia-Luna, S.; Martin, E.; Borucki, M.K.; Frank, M.; Estrada-Franco, J.G.; Rodríguez-Pérez, M.A.; Fernández-Santos, N.A.; et al. High Rate of Non-Human Feeding by Aedes aegypti Reduces Zika Virus Transmission in South Texas. Viruses 2020, 12, 453. https://doi.org/10.3390/v12040453
Olson MF, Ndeffo-Mbah ML, Juarez JG, Garcia-Luna S, Martin E, Borucki MK, Frank M, Estrada-Franco JG, Rodríguez-Pérez MA, Fernández-Santos NA, et al. High Rate of Non-Human Feeding by Aedes aegypti Reduces Zika Virus Transmission in South Texas. Viruses. 2020; 12(4):453. https://doi.org/10.3390/v12040453
Chicago/Turabian StyleOlson, Mark F., Martial L. Ndeffo-Mbah, Jose G. Juarez, Selene Garcia-Luna, Estelle Martin, Monica K. Borucki, Matthias Frank, José Guillermo Estrada-Franco, Mario A. Rodríguez-Pérez, Nadia A. Fernández-Santos, and et al. 2020. "High Rate of Non-Human Feeding by Aedes aegypti Reduces Zika Virus Transmission in South Texas" Viruses 12, no. 4: 453. https://doi.org/10.3390/v12040453