The Two Prevalent Genotypes of an Emerging Infectious Disease, Deformed Wing Virus, Cause Equally Low Pupal Mortality and Equally High Wing Deformities in Host Honey Bees
Abstract
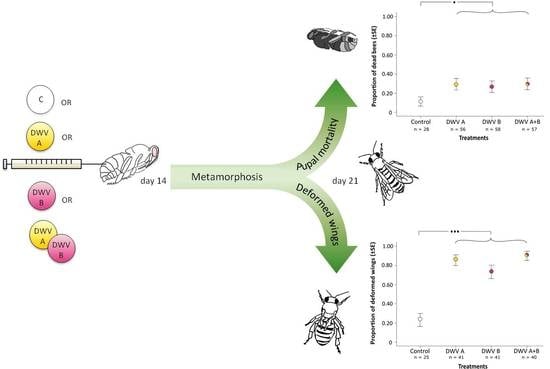
:1. Introduction
2. Materials and Methods
2.1. Source of Honey Bees
2.2. Virus Propagation and Assessment of Inocula
2.3. Viral Inoculation and Virulence
2.4. RNA Extraction and Detection of Virus
- (i)
- when testing whether honey bee source colonies were free of virus,
- (ii)
- when screening pupal homogenates (our inocula) for RNA viruses and quantifying DWV titer in them,
- (iii)
- when quantifying viral titers in adult worker bees arising from inoculation experiments, and
- (iv)
- when quantifying viral titers in pupae at zero and three days p.i.
2.5. Statistical Analyses
3. Results
3.1. Analysis of Inocula
3.2. Impact of Inoculation on Honey Bee Pupae
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
References
- Klein, A.-M.; Vaissiére, B.E.; Cane, J.H.; Steffan-Dewenter, I.; Cunningham, S.A.; Kremen, C.; Tscharntke, T. Importance of pollinators in changing landscapes for world crops. Proc. R. Soc. Lond. B 2007, 274, 303–314. [Google Scholar] [CrossRef]
- Hung, K.-L.J.; Kingston, J.M.; Albrecht, M.; Holway, D.A.; Kohn, J.R. The worldwide importance of honey bees as pollinators in natural habitats. Proc. R. Soc. B Biol. Sci. 2018, 285, 20172140. [Google Scholar] [CrossRef] [Green Version]
- Aizen, M.A.; Harder, L.D. The global stock of domesticated honey bees is growing slower than agricultural demand for pollination. Curr. Biol. 2009, 19, 915–918. [Google Scholar] [CrossRef]
- Potts, S.G.; Imperatriz-Fonseca, V.; Ngo, H.T.; Aizen, M.A.; Biesmeijer, J.C.; Breeze, T.D.; Dicks, L.V.; Garibaldi, L.A.; Hill, R.; Settele, J.; et al. Safeguarding pollinators and their values to human well-being. Nature 2016, 540, 220–229. [Google Scholar] [CrossRef] [Green Version]
- Oldroyd, B.P. What’s killing American honey bees? PLoS Biol. 2007, 5, e168. [Google Scholar] [CrossRef]
- Watanabe, M.E. What’s new with honeybees? Bioscience 2009, 59, 1010. [Google Scholar] [CrossRef]
- Neumann, P.; Carreck, N.L. Honey bee colony losses. J. Apic. Res. 2010, 49, 1–6. [Google Scholar] [CrossRef]
- Vanbergen, A.J.; Initiative, I.P. Threats to an ecosystem service: Pressures on pollinators. Front. Ecol. Environ. 2013, 11, 251–259. [Google Scholar] [CrossRef]
- Goulson, D.; Nicholls, E.; Botías, C.; Rotheray, E.L. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 2015, 347, 1255957. [Google Scholar] [CrossRef]
- Highfield, A.C.; El Nagar, A.; Mackinder, L.C.M.; Noël, L.M.-L.J.; Hall, M.J.; Martin, S.J.; Schroeder, D.C. Deformed wing virus implicated in overwintering honeybee colony losses. Appl. Environ. Microbiol. 2009, 75, 7212–7220. [Google Scholar] [CrossRef]
- Dainat, B.; Evans, J.D.; Chen, Y.P.; Gauthier, L.; Neumann, P. Dead or alive: Deformed wing virus and Varroa destructor reduce the life span of winter honeybees. Appl. Environ. Microbiol. 2012, 78, 981–987. [Google Scholar] [CrossRef]
- Nazzi, F.; Brown, S.P.; Annoscia, D.; Del Piccolo, F.; Di Prisco, G.; Varricchio, P.; Della Vedova, G.; Cattonaro, F.; Caprio, E.; Pennacchio, F. Synergistic parasite-pathogen interactions mediated by host immunity can drive the collapse of honeybee colonies. PLoS Pathog. 2012, 8, e1002735. [Google Scholar] [CrossRef]
- Francis, R.M.; Nielsen, S.L.; Kryger, P. Varroa-virus interaction in collapsing honey bee colonies. PLoS ONE 2013, 8, e57540. [Google Scholar] [CrossRef]
- Nguyen, B.K.; Ribière, M.; vanEngelsdorp, D.; Snoeck, C.; Saegerman, C.; Kalkstein, A.L.; Schurr, F.; Brostaux, Y.; Faucon, J.-P.; Haubruge, E. Effects of honey bee virus prevalence, Varroa destructor load and queen condition on honey bee colony survival over the winter in Belgium. J. Apic. Res. 2011, 50, 195–202. [Google Scholar] [CrossRef]
- Budge, G.E.; Pietravalle, S.; Brown, M.; Laurenson, L.; Jones, B.; Tomkies, V.; Delaplane, K.S. Pathogens as predictors of honey bee colony strength in England and Wales. PLoS ONE 2015, 10, e0133228. [Google Scholar] [CrossRef]
- Di Prisco, G.; Annoscia, D.; Margiotta, M.; Ferrara, R.; Varricchio, P.; Zanni, V.; Caprio, E.; Nazzi, F.; Pennacchio, F. A mutualistic symbiosis between a parasitic mite and a pathogenic virus undermines honey bee immunity and health. Proc. Natl. Acad. Sci. USA 2016, 113, 3203–3208. [Google Scholar] [CrossRef]
- McMahon, D.P.; Natsopoulou, M.E.; Doublet, V.; Fürst, M.; Weging, S.; Brown, M.J.F.; Gogol-Döring, A.; Paxton, R.J. Elevated virulence of an emerging viral genotype as a driver of honeybee loss. Proc. R. Soc. Lond. B Biol. Sci. 2016, 283, 20160811. [Google Scholar] [CrossRef] [Green Version]
- Natsopoulou, M.E.; McMahon, D.P.; Doublet, V.; Frey, E.; Rosenkranz, P.; Paxton, R.J. The virulent, emerging genotype B of Deformed wing virus is closely linked to overwinter honeybee worker loss. Sci. Rep. 2017, 7, 5242. [Google Scholar] [CrossRef]
- Martin, S.J.; Highfield, A.C.; Brettell, L.; Villalobos, E.M.; Budge, G.E.; Powell, M.; Nikaido, S.; Schroeder, D.C. Global honey bee viral landscape altered by a parasitic mite. Science 2012, 336, 1304–1306. [Google Scholar] [CrossRef]
- Mondet, F.; de Miranda, J.R.; Kretzschmar, A.; Le Conte, Y.; Mercer, A.R. On the front line: Quantitative virus dynamics in honeybee (Apis mellifera L.) colonies along a new expansion front of the parasite Varroa destructor. PLoS Pathog. 2014, 10, e1004323. [Google Scholar] [CrossRef]
- Bailey, L.; Ball, B.V. Honey Bee Pathology, 2nd ed.; Academic Press: Cambridge, MA, USA, 1991. [Google Scholar]
- Chen, Y.; Pettis, J.S.; Feldlaufer, M.F. Detection of multiple viruses in queens of the honey bee Apis mellifera L. J. Invertebr. Pathol. 2005, 90, 118–121. [Google Scholar] [CrossRef]
- Chen, Y.; Evans, J.D.; Feldlaufer, M. Horizontal and vertical transmission of viruses in the honey bee, Apis mellifera. J. Invertebr. Pathol. 2006, 92, 152–159. [Google Scholar] [CrossRef]
- Yue, C.; Genersch, E. RT-PCR analysis of Deformed wing virus in honeybees (Apis mellifera) and mites (Varroa destructor). J. Gen. Virol. 2005, 86, 3419–3424. [Google Scholar] [CrossRef]
- Yue, C.; Schröder, M.; Gisder, S.; Genersch, E. Vertical-transmission routes for Deformed wing virus of honeybees (Apis mellifera). J. Gen. Virol. 2007, 88, 2329–2336. [Google Scholar] [CrossRef]
- De Miranda, J.R.; Fries, I. Venereal and vertical transmission of Deformed wing virus in honeybees (Apis mellifera L.). J. Invertebr. Pathol. 2008, 98, 184–189. [Google Scholar] [CrossRef]
- De Miranda, J.R.; Genersch, E. Deformed wing virus. J. Invertebr. Pathol. 2010, 103, S48–S61. [Google Scholar] [CrossRef]
- Ball, B.V. Varroa jacobsoni as a virus vector. In Present Status of Varroatosis in Europe and Progress in the Varroa Mite Control; Cavalloro, R., Ed.; ECSC-EEC-EAEC of the European Economic Commission: Luxembourg, 1989; pp. 241–244. [Google Scholar]
- Nordström, S. Distribution of Deformed wing virus within honey bee (Apis mellifera) brood cells infested with the ectoparasitic mite Varroa destructor. Exp. Appl. Acarol. 2003, 29, 293–302. [Google Scholar] [CrossRef]
- Gisder, S.; Aumeier, P.; Genersch, E. Deformed wing virus: Replication and viral load in mites (Varroa destructor). J. Gen. Virol. 2009, 90, 463–467. [Google Scholar] [CrossRef]
- Möckel, N.; Gisder, S.; Genersch, E. Horizontal transmission of Deformed wing virus: Pathological consequences in adult bees (Apis mellifera) depend on the transmission route. J. Gen. Virol. 2011, 92, 370–377. [Google Scholar] [CrossRef]
- Annoscia, D.; Brown, S.; Di Prisco, G.; De Paoli, E.; Del Fabbro, S.; Zanni, V.; Galbraith, D.; Caprio, E.; Grozinger, C.M.; Pennacchio, F.; et al. Haemolymph removal by the parasite Varroa destructor can trigger the proliferation of the Deformed wing virus in mite infested bees (Apis mellifera), contributing to enhanced pathogen virulence. bioRxiv 2018. [Google Scholar] [CrossRef]
- Ongus, J.R.; Peters, D.; Bonmatin, J.M.; Bengsch, E.; Vlak, J.M.; van Oers, M.M. Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor. J. Gen. Virol. 2004, 85, 3747–3755. [Google Scholar] [CrossRef] [PubMed]
- Aupperle, H.; Genersch, E. Diagnostischer Farbatlas der Bienenepathologie. Diagnostic Color Atlas of Bee Pathology; Verlag LABOKLIN: Bad Kissingen, Germany, 2016; p. 191. [Google Scholar]
- Erban, T.; Harant, K.; Hubalek, M.; Vitamvas, P.; Kamler, M.; Poltronieri, P.; Tyl, J.; Markovic, M.; Titera, D. In-depth proteomic analysis of Varroa destructor: Detection of DWV-complex, ABPV, VdMLV and honeybee proteins in the mite. Sci. Rep. 2015, 5, 13907. [Google Scholar] [CrossRef]
- Ball, B.V.; Allen, M.F. The prevalence of pathogens in honey bee (Apis mellifera) colonies infested with the parasitic mite Varroa jacobsoni. Ann. Appl. Biol. 1988, 113, 237–244. [Google Scholar] [CrossRef]
- Shen, M.; Cui, L.; Ostiguy, N.; Cox-Foster, D. Intricate transmission routes and interactions between picorna-like viruses (Kashmir bee virus and Sacbrood virus) with the honeybee host and the parasitic varroa mite. J. Gen. Virol. 2005, 86, 2281–2289. [Google Scholar] [CrossRef] [PubMed]
- Rosenkranz, P.; Aumeier, P.; Ziegelmann, B. Biology and control of Varroa destructor. J. Invertebr. Pathol. 2010, 103, S96–S119. [Google Scholar] [CrossRef]
- Natsopoulou, M.E.; McMahon, D.P.; Paxton, R.J. Parasites modulate within-colony activity and accelerate the temporal polyethism schedule of a social insect, the honey bee. Behav. Ecol. Sociobiol. 2016, 70, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Benaets, K.; Van Geystelen, A.; Cardoen, D.; De Smet, L.; de Graaf, D.C.; Schoofs, L.; Larmuseau, M.H.D.; Brettell, L.E.; Martin, S.J.; Wenseleers, T. Covert Deformed wing virus infections have long-term deleterious effects on honeybee foraging and survival. Proc. R. Soc. B Biol. Sci. 2017, 284, 20162149. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.; Ball, B. The incidence and world distribution of honey bee viruses. Bee World 1996, 77, 141–162. [Google Scholar] [CrossRef]
- Lanzi, G.; de Miranda, J.R.; Boniotti, M.B.; Cameron, C.E.; Lavazza, A.; Capucci, L.; Camazine, S.M.; Rossi, C. Molecular and biological characterization of Deformed wing virus of honeybees (Apis mellifera L.). J. Virol. 2006, 80, 4998–5009. [Google Scholar] [CrossRef]
- Škubník, K.; Nováček, J.; Füzik, T.; Přidal, A.; Paxton, R.J.; Plevka, P. Structure of Deformed wing virus, a major honey bee pathogen. Proc. Natl. Acad. Sci. USA 2017, 114, 3210–3215. [Google Scholar] [CrossRef] [PubMed]
- Lamp, B.; Url, A.; Seitz, K.; Eichhorn, J.; Riedel, C.; Sinn, L.J.; Indik, S.; Köglberger, H.; Rümenapf, T. Construction and rescue of a molecular clone of Deformed wing virus (DWV). PLoS ONE 2016, 11, e0164639. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.; Revill, P. Overview of Hepatitis B viral replication and genetic variability. J. Hepatol. 2016, 64, S4–S16. [Google Scholar] [CrossRef] [PubMed]
- Mordecai, G.J.; Brettell, L.E.; Martin, S.J.; Dixon, D.; Jones, I.M.; Schroeder, D.C. Superinfection exclusion and the long-term survival of honey bees in Varroa-infested colonies. ISME J. 2015, 10, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Mordecai, G.J.; Wilfert, L.; Martin, S.J.; Jones, I.M.; Schroeder, D.C. Diversity in a honey bee pathogen: First report of a third master variant of the Deformed wing virus quasispecies. ISME J. 2016, 10, 1264–1273. [Google Scholar] [CrossRef]
- Kevill, J.; Highfield, A.; Mordecai, G.; Martin, S.; Schroeder, D. ABC assay: Method development and application to quantify the role of three DWV master variants in overwinter colony losses of European honey bees. Viruses 2017, 9, 314. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.; Jironkin, A.; Chandler, D.; Burroughs, N.; Evans, D.J.; Ryabov, E.V. Recombinants between Deformed wing virus and Varroa destructor virus-1 may prevail in Varroa destructor-infested honeybee colonies. J. Gen. Virol. 2011, 92, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Zioni, N.; Soroker, V.; Chejanovsky, N. Replication of Varroa destructor virus 1 (VDV-1) and a Varroa destructor virus 1–Deformed wing virus recombinant (VDV-1–DWV) in the head of the honey bee. Virology 2011, 417, 106–112. [Google Scholar] [CrossRef]
- Wang, H.; Xie, J.; Shreeve, T.G.; Ma, J.; Pallett, D.W.; King, L.A.; Possee, R.D. Sequence recombination and conservation of Varroa destructor virus-1 and Deformed wing virus in field collected honey bees (Apis mellifera). PLoS ONE 2013, 8, e74508. [Google Scholar] [CrossRef]
- Ryabov, E.V.; Wood, G.R.; Fannon, J.M.; Moore, J.D.; Bull, J.C.; Chandler, D.; Mead, A.; Burroughs, N.; Evans, D.J. A virulent strain of Deformed wing virus (DWV) of honeybees (Apis mellifera) prevails after Varroa destructor-mediated, or in vitro, transmission. PLoS Pathog. 2014, 10, e1004230. [Google Scholar] [CrossRef]
- Cornman, R.S. Relative abundance of Deformed wing virus, Varroa destructor virus 1, and their recombinants in honey bees (Apis mellifera) assessed by kmer analysis of public RNA-seq data. J. Invertebr. Pathol. 2017, 149, 44–50. [Google Scholar] [CrossRef]
- Dalmon, A.; Desbiez, C.; Coulon, M.; Thomasson, M.; Le Conte, Y.; Alaux, C.; Vallon, J.; Moury, B. Evidence for positive selection and recombination hotspots in Deformed wing virus (DWV). Sci. Rep. 2017, 7, 41045. [Google Scholar] [CrossRef] [PubMed]
- Fürst, M.A.; McMahon, D.P.; Osborne, J.L.; Paxton, R.J.; Brown, M.J.F. Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature 2014, 506, 364–366. [Google Scholar] [CrossRef] [PubMed]
- McMahon, D.P.; Fürst, M.A.; Caspar, J.; Theodorou, P.; Brown, M.J.F.; Paxton, R.J. A sting in the spit: Widespread cross-infection of multiple RNA viruses across wild and managed bees. J. Anim. Ecol. 2015, 84, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Radzevičiūtė, R.; Theodorou, P.; Husemann, M.; Japoshvili, G.; Kirkitadze, G.; Zhusupbaeva, A.; Paxton, R.J. Replication of honey bee-associated RNA viruses across multiple bee species in apple orchards of Georgia, Germany and Kyrgyzstan. J. Invertebr. Pathol. 2017, 146, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Wilfert, L.; Long, G.; Leggett, H.C.; Schmid-Hempel, P.; Butlin, R.; Martin, S.J.M.; Boots, M. Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science 2016, 351, 594–597. [Google Scholar] [CrossRef]
- Ongus, J.R.; Fombong, A.T.; Irungu, J.; Masiga, D.; Raina, S. Prevalence of common honey bee pathogens at selected apiaries in Kenya, 2013/2014. Int. J. Trop. Insect Sci. 2018, 38, 58–70. [Google Scholar] [CrossRef]
- Strauss, U.; Human, H.; Gauthier, L.; Crewe, R.M.; Dietemann, V.; Pirk, C.W.W. Seasonal prevalence of pathogens and parasites in the savannah honeybee (Apis mellifera scutellata). J. Invertebr. Pathol. 2013, 114, 45–52. [Google Scholar] [CrossRef]
- Ryabov, E.V.; Childers, A.K.; Chen, Y.; Madella, S.; Nessa, A.; van Engelsdorp, D.; Evans, J.D. Recent spread of Varroa destructor virus-1, a honey bee pathogen, in the United States. Sci. Rep. 2017, 7, 17447. [Google Scholar] [CrossRef]
- Roberts, J.M.K.; Anderson, D.L.; Durr, P.A. Absence of Deformed wing virus and Varroa destructor in Australia provides unique perspectives on honeybee viral landscapes and colony losses. Sci. Rep. 2017, 7, 6925. [Google Scholar] [CrossRef]
- Schmid-Hempel, P. Evolutionary Parasitology; Oxford University Press: Oxford, UK, 2011; p. 516. [Google Scholar]
- Gisder, S.; Möckel, N.; Eisenhardt, D.; Genersch, E. In vivo evolution of viral virulence: Switching of Deformed wing virus between hosts results in virulence changes and sequence shifts. Environ. Microbiol. 2018. [Google Scholar] [CrossRef]
- Winston, M.L. The Biology of the Honey Bee; Harvard University Press: Cambridge, MA, USA, 1987; p. 281. [Google Scholar]
- McMahon, D.P.; Wilfert, L.; Paxton, R.J.; Brown, M.J.F. Chapter Eight—Emerging viruses in bees: From molecules to ecology. In Advances in Virus Research; Malmstrom, C.M., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 101, pp. 251–291. [Google Scholar]
- Locke, B.; Forsgren, E.; Fries, I.; de Miranda, J.R. Acaricide treatment affects viral dynamics in Varroa destructor-infested honey bee colonies via both host physiology and mite control. Appl. Environ. Microbiol. 2012, 78, 227–235. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, J.R.; Bailey, L.; Ball, B.V.; Blanchard, P.; Budge, G.; Chejanovsky, N.; Chen, Y.-P.; Gauthier, L.; Genersch, E.; de Graaf, D.; et al. Standard methods for virus research in Apis mellifera. J. Apic. Res. 2013, 52, 1–56. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunt, M.; Gall, A.; Ong, S.H.; Brener, J.; Ferns, B.; Goulder, P.; Nastouli, E.; Keane, J.A.; Kellam, P.; Otto, T.D. IVA: Accurate de novo assembly of RNA virus genomes. Bioinformatics 2015, 31, 2374–2376. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Levin, S.; Galbraith, D.; Sela, N.; Erez, T.; Grozinger, C.M.; Chejanovsky, N. Presence of Apis rhabdovirus-1 in populations of pollinators and their parasites from two Continents. Front. Microbiol. 2017, 8, 2482. [Google Scholar] [CrossRef]
- Remnant, E.J.; Shi, M.; Buchmann, G.; Blacquière, T.; Holmes, E.C.; Beekman, M.; Ashe, A. A diverse range of novel RNA viruses in geographically distinct honey bee populations. J. Virol. 2017, 91, e00158-17. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 1, 1–48. [Google Scholar] [CrossRef]
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous inference in general parametric models. Biom. J. 2008, 50, 346–363. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Martin, S.J. The role of Varroa and viral pathogens in the collapse of honeybee colonies: A modelling approach. J. Appl. Ecol. 2001, 38, 1082–1093. [Google Scholar] [CrossRef]
- Sumpter, D.J.T.; Martin, S.J. The dynamics of virus epidemics in varroa-infested honey bee colonies. J. Anim. Ecol. 2004, 73, 51–63. [Google Scholar] [CrossRef]
- Brutscher, L.M.; Daughenbaugh, K.F.; Flenniken, M.L. Antiviral defense mechanisms in honey bees. Curr. Opin. Insect Sci. 2015, 10, 71–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gullan, P.J.; Cranston, P.S. The Insects: An Outline in Entomology, 5th ed.; Blackwell: Oxford, UK, 2014; p. 624. [Google Scholar]
- Brettell, L.; Mordecai, G.; Schroeder, D.; Jones, I.; da Silva, J.; Vicente-Rubiano, M.; Martin, S. A comparison of Deformed wing virus in deformed and asymptomatic honey bees. Insects 2017, 8, 28. [Google Scholar] [CrossRef]
- Neumann, P.; Yañez, O.; Fries, I.; de Miranda, J.R. Varroa invasion and virus adaptation. Trends Parasitol. 2012, 28, 353–354. [Google Scholar] [CrossRef] [PubMed]
- Forsgren, E.; Locke, B.; Semberg, E.; Laugen, A.T.; Miranda, J.R.D. Sample preservation, transport and processing strategies for honeybee RNA extraction: Influence on RNA yield, quality, target quantification and data normalization. J. Virol. Methods 2017. [Google Scholar] [CrossRef]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.M.; Birol, İ. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Hyatt, D.; LoCascio, P.F.; Hauser, L.J.; Uberbacher, E.C. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 2012, 28, 2223–2230. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tehel, A.; Vu, Q.; Bigot, D.; Gogol-Döring, A.; Koch, P.; Jenkins, C.; Doublet, V.; Theodorou, P.; Paxton, R. The Two Prevalent Genotypes of an Emerging Infectious Disease, Deformed Wing Virus, Cause Equally Low Pupal Mortality and Equally High Wing Deformities in Host Honey Bees. Viruses 2019, 11, 114. https://doi.org/10.3390/v11020114
Tehel A, Vu Q, Bigot D, Gogol-Döring A, Koch P, Jenkins C, Doublet V, Theodorou P, Paxton R. The Two Prevalent Genotypes of an Emerging Infectious Disease, Deformed Wing Virus, Cause Equally Low Pupal Mortality and Equally High Wing Deformities in Host Honey Bees. Viruses. 2019; 11(2):114. https://doi.org/10.3390/v11020114
Chicago/Turabian StyleTehel, Anja, Quynh Vu, Diane Bigot, Andreas Gogol-Döring, Peter Koch, Christina Jenkins, Vincent Doublet, Panagiotis Theodorou, and Robert Paxton. 2019. "The Two Prevalent Genotypes of an Emerging Infectious Disease, Deformed Wing Virus, Cause Equally Low Pupal Mortality and Equally High Wing Deformities in Host Honey Bees" Viruses 11, no. 2: 114. https://doi.org/10.3390/v11020114