Whole Genome Analysis of Cyclin Dependent Kinase (CDK) Gene Family in Cotton and Functional Evaluation of the Role of CDKF4 Gene in Drought and Salt Stress Tolerance in Plants
Abstract
:1. Introduction
2. Results
2.1. Identification and Sequence Analysis of CDK Proteins in Cotton Genome
2.2. Gene Structure and Amino Acid Motif Analysis of the CDK Genes in Cotton
2.3. Phylogenetic Analyses and Protein Alignments of the CDK-Proteins in Cotton with Other Plants
2.4. Chromosomal Distribution of Cotton Genes Encoding CDK Proteins
2.5. Gene Duplication, Orthologs, Paralogs, and Selection Type of the CDK Genes
2.6. Promoter (Cis-Element) Analysis
2.7. RNA Sequence Expression Profiling of the CDK Genes under Drought and Salt Stress Condition
2.8. RT-qPCR Analysis of the Cotton CDK Genes under Drought and Salt Stress
2.9. RT-qPCR Validation of the Highly Upregulated CDK Genes in Tisues of G. tomentosum (Tolerant) and G. hirsutum (Sensitive) to Drought and Salt Stresses
2.10. Experimental Determination of the Subcellular Localization of Gh_D12G2017 (CDKF4) Protein
2.11. RT-qPCR Analysis of the Gh_D12G2017 (CDKF4) Gene in Upland Cotton Tissues and Confirmation of the Transfomed Lines of the Model Plant
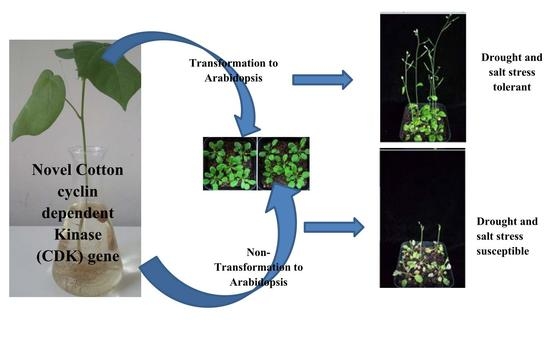
2.12. Response of the Overexpressed Lines and the Wild Type under Salt and Drought Stress Conditions
2.13. Evaluation of Stress Responsive Genes on the Tissues of Transgenic and Wild Arapidopsis Plants Under Salt and Drought Condition
3. Discussion
4. Materials and Methods
4.1. Identification and Sequence Analysis of CDK Proteins in Cotton Genome
4.2. Chromosomal Location and Gene Duplication of CDK Genes in Upland Cotton
4.3. Phylogenetic Analysis, Gene Structure, Motif and Functional Classification of CDK Proteins in Cotton
4.4. Signal Peptide and Promoter Cis-Element Analysis of the Cotton CDK Genes
4.5. Analysis of the Expression Patterns of Cotton CDK Genes in Different Tissues under Drought Stress Using RNA-seq Data
4.6. Plant Materials and Stress Treatments
4.7. RNA Isolation and RT-qPCR analysis
4.8. Validation of the Ortholog and Key CDK Genes in Cotton under Drought Stress
4.9. Clonining of the Highly Expressed Gene under Salt and Drought Stress, Gh_D12G2017 (CDKF4) in Arabidopsis thaliana (Ecotype Colombia-0) Lines
4.10. Response of Overexpressed Gh_D12G2017 (CDKF4) Transgenic Arabidopsis Lines to Drought and Salt Stress Condition
4.11. RT-qPCR Analysis of the Expression of Abiotics Stress Responsive Genes in Transgenic and Wild Type Lines
4.12. Experimental Determination of the Subcellular Location of the Transformed Gene by Use of Onion Epidermal Cell
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CDK | Cyclin Dependent Kinase |
| SOD | Superoxide Dismutase |
| ABRE 4 | Abscisic acid (ABA)-Responsive Element (ABRE)-Binding Transcription Factors 4 |
| LEA | Late Embryogenesis Abundant |
| RD29A | Responsive Drought 29A |
| POD | Peroxidase |
| CBL1 | Calcineurin B-like protein 1 |
| CAT | Catalase |
References
- Queralt, E.; Uhlmann, F. Cdk-counteracting phosphatases unlock mitotic exit. Curr. Opin. Cell Biol. 2008, 20, 661–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malumbres, M. Cyclin-dependent kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loyer, P.; Trembley, J.H.; Katona, R.; Kidd, V.J.; Lahti, J.M. Role of CDK/cyclin complexes in transcription and RNA splicing. Cell. Signal. 2005, 17, 1033–1051. [Google Scholar] [CrossRef] [PubMed]
- Morgan, D.O. CYCLIN-DEPENDENT KINASES: Engines, Clocks, and Microprocessors. Annu. Rev. Cell Dev. Biol. 1997, 13, 261–291. [Google Scholar] [CrossRef] [PubMed]
- Campo, S.; Baldrich, P.; Messeguer, J.; Lalanne, E.; Coca, M.; San Segundo, B. Overexpression of a Calcium-Dependent Protein Kinase Confers Salt and Drought Tolerance in Rice by Preventing Membrane Lipid Peroxidation. Plant Physiol. 2014, 165, 688–704. [Google Scholar] [CrossRef] [PubMed]
- Bartels, D.; Sunkar, R. Drought and salt tolerance in plants. CRC Crit. Rev. Plant Sci. 2005, 24, 23–58. [Google Scholar] [CrossRef]
- Zhou, M.; Sun, G.; Sun, Z.; Tang, Y.; Wu, Y. Cotton proteomics for deciphering the mechanism of environment stress response and fiber development. J. Proteom. 2014, 105, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Pandey, N.; Kumar, A.; Shirke, P.A. Physiological performance and differential expression profiling of genes associated with drought tolerance in root tissue of four contrasting varieties of two Gossypium species. Protoplasma 2016, 253, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Kuppu, S.; Mishra, N.; Hu, R.; Sun, L.; Zhu, X.; Shen, G.; Blumwald, E.; Payton, P.; Zhang, H. Water-Deficit Inducible Expression of a Cytokinin Biosynthetic Gene IPT Improves Drought Tolerance in Cotton. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Mir, R.R.; Zaman-Allah, M.; Sreenivasulu, N.; Trethowan, R.; Varshney, R.K. Integrated genomics, physiology and breeding approaches for improving drought tolerance in crops. Theor. Appl. Genet. 2012, 125, 625–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weston, D.J.; Gunter, L.E.; Rogers, A.; Wullschleger, S.D. Connecting Genes, Coexpression Modules, and Molecular Signatures to Environmental Stress Phenotypes in Plants. BMC Syst. Biol. 2008, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Sofia, A.; de Almeida, A.M.; da Silva, A.B.; da Silva, J.M.; Paula, A.; Santos, D.; Fevereiro, P.; Sousa Araujo, S. de Abiotic Stress Responses in Plants: Unraveling the Complexity of Genes and Networks to Survive. Abiotic Stress Plant Responses Appl. Agric. 2013, 49–101. [Google Scholar] [CrossRef]
- Sakai, S.; Harada, Y.; Biology, E.; Sakai, S.; Harada, Y. Sink-limitation and the size-number trade-off of organs: Production of organs using a fixed amount of reserves. Evolution 2001, 55, 467–476. [Google Scholar] [CrossRef]
- Tank, J.G.; Thaker, V.S. Cyclin dependent kinases and their role in regulation of plant cell cycle. Biol. Plant. 2011, 55, 201–212. [Google Scholar] [CrossRef]
- Ma, X.; Qiao, Z.; Chen, D.; Yang, W.; Zhou, R.; Zhang, W.; Wang, M. CYCLIN-DEPENDENT KINASE G2 regulates salinity stress response and salt mediated flowering in Arabidopsis thaliana. Plant Mol. Biol. 2015, 88, 287–299. [Google Scholar] [CrossRef] [PubMed]
- De Veylder, L.; Joubès, J.; Inzé, D. Plant cell cycle transitions. Curr. Opin. Plant Biol. 2003, 6, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Hayles, J.; Beach, D.; Durkacz, B.; Nurse, P. The fission yeast cell cycle control gene cdc2: Isolation of a sequence suc1 that suppresses cdc2 mutant function. Mol. Gen. Genet. 1986, 202, 291–293. [Google Scholar] [CrossRef] [PubMed]
- Hadwiger, J.A.; Wittenberg, C.; Mendenhall, M.D.; Reed, S.I. The Saccharomyces cerevisiae CKS1 gene, a homolog of the S. pombe suc1 gene, encodes a subunit of the cdc28 protein kinase complex. Mol. Cell. Biol. 1989, 9, 2034–2041. [Google Scholar] [CrossRef] [PubMed]
- De Veylder, L.; Segers, G.; Glab, N.; Casteels, P.; Van Montagu, M.; Inzé, D. The arabidopsis Cks1At protein binds the cyclin-dependent kinases Cdc2aAt and Cdc2bAt. FEBS Lett. 1997, 412, 446–452. [Google Scholar] [CrossRef]
- De Veylder, L.; Beemster, G.T.S.; Beeckman, T.; Inzé, D. CKS1At overexpression in Arabidopsis thaliana inhibits growth by reducing meristem size and inhibiting cell-cycle progression. Plant J. 2001, 25, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Sacks, M.M.; Silk, W.K.; Burman, P. Effect of Water Stress on Cortical Cell Division Rates within the Apical Meristem of Primary Roots of Maize. Plant Physiol. 1997, 114, 519–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuppler, U.; He, P.; John, P.; Munns, R. Effect of water stress on cell division and cell-division-cycle 2-like cell-cycle kinase activity in wheat leaves. Plant Physiol. 1998, 117, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.F.; Jin, Y.H.; Yoo, C.Y.; Lin, X.-L.; Kim, W.-Y.; Yun, D.-J.; Bressan, R.A.; Hasegawa, P.M.; Jin, J.B. CYCLIN H;1 Regulates Drought Stress Responses and Blue Light-Induced Stomatal Opening by Inhibiting Reactive Oxygen Species Accumulation in Arabidopsis. Plant Physiol. 2013, 162, 1030–1041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Li, Y.; Xie, Q.; Wu, Y. Loss of CDKC;2 increases both cell division and drought tolerance in Arabidopsis thaliana. Plant J. 2017, 91, 816–828. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39. [Google Scholar] [CrossRef] [PubMed]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Dong, Q.; Hu, Y.; Zhou, Z.; Cai, X.; Wang, X.; Hou, Y.; Wang, K.; et al. Cotton late embryogenesis abundant (lea2) genes promote root growth and confers drought stress tolerance in transgenic Arabidopsis Thaliana. G3 (Bethesda) 2018, 8, g3.200423.2018. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Genome-Wide Analysis of the Cyclin Family in Arabidopsis and Comparative Phylogenetic Analysis of Plant Cyclin-Like Proteins. Plant Physiol. 2004, 135, 1084–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuxreiter, M.; Simon, I.; Friedrich, P.; Tompa, P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J. Mol. Biol. 2004, 338, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Qi, G.; Kong, Y.; Kong, D.; Gao, Q.; Zhou, G. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 2010, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Stamatoyannopoulos, J.A.; Bailey, T.L.; Noble, W.S. Quantifying similarity between motifs. Genome Biol. 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Gu, Y.H.; Wang, H.Y.; Zheng, W.; Li, X.; Zhao, C.W.; Zhang, Y.Z. Digital gene expression analysis based on integrated de novo transcriptome assembly of sweet potato [Ipomoea batatas (L.) Lam]. PLoS ONE 2012, 7, e36234. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-K.; Zhang, Y.; Zhang, M.; Goebel, M.; Kim, H.J.; Triplett, B.A.; Stelly, D.M.; Zhang, H.-B. Construction of a plant-transformation-competent BIBAC library and genome sequence analysis of polyploid Upland cotton (Gossypium hirsutum L.). BMC Genom. 2013, 14, 208. [Google Scholar] [CrossRef] [PubMed]
- Salama, S.R.; Hendricks, K.B.; Thorner, J. G1 cyclin degradation: The PEST motif of yeast Cln2 is necessary, but not sufficient, for rapid protein turnover. Mol. Cell. Biol. 1994, 14, 7953–7966. [Google Scholar] [CrossRef] [PubMed]
- Bies-Ethève, N.; Gaubier-Comella, P.; Debures, A.; Lasserre, E.; Jobet, E.; Raynal, M.; Cooke, R.; Delseny, M. Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol. Biol. 2008, 67, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Philippe, R.; Courtois, B.; McNally, K.L.; Mournet, P.; El-Malki, R.; Le Paslier, M.C.; Fabre, D.; Billot, C.; Brunel, D.; Glaszmann, J.C.; et al. Structure, allelic diversity and selection of Asr genes, candidate for drought tolerance, in Oryza sativa L. and wild relatives. Theor. Appl. Genet. 2010, 121, 769–787. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Adams, K.L. Expression Partitioning between Genes Duplicated by Polyploidy under Abiotic Stress and during Organ Development. Curr. Biol. 2007, 17, 1669–1674. [Google Scholar] [CrossRef] [PubMed]
- Salih, H.; Gong, W.; He, S.; Sun, G.; Sun, J.; Du, X. Genome-wide characterization and expression analysis of MYB transcription factors in Gossypium hirsutum. BMC Genet. 2016, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Li, X. Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum). Planta 2014, 241, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Hundertmark, M.; Hincha, D.K. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Zhang, Q.; Cheng, T.; Pan, H.; Yang, W.; Sun, L. Genome-wide identification and analysis of late embryogenesis abundant (LEA) genes in Prunus mume. Mol. Biol. Rep. 2013, 40, 1937–1946. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ng, C.K.Y.; Fan, L.M. MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 2014, 114, 80–91. [Google Scholar] [CrossRef]
- De Azevedo Neto, A.D.; Prisco, J.T.; Enéas-Filho, J.; Abreu, C.E.B.D.; Gomes-Filho, E. Effect of salt stress on antioxidative enzymes and lipid peroxidation in leaves and roots of salt-tolerant and salt-sensitive maize genotypes. Environ. Exp. Bot. 2006, 56, 87–94. [Google Scholar] [CrossRef]
- Tamirisa, S.; Vudem, D.R.; Khareedu, V.R. A Cyclin Dependent Kinase Regulatory Subunit (CKS) Gene of Pigeonpea Imparts Abiotic Stress Tolerance and Regulates Plant Growth and Development in Arabidopsis. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tian, L.-H.; Zhao, J.-F.; Song, Y.; Zhang, C.-J.; Guo, Y. Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol. 2009, 149, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Zörb, C.; Schmitt, S.; Neeb, A.; Karl, S.; Linder, M.; Schubert, S. The biochemical reaction of maize (Zea mays L.) to salt stress is characterized by a mitigation of symptoms and not by a specific adaptation. Plant Sci. 2004, 167, 91–100. [Google Scholar] [CrossRef]
- Xu, C.; Sibicky, T.; Huang, B. Protein profile analysis of salt-responsive proteins in leaves and roots in two cultivars of creeping bentgrass differing in salinity tolerance. Plant Cell Rep. 2010, 29, 595–615. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi, M.R.; Corral-Martínez, P.; Mousavi, A.; Salmanian, A.H.; Moieni, A.; Seguí-Simarro, J.M. An efficient method for transformation of pre-androgenic, isolated Brassica napus microspores involving microprojectile bombardment and Agrobacterium-mediated transformation. Acta Physiol. Plant. 2009, 31, 1313–1317. [Google Scholar] [CrossRef]
- Kurusu, T.; Nishikawa, D.; Yamazaki, Y.; Gotoh, M.; Nakano, M.; Hamada, H.; Yamanaka, T.; Iida, K.; Nakagawa, Y.; Saji, H.; et al. Plasma membrane protein OsMCA1 is involved in regulation of hypo-osmotic shock-induced Ca2+ influx and modulates generation of reactive oxygen species in cultured rice cells. BMC Plant Biol. 2012, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganança, J.F.T.; Freitas, J.G.R.; Nóbrega, H.G.M.; Rodrigues, V.; Antunes, G.; Gouveia, C.S.S.; Rodrigues, M.; Chaïr, H.; De Carvalho, M.Â.A.P.; Lebot, V. Screening for drought tolerance in thirty three taro cultivars. Not. Bot. Horti Agrobot. Cluj-Napoca 2018, 46, 65–74. [Google Scholar] [CrossRef]
- Pandey, H.C.; Baig, M.J.; Chandra, A.; Bhatt, R.K. Drought stress induced changes in lipid peroxidation and antioxidant system in genus Avena. J. Environ. Biol. 2010, 31, 435–440. [Google Scholar] [PubMed]
- Lu, P.; Magwanga, R.O.; Lu, H.; Kirungu, J.N.; Wei, Y.; Dong, Q.; Wang, X.; Cai, X.; Zhou, Z.; Wang, K.; et al. A novel G-protein-coupled receptors gene from upland cotton enhances salt stress tolerance in transgenic Arabidopsis. Genes (Basel) 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Lu, H.; Wang, X.; Cai, X.; Zhou, Z.; Zhang, Z.; Salih, H.; Wang, K.; et al. Characterization of the late embryogenesis abundant (LEA) proteins family and their role in drought stress tolerance in upland cotton. BMC Genet. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Rong, J.; Feltus, F.A.; Liu, L.; Lin, L.; Paterson, A.H. Gene copy number evolution during tetraploid cotton radiation. Heredity (Edinb) 2010, 105, 463–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Song, J.; Wang, F.; Zhang, X.S. Genome-wide identification and expression analysis of rice cell cycle genes. Plant Mol. Biol. 2007, 64, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Lehti-Shiu, M.D.; Thomashow, M.; Shiu, S.-H. Evolution of stress-regulated gene expression in duplicate genes of Arabidopsis thaliana. PLoS Genet. 2009, 5, e1000581. [Google Scholar] [CrossRef] [PubMed]
- Flagel, L.E.; Wendel, J.E. Gene duplication and evolutionaary novelty in plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanc, G.; Hokamp, K.; Wolfe, K.H. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003, 13, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Vandepoele, K.; Raes, J.; De Veylder, L.; Rouze, P.; Rombauts, S.; Inze, D. Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 2002, 14, 903–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dossa, K.; Diouf, D.; Cissé, N. Genome-Wide Investigation of Hsf Genes in Sesame Reveals Their Segmental Duplication Expansion and Their Active Role in Drought Stress Response. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Grant, D.; Cregan, P.; Shoemaker, R.C. Genome organization in dicots: Genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc. Natl. Acad. Sci. USA 2000, 97, 4168–4173. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wang, X.; Lu, Y.; Cai, X.; Ye, Z.; Zhang, J. Genome-wide analysis of the cyclin gene family in tomato. Int. J. Mol. Sci. 2013, 15, 120–140. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Gao, J.; Zeng, Q.Y. Genome-wide analysis of the LEA (late embryogenesis abundant) protein gene family in Populus trichocarpa. Tree Genet. Genomes 2013, 9, 253–264. [Google Scholar] [CrossRef]
- Wahl, M.C.; Will, C.L.; Lührmann, R. The Spliceosome: Design Principles of a Dynamic RNP Machine. Cell 2009, 136, 701–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guimarães-Dias, F.; Neves-Borges, A.C.; Viana, A.A.B.; Mesquita, R.O.; Romano, E.; de Fátima Grossi-de-Sá, M.; Nepomuceno, A.L.; Loureiro, M.E.; Alves-Ferreira, M. Expression analysis in response to drought stress in soybean: Shedding light on the regulation of metabolic pathway genes. Genet. Mol. Biol. 2012, 35, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Hajiboland, R. Chapter 1—Reactive Oxygen Species and Photosynthesis. In Oxidative Damage to Plants; Parvaiz, A., Ed.; Elsevier: Waltham, MA, USA, 2014; pp. 1–63. ISBN 978-0-12-799963-0. [Google Scholar]
- Jo, B.H.; Van Lerberghe, L.M.; Motsegood, K.M.; Beebe, D.J.; Fuchs, T.A.; Abed, U.; Goosmann, C.; Hurwitz, R.; Schulze, I.; Wahn, V.; et al. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 2010, 2, 231–241. [Google Scholar] [CrossRef]
- Msanne, J.; Lin, J.; Stone, J.M.; Awada, T. Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 2011, 234, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Hao, R.; Cheng, T.; Pan, H.; Yang, W.; Wang, J.; Zhang, Q. Genome-Wide Analysis of the AP2/ERF Gene Family in Prunus mume. Plant Mol. Biol. Rep. 2013, 31, 741–750. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R Software; R Foundation for Statistical Computing: Vienna, Austria, 2008; Volume 739, p. 409. [Google Scholar]
- Hoagland, D.R.; Arnon, D.I. The Water-Culture Method for Growing Plants Without Soil, 2nd ed.; CABI: California, CA, USA, 1950; Volume 347, ISBN 19500302257. [Google Scholar]
- Parent, B.; Suard, B.; Serraj, R.; Tardieu, F. Rice leaf growth and water potential are resilient to evaporative demand and soil water deficit once the effects of root system are neutralized. Plant Cell Environ. 2010, 33, 1256–1267. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Benoit, R.M.; Ostermeier, C.; Geiser, M.; Li, J.S.Z.; Widmer, H.; Auer, M. Seamless insert-plasmid assembly at high efficiency and low cost. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Cao, Z.; Li, Y.; Zhao, Y.; Zhang, H. A simple and effective method for protein subcellular localization using Agrobacterium-mediated transformation of onion epidermal cells. Biologia (Bratisl) 2007, 62, 529–532. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.Y.; Yu, H.Y.; Yang, M.M.; Kong, D.S.; Zhang, Y.J. Effect of Drought Stress on Lipid Peroxidation, Osmotic Adjustment and Antioxidant Enzyme Activity of Leaves and Roots of Lycium ruthenicum Murr. Seedling. Russ. J. Plant Physiol. 2018, 65, 244–250. [Google Scholar] [CrossRef]
- de BritoI, G.G.; Sofiatti, V.; de Andrade Lima, M.M.; de Carvalho, L.P.; da Silva Filho, J.L. Physiological traits for drought phenotyping in cotton. Acta Sci. Agron. 2011, 33, 117–125. [Google Scholar] [CrossRef]











| Cotton Species | Gene ID | Gene Name | Description | Chro. | Start | End | Strand | Length (bp) |
|---|---|---|---|---|---|---|---|---|
| G. hirsutum (AD) | Gh_A09G0392 | CDKB1-2 | Cyclin-dependent kinase B1-2 | A09 | 25,358,935 | 25,360,412 | + | 1478 |
| Gh_D04G0378 | CDKB1-2 | Cyclin-dependent kinase B1-2 | D04 | 5,886,575 | 5,888,627 | + | 2053 | |
| Gh_Sca005019G02 | CDKB1-2 | Cyclin-dependent kinase B1-2 | scaffold | 8,409 | 9,886 | − | 1478 | |
| Gh_A08G1333 | CDKC-1 | Cyclin-dependent kinase C-1 | A08 | 86,315,556 | 86,319,915 | − | 4360 | |
| Gh_D08G1628 | CDKC-2 | Cyclin-dependent kinase C-2 | D08 | 51,109,124 | 51,113,530 | − | 4407 | |
| Gh_A09G0498 | CDKD-1 | Cyclin-dependent kinase D-1 | A09 | 39,565,295 | 39,569,539 | − | 4245 | |
| Gh_A09G1581 | CDKD-1 | Cyclin-dependent kinase D-1 | A09 | 69,169,679 | 69,177,142 | + | 7464 | |
| Gh_D09G0505 | CDKD-1 | Cyclin-dependent kinase D-1 | D09 | 24,483,522 | 24,487,751 | − | 4230 | |
| Gh_D09G1668 | CDKD-1 | Cyclin-dependent kinase D-1 | D09 | 44,329,951 | 44,337,731 | + | 7781 | |
| Gh_A09G1688 | CDKE-1 | Cyclin-dependent kinase E-1 | A09 | 70,391,997 | 70,393,439 | + | 1443 | |
| Gh_A13G0098 | CDKE-1 | Cyclin-dependent kinase E-1 | A13 | 1,179,217 | 1,180,641 | + | 1425 | |
| Gh_D09G1794 | CDKE-1 | Cyclin-dependent kinase E-1 | D09 | 45,602,368 | 45,603,810 | + | 1443 | |
| Gh_D13G0113 | CDKE-1 | Cyclin-dependent kinase E-1 | D13 | 1,148,889 | 1,150,328 | + | 1440 | |
| Gh_A05G0178 | CDKF-1 | Cyclin-dependent kinase F-1 | A05 | 1,861,427 | 1,863,742 | − | 2316 | |
| Gh_A07G0040 | CDKF-1 | Cyclin-dependent kinase F-1 | A07 | 573,679 | 576,822 | + | 3144 | |
| Gh_D05G0242 | CDKF-1 | Cyclin-dependent kinase F-1 | D05 | 2,194,647 | 2,196,286 | − | 1640 | |
| Gh_D07G0069 | CDKF-1 | Cyclin-dependent kinase F-1 | D07 | 683,455 | 686,592 | − | 3138 | |
| Gh_A03G1965 | CDKF-4 | Cyclin-dependent kinase F-4 | A03 | 164,071 | 171,343 | + | 7273 | |
| Gh_A08G1357 | CDKF-4 | Cyclin-dependent kinase F-4 | A08 | 87,774,325 | 87,778,918 | + | 4594 | |
| Gh_A12G1847 | CDKF-4 | Cyclin-dependent kinase F-4 | A12 | 81,080,441 | 81,083,558 | + | 3118 | |
| Gh_D03G1838 | CDKF-4 | Cyclin-dependent kinase F-4 | D03 | 28,784 | 31,900 | − | 3117 | |
| Gh_D08G1653 | CDKF-4 | Cyclin-dependent kinase F-4 | D08 | 52,031,672 | 52,034,687 | + | 3016 | |
| Gh_D12G2017 | CDKF-4 | Cyclin-dependent kinase F-4 | D12 | 53,176,398 | 53,179,530 | + | 3133 | |
| Gh_A03G1115 | CDKG-2 | Cyclin-dependent kinase G-2 | A03 | 81,001,450 | 81,004,726 | − | 3277 | |
| Gh_A04G1202 | CDKG-2 | Cyclin-dependent kinase G-2 | A04 | 62,205,596 | 62,208,956 | + | 3361 | |
| Gh_A07G0469 | CDKG-2 | Cyclin-dependent kinase G-2 | A07 | 6,070,719 | 6,072,909 | − | 2191 | |
| Gh_A12G1705 | CDKG-2 | Cyclin-dependent kinase G-2 | A12 | 78,774,164 | 78,776,378 | + | 2215 | |
| Gh_D02G1543 | CDKG-2 | Cyclin-dependent kinase G-2 | D02 | 53,453,957 | 53,457,227 | − | 3271 | |
| Gh_D04G1812 | CDKG-2 | Cyclin-dependent kinase G-2 | D04 | 50,433,225 | 50,436,586 | + | 3362 | |
| Gh_D07G0534 | CDKG-2 | Cyclin-dependent kinase G-2 | D07 | 6,034,826 | 6,036,921 | − | 2096 | |
| Gh_D12G1867 | CDKG-2 | Cyclin-dependent kinase G-2 | D12 | 51,280,460 | 51,282,677 | + | 2218 | |
| G. raimondii (DD) | Gorai.012G047600 | CDKB1-2 | Cyclin-dependent kinase B1-2 | Chr12 | 5,984,971 | 5,987,585 | + | 2615 |
| Gorai.004G175700 | CDKC-1 | Cyclin-dependent kinase C-1 | Chr04 | 47,893,134 | 47,898,215 | − | 5082 | |
| Gorai.006G057400 | CDKD-1 | Cyclin-dependent kinase D-1 | Chr06 | 21,087,092 | 21,092,990 | + | 5899 | |
| Gorai.006G193300 | CDKD-1 | Cyclin-dependent kinase D-1 | Chr06 | 45,020,096 | 45,028,617 | + | 8522 | |
| Gorai.006G206400 | CDKE-1 | Cyclin-dependent kinase E-1 | Chr06 | 46,208,009 | 46,210,865 | + | 2857 | |
| Gorai.013G013100 | CDKE-1 | Cyclin-dependent kinase E-1 | Chr13 | 885,517 | 888,039 | + | 2523 | |
| Gorai.001G006600 | CDKF-1 | Cyclin-dependent kinase F-1 | Chr01 | 593,026 | 596,558 | − | 3533 | |
| Gorai.009G026100 | CDKF-1 | Cyclin-dependent kinase F-1 | Chr09 | 1,996,952 | 1,999,805 | − | 2,854 | |
| Gorai.003G187100 | CDKF-4 | Cyclin-dependent kinase F-4 | Chr03 | 45,739,175 | 45,742,291 | + | 3117 | |
| Gorai.004G178800 | CDKF-4 | Cyclin-dependent kinase F-4 | Chr04 | 48,759,184 | 48,764,623 | + | 5440 | |
| Gorai.008G220700 | CDKF-4 | Cyclin-dependent kinase F-4 | Chr08 | 50,729,473 | 50,732,623 | + | 3151 | |
| Gorai.001G060800 | CDKG-2 | Cyclin-dependent kinase G-2 | Chr01 | 6,021,347 | 6,025,975 | − | 4629 | |
| Gorai.005G170300 | CDKG-2 | Cyclin-dependent kinase G-2 | Chr05 | 49,845,864 | 49,852,364 | − | 6501 | |
| Gorai.008G205000 | CDKG-2 | Cyclin-dependent kinase G-2 | Chr08 | 49,026,157 | 49,029,075 | + | 2919 | |
| Gorai.012G174900 | CDKG-2 | Cyclin-dependent kinase G-2 | Chr12 | 34,446,565 | 34,450,903 | + | 4339 | |
| G. arboretum (AA) | Cotton_A_13039 | CDKD-1 | Cyclin-dependent kinase D-1 | Chr11 | 62,348,669 | 62,353,166 | + | 4498 |
| Cotton_A_13038 | CDKD-1 | Cyclin-dependent kinase D-1 | Chr11 | 62,353,469 | 62,360,835 | + | 7367 | |
| Cotton_A_11170 | CDKF-1 | Cyclin-dependent kinase F-1 | Chr11 | 56,386,909 | 56,389,193 | + | 2285 | |
| Cotton_A_07964 | CDKF-4 | Cyclin-dependent kinase F-4 | Chr06 | 49,941,928 | 49,944,975 | + | 3048 | |
| Cotton_A_25379 | CDKF-4 | Cyclin-dependent kinase F-4 | Chr10 | 112,331,167 | 112,334,076 | − | 2910 | |
| Cotton_A_14275 | CDKF-4 | Cyclin-dependent kinase F-4 | Chr07 | 19,382,779 | 19,385,783 | + | 3005 | |
| Cotton_A_01035 | CDKE-1 | Cyclin-dependent kinase E-1 | Chr13 | 75,011,456 | 75,013,871 | − | 2416 | |
| Cotton_A_10347 | CDKF-1 | Cyclin-dependent kinase F-1 | Chr01 | 768,252 | 771,750 | − | 3499 | |
| Cotton_A_13019 | CDKC-2 | Cyclin-dependent kinase C-2 | Chr03 | 85,375,450 | 85,379,809 | − | 4360 | |
| Cotton_A_19907 | CDKG-2 | Cyclin-dependent kinase G-2 | Chr01 | 90,376,726 | 90,378,745 | + | 2020 | |
| Cotton_A_08058 | CDKG-2 | Cyclin-dependent kinase G-2 | Chr06 | 114,700,563 | 114,702,777 | + | 2215 | |
| Cotton_A_14138 | CDKG-2 | Cyclin-dependent kinase G-2 | Chr12 | 122,143,663 | 122,147,697 | + | 4035 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Cai, X.; Zhou, Z.; Wang, X.; Diouf, L.; Xu, Y.; Hou, Y.; Hu, Y.; et al. Whole Genome Analysis of Cyclin Dependent Kinase (CDK) Gene Family in Cotton and Functional Evaluation of the Role of CDKF4 Gene in Drought and Salt Stress Tolerance in Plants. Int. J. Mol. Sci. 2018, 19, 2625. https://doi.org/10.3390/ijms19092625
Magwanga RO, Lu P, Kirungu JN, Cai X, Zhou Z, Wang X, Diouf L, Xu Y, Hou Y, Hu Y, et al. Whole Genome Analysis of Cyclin Dependent Kinase (CDK) Gene Family in Cotton and Functional Evaluation of the Role of CDKF4 Gene in Drought and Salt Stress Tolerance in Plants. International Journal of Molecular Sciences. 2018; 19(9):2625. https://doi.org/10.3390/ijms19092625
Chicago/Turabian StyleMagwanga, Richard Odongo, Pu Lu, Joy Nyangasi Kirungu, Xiaoyan Cai, Zhongli Zhou, Xingxing Wang, Latyr Diouf, Yanchao Xu, Yuqing Hou, Yangguang Hu, and et al. 2018. "Whole Genome Analysis of Cyclin Dependent Kinase (CDK) Gene Family in Cotton and Functional Evaluation of the Role of CDKF4 Gene in Drought and Salt Stress Tolerance in Plants" International Journal of Molecular Sciences 19, no. 9: 2625. https://doi.org/10.3390/ijms19092625