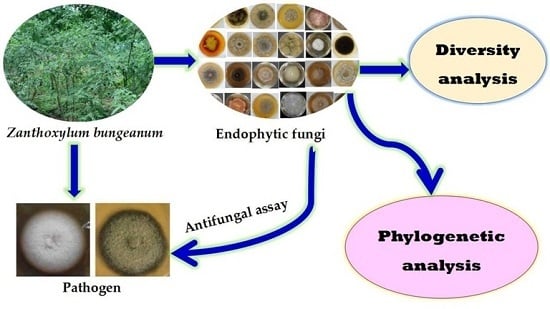
1. Introduction
Endophytic fungi have been reported as novel sources of bioactive compounds to be applied in the agricultural field. It has been frequently reported that endophytic fungi can protect host plants against pests by producing protective compounds, conferring the resistances of host plants to biotic or abiotic stresses by enhancing defensive system and improving the growth and product yield of plants directly or indirectly [
1,
2,
3]. Given the sideeffects of indiscriminate use of conventional chemical fungicides, i.e., contaminating environment, damaging human health, inducing pathogen resistance to fungicides, and causing resurgence of plant disease, the exploitation of natural bio-control agents has become an overwhelming trend in integrated pest management [
4,
5,
6]. Plant endophytic fungi are just such natural resources of bio-control agents. Substantial renewed attention has been paid to the inhibitory activity of endophytic fungi against pathogenic fungi and their potential in the biological control of plant diseases [
7,
8].
Zanthoxylum bungeanum is an aromatic plant of the family Rutaceae and is native to southwestern China. It has a long history as a pungent foodstuff and seasoning in Korea, China, and other East Asian countries [
9]. Phytochemical studies of
Z. bungeanum have been carried out in recent years including compound isolation, structure elucidation, extraction of essential oils and its pharmacological activities, and extraction optimization of polysaccharide and antioxidant activities, which demonstrated the importance of this plant due to its huge economic value [
10,
11,
12,
13]. However, different pathogenic plant fungi frequently infect
Z. bungeanum during its growth process, which causes serious effects on yield and quality [
14]. The most reported pathogenic fungi isolated from
Z. bungeanum were
Pseudocercospora zanthoxyli and
Fusarium sambucinum in the Shaanxi and Gansu districts, which resulted in prickly ash leaf mold and stem dry rot, respectively [
15,
16]. The use of chemical pesticides is the current main method of controlling the aforementioned pathogenic fungi [
17]. Considering the sideeffects of chemical fungicides, it is necessary to explore new nontoxic and efficient alternatives to synthetic pesticides to control plant pathogenic fungi.
Until recently, there have been few reports about the endophytic fungi of
Z. bungeanum. One study about endophytic fungi from pericarpium zanthoxyli was reported by Liu et al. [
18], in which 12 endophytic fungal isolates were obtained from pericarpium zanthoxyli and found one isolate that could produce a volatile, fragrant metabolite. However, the endophytic fungi from pericarpium zanthoxyli in this report were not identified, and the variety and development stage of zanthoxyli were also not introduced clearly, both of which are considered important factors affecting the number and species of endophytic fungi obtained from plant tissues [
18,
19]. Hence, it is critical to conduct systematic studies on the biodiversity analysis and antifungal activities of endophytic fungi from
Z. bungeanum. The objectives of this research were to explore the α diversity and phylogenetic relationships of endophytic fungi associated with
Z. bungeanum different tissues and to screen for isolates with obvious antifungal activity against the pathogenic fungi
P. zanthoxyli and
F. sambucinum. Our present research is aimed at further investigating the evolution of endophytic fungi communities in plant micro-ecological systems and providing valuable information for the exploitation of effective natural bio-control agents for
Z. bungeanum diseases.
3. Discussion
Plant endophytic fungi are highly taxonomically diverse and are also demonstrated to adjust the morphological and physiological functions of the host plant through multiple mechanisms, including stimulating its resistance to biotic and abiotic stresses [
24]. It is important to explore endophytic fungi from different plants to obtain many natural resources as well as to understand the biodiversity of endophytic fungal community in a symbiotic relationship. Considering the fragrant specificity of
Z. bungeanum, there is a possibility to isolate novel endophytic fungi that possess special functions. Isolating and identifying new endophytic fungi from
Z. bungeanum might lead to the discovery of new and unusual compounds with biotechnological and pharmaceutical applications. In our present study, 940 endophytic fungal isolates were obtained from the roots, stems, leaves, fruits, and thorns of
Z. bungeanum by a culture-dependent method, which were subsequently categorized into 93 morphotypes, 43 species, 23 genera, eight orders, four classes, and two phyla. A high number of endophytic fungal species were encountered during this relatively small survey, despite the fact that the methodology employed in our research is culture-specific and slow growing, and some non-culturable species are likely to be missed [
25]. Diversity analysis shows that endophytic fungi residing in
Z. bungeanum are highly diverse. It has been widely reported that culture-dependent methods for isolating microbes from surface-sterilized plant tissues result in a large quantity of endophytic fungi, and researchers have analyzed their biodiversity [
24,
26]. Specifically, the endophytic fungi from the roots, stems, leaves, fruits, and thorns of
Z. bungeanum were separately preserved and identified, which exhibited obvious tissue specificity. The endophytic fungi tissue specificity in different parts of
Z. bungeanum may be caused by differences in the plant tissue microenvironment. Host plant identity and tissues sampled are major driving factors for the endophytic fungal community composition and dynamics. Similar results on the microecological distribution of endophytes in different tissues in
Angelica sinensis and
Azadirachta indica were also observed, which also demonstrated that endophytes have tissue specificity [
19,
27]. The abundance, richness, species composition, and diversity of endophytic assemblages of
Z. bungeanum were found to be significantly dependent on the sample tissue. Earlier studies have proposed that possible reasons for the diversity are the physiology and chemistry of the colonized tissues, different plant inhabitants or a different environment might influence endophyte recruitment [
28,
29].
All of endophytic fungi isolated from
Z. bungeanum belonged mainly to the Ascomycota, within in the classes of Sordariomycetes and Dothideomycetes by morphological and ITS sequence identification. Other researchers also reported that Sordariomycetes and Dothideomycetes are the main groups of endophytic fungi from other plants [
26,
30]. Endophytic fungal species abundance distribution was widely reported to be skewed, with many frequent species and several incidental species, which might be related to the sampling size and method [
20]. There is obvious tissue specificity of endophytic fungi genera in
Z. bungeanum. Similar results have also been widely reported in other plants [
7]. The genera of
Fusarium and
Alternaria are common in the stems, roots, leaves, fruits, and thorns of
Z. bungeanum. The genera
Bionectria,
Rosellinia,
Paraphoma,
Rhizopycnis, and
Acrocalymma were only isolated from the roots. Seven genera,
Nectria,
Clonostachys,
Sarocladium,
Leptosphaerulina,
Epicoccum,
Botryosphaeria, and
Irpex, were specific to the stems. The genus
Auricularia was only isolated from the leaves. Other genera can be isolated from two or three different tissues of
Z. bungeanum. In the present study,
Alternaria,
Fusarium, and
Phoma were frequently isolated species with high relative abundances of 30.85%, 13.71%, and 12.77%, respectively (
Figure 3F), all of which have a cosmopolitan distribution and are found in association with a wide variety of host plants [
31,
32,
33].
Alternaria and
Fusarium are reported to be the most frequent and common genera of endophytic fungi from different plant species as well as various environmental conditions [
21,
34]. Several other genera were also isolated from
Z. bungeanum, including
Gibberella,
Nectria,
Clonostachys,
Bionectria,
Phomopsis,
Cytospora,
Diaporthe,
Rosellinia,
Sarocladium,
Peyronellaea,
Leptosphaerulina,
Epicoccum,
Paraphoma,
Rhizopycnis,
Acrocalymma,
Dothiorella,
Botryosphaeria,
Aspergillus,
Irpex, and
Auricularia. Although several endophytic fungal genera were isolated with low relative abundance, those minor genera may have an important ecological role for their host plants or could be capable of synthesizing bioactive compounds [
34]. Some of the above endophytic fungal genera are reported to be commonly associated with plant disease symptoms in several plants. For example,
F. proliferatum is a common pathogen of numerous crops and an agent of wilt, blight, and diebacks of palm trees [
35]. The
Diaporthe and
Phomopsis complex are causal agents of seed decay and cause soybean blight and canker diseases [
36].
Botryosphaeria and its anamorph complex are especially responsible for symptoms such as fruit rot, shoot blight, dieback, and canker of numerous woody hosts [
37]. However, it is not inconsistent that endophytic fungi could also be pathogens because plant pathogens and endophytes might convert mutually by influencing a favorable outer environment or plant disease conditions [
38].
In recent years, there has been an increasing demand for identifying new antimicrobial agents due to the development of pathogen resistance to available pesticides. Although many chemically synthesized pesticides have been generated, their side effects have also been frequently reported, including pesticide residue, pathogenic resistance to pesticide, or resurgence of pests [
4]. Endophytic fungi have been considered a novel resource of natural antimicrobial compounds with efficient and environmentally friendly characteristics [
39,
40]. It was observed that the ethyl acetate (EtOAc) extract of the endophytic fungus
Trichoderma harzianum offered excellent control of the tomato gray mold caused by
Botrytis cinerea without fungicide resistance and in an environmentally friendly manner [
41]. Santiago et al. [
42] also found one endophytic fungal isolate from
Cinnamomum mollissimum that possessed efficient killing ability against the pathogenic fungus
Aspergillus niger. Pan et al. [
43] found that an EtOAc extract of the endophytic fungus
Chaetomium globosum from
Houttuynia cordata showed a wide antifungal spectrum. The specific secondary metabolites of endophytic fungi, such as helvolic acid, fumitremorgin B, verruculogen, and spirobisnaphthalenes, were also found to exhibit strong antifungal activity against multifarious plant pathogenic fungi [
44,
45,
46]. In the present work, we investigated the inhibitory effects of the crude EtOAc extracts of endophytic fungi from
Z. bungeanum on host-specific pathogenic fungi
F. sambucinum and
P. zanthoxyli. Several endophytic fungi were observed with inhibitory effects on both of the pathogens. However, there were only five endophytic isolates that showed obvious strong inhibitory effects (
IR > 50%) on
F. sambucinum: Zbf-S11 (
Epicoccum nigrum), Zbf-S27 (
Diaporthe sp.), Zbf-S47 (
Peyronellaea glomerata), Zbf-S48 (
Phomopsis sp.), and Zbf-S49 (
Phomopsis vaccinii) (
Figure 6A and
Figure 7A). There were four endophytic fungal isolates that exhibited strong inhibitory effects on
P. zanthoxyli: Zbf-S1 (
Fusarium sp.), Zbf-S11 (
Epicoccum nigrum), Zbf-R1 (
Fusarium sp.), and Zbf-T3 (
Diaporthe cotoneastri) (
Figure 6B and
Figure 7B). The endophytic isolate Zbf-S11
E. Nigrum has excellent inhibitory effects on both pathogens and is worth further investigation. Inhibitory rate dynamics of all eight endophytic fungi with strong inhibitory effects showed the characteristic of long-lasting efficiency, which might be attributed to the existence of antifungal compound produced by endophytic fungi. For example, two polyketides with prominent inhibitory activity were isolated from the endophytic fungus
Cryptosporiopsis sp. obtained from
Zanthoxylum leprieurii [
47]. Antimicrobial fusaruside was characterized from the chloroform-methanol extract of endophytic
Fusarium sp. IFB-121 of
Quercus variabilis [
48]. It is worth further isolating and characterizing secondary metabolites of
Z. bungeanum endophytic fungi and establishing more bioactivity testing models to explore natural resources. It may be possible to utilize the endophytic fungi of
Z. bungeanum as biocontrol agents to control its pathogenic fungi.
The present study is the first report to systematically analyze the biodiversity and antifungal activity of endophytic fungi isolated from
Z. bungeanum using culture-dependent methods. This study demonstrated the tissue specificity of endophytic fungi in different parts of
Z. bungeanum. All of the endophytic fungi of
Z. bungeanum were identified by morphological observation and rDNA ITS identification. Moreover, the identification of endophytic fungi can be confirmed by the in-depth physiological metabolism, biochemical function detection, and sequence analyses of multiple gene regions in the future [
49,
50]. Using a culture-dependent method might miss some unculturable endophytic fungi, which might influence the endophytic fungi diversity results. Nevertheless, we have obtained numerous endophytic fungal isolates from
Z. bungeanum. It is important to directly study the composition and structure of microbial populations at the genetic level by constructing the clone library and bypassing the step of strain isolation and plate cultivation, which is convenient, efficient, and more suitable for fungal diversity analysis due to the higher richness. We are carrying out the research using the method of clone libraries and rDNA ITS sequencing to systematically analyze the diversity of endophytic fungi of
Z. bungeanum. It has also been reported that some genera might be excluded from clone libraries but could be isolated by pure cultivation [
23]. The combination of the two methods would be complementary in achieving a better understanding of the diversity of fungal communities of
Z. bungeanum. Eight endophytic fungal isolates from
Z. bungeanum, especially Zbf-S11
E. nigrum, exhibited significant inhibitory effects on its host plant pathogenic fungi
F. sambucinum and
P. zanthoxyli. Although we only investigated the antifungal activity of the EtOAc extracts of endophytic fungi, we screened out several endophytic isolates with strong antifungal activity. If further experiments are carried out to isolate pure compounds and determine their biological activities, we might obtain many novel natural compounds with promising activity from endophytic fungi of
Z. bungeanum. Presently, the systematical chemical analyses of secondary metabolites of the eight endophytic fungal isolates (Zbf-S11, Zbf-S27, Zbf-S47, Zbf-S48, Zbf-S49, Zbf-S1, Zbf-R1, Zbf-T3) are being carried out. The present research offers a framework for further investigation and utilization of endophytic fungi associated with
Z. bungeanum.
4. Materials and Methods
4.1. Plant Material, Pathogenic Fungi and Chemicals
Ten healthy and asymptomatic three-year-old Zanthoxylum bungeanum (cultivar: Dahongpao) plants were randomly selected in July 2015, which covered the whole planting area of Z. bungeanum in the nursery garden of Northwest A&F University (34°16′ N; 108°4′ W) located in the Yangling District of Shaanxi province (China). The roots, stems, leaves, fruits, and thorns of each plant were collected and then immediately brought to the laboratory. Ten samples of every tissue from each plant were chosen randomly, and then we combined all samples of every tissue from 10plants. Finally, 100 samples of each tissue were obtained and stored at 4 °C in the refrigerator. All of the samples were used to isolate endophytic fungi within 24 h after collection.
Our research team obtained the pathogenic fungi
Fusarium sambucinum and
Pseudocercospora zanthoxyli in previous studies [
16,
17]. Both of these strains were maintained on potato dextrose agar (PDA) slants in cryovials at 4 °C.
All the chemicals were purchased from Jie Cheng Chemical and Glass Company (Yangling, China) except that those were peculiarly explained where they were bought.
4.2. Isolation and Preservation of Endophytic Fungi
All of the samples from each tissue were washed separately by running tap water to remove dust or other residues on the surface. For roots and stems, each sample was cut into approximate 1.0 cm × 1.0 cm segments by an autoclaved pinch cutter. For leaves, each sample was cut into a small disc with a diameter of approximately 1.0 cm. For fruits and thorns, each sample was cut with an incision. The samples from every tissue were disinfected by soaking in 75% ethanol for 2 min, then soaked in a 0.2% mercuric chloride solution for 10 min and then washed three times with autoclaved water. The 0.2% mercuric chloride disinfectant solution was recovered. After that, the samples were transferred onto dried sterile filter paper to remove the liquid from the surface of samples. Subsequently, each sample was placed on a potato dextrose agar (PDA) plate and kept at 25 °C in the incubator for 20 days. During the incubation period, all of the plant samples were observed every day, and any newly emerged fungal spot was immediately picked out by autoclaved toothpicks and transferred to another fresh PDA plate. The resulting fungal isolates were further purified and then maintained on PDA slants in cryovials at 4 °C and −80 °C. All operations were carried out under sterile conditions.
4.3. Identification of Endophytic Fungi
Initially, the purified isolates were grouped based on their morphological characteristics including colony color, hyphal shape and structure, growth rate, spore morphology and color, and exudatecolor. All of the endophytic fungi were first categorized according to their morphological characteristics. We defined the endophyic fungal isolates as being of the same morphotype if they possessed the same characteristics of colony, mycelia, and spore. In the study, 93 morphotypes were obtained. After that, one isolate representing one morphotype was selected for molecular identification. In our study, 93 isolates were then subjected to molecular identification by analyzing the internal transcribed spacer (ITS) region of the nuclear ribosomal DNA. All included steps were as follows: genomic DNA extraction, polymerase chain reaction (PCR) amplification, ITS sequencing, and analysis by basic local alignment search tool (BLAST).
Endophytic fungi were cultured on PDA plates before DNA extraction. When the colonies of endophytic fungi reached enough mass for DNA extraction, mycelia were scraped from the surface of the PDA plate using sterile toothpicks. Collected mycelia were ground into powder in liquid nitrogen using an autoclaved mortar. Then, 500 mg of mycelia powder was subjected to genomic DNA extraction using a TaKaRa MiniBEST Plant Genomic DNA Extraction Kit (Takara Biotechnology Co., Ltd., Dalian, China, Code No. 9768). The extraction process was carried out according to the manufacturer’s instructions. The extracted DNA was dissolved in 100 μL distilled water and stored at 4 °C until further use.
The total DNA was amplified by PCR using primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) [
43]. Amplification was conducted in 30 μL of PCR mixture containing 15 μL Premix TaqTM (Takara Biotechnology Co., Ltd., Dalian, China No. RR003A), 0.5μL template DNA, 1.0 μL ITS1 primer, 1.0 μL ITS4 primer, and 12.5 μL distilled water. PCR amplifications were performed in a thermal cycler with an initial denaturing step at 94 °C for 3 min, followed by 34 amplification cycles of 30 s denaturation at 94 °C, 30 s primers annealing at 54 °C, 45 s extension at 72 °C, and then a final elongation step of 10 min at 72 °C. PCR products were analyzed by electrophoresis using a 1% agarose gel (
w/
v) containing 0.01% (
v/
v) GoldView nucleic acid stain. Visual confirmation of the ITS region under the impact of UV light was performed by an image capture device.
All PCR products of endophytic fungi were sent to Sangon Biotech Co., Ltd. (Shanghai, China) for sequencing. The raw obtained sequences were aligned using MEGA7 [
51], edited manually, and then BLAST (Basic Local Alignment Search Tool) was used to search for the best match in the National Center for Biotechnology Information (NCBI) GenBank database (
http://www.ncbi.nlm.nih.gov/) to identify endophytic fungi. Sequences with similarity over 94% belonged to the same genus, and those with similarity over 97% belonged to the same species [
23]. The consensus sequence data of 93 endophytic fungal isolates were summarized by SEQUIN and then submitted to NCBI and GenBank accession numbers were assigned (
Table 1).
4.4. Phylogenetic Analyses of the Endophytic Fungi
Based on morphological and molecular identification results, the 93 endophytic fungal isolates in this study were classified into 14 families (
Table 2). The endophytic fungi belonging to the same family were analyzed in the same phylogenetic tree. We selected a neighbor-joining (NJ) method to analyze the phylogenetic relationships of the
Z. Bungeanum endophytic fungi. Each NJ tree was constructed by MEGA7 for the endophytic fungi belonging to the same family. The fungi used for each NJ tree alignment included tested endophytic fungi belonging to the family, several homologous fungal strains, and one exogenous fungal strain belonging to the same family but not the same genus. The ITS sequences of homologous and exogenous fungal strains were retrieved from NCBI. All sequence datasets were processed by MEGA7. The evolutionary history was inferred using the NJ method with 1000 Bootstrap replications. The phylogenetic tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distance used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. All positions containing gaps and missing date were deleted. Finally, we constructed 14 NJ trees in our present research that were marked as family group A to N, successively.
4.5. Diversity Analysis of the Endophytic Fungi
Using species as the statistical unit, we counted the number of isolates (
N) and calculated the isolation frequency (
IF) for each endophytic fungal species in different tissues or the total plant (
Table 3). The isolation frequency was calculated according to Equation (1). The species richness was evaluated by the species richness index (
S) and Margalef index (
D′), which are two important parameters for alpha diversity analysis [
26]. Species richness index (
S) was obtained by counting the number of endophytic fungal species in each tissue or total plant. The Margalef index (
D′) was calculated by Equation (2).The species diversity was evaluated by the Shannon–Wiener index (
H′), Simpson’s diversity index (
Ds), and Simpson’s dominant index (
λ) [
26,
52]. The Shannon–Wiener index (
H), Simpson’s diversity index (
Ds), and Simpson’s dominant index (
λ) were calculated by Equations (3)–(5), respectively. The probability of interspecific encounter (
PIE) index was used to evaluate the encountering probability of the individuals belonging to different species [
53].
PIE index was calculated by Equation (6). Species Evenness was evaluated by Pielou’s evenness index (
J) [
34], which was calculated by Equation (7). The relative abundance (
RA) for each genus was also calculated by Equation (8):
where
Ni is the number of isolates belonging to the
ith species,
Nt is the total number of endophytic fungal isolates in each tissue or total plant,
S is the number of total species in each tissue or total plant, and
N′ is the number of endophytic fungal isolates from each class, order, or genus in each tissue or total plant.
4.6. Metabolites Extraction from Endophytic Fungi
The metabolites of 93 endophytic fungal isolates, representing 93 morphotypes, were extracted by ethyl acetate (EtOAc) for antifungal assays. The endophytic fungi preserved at 4 °C in the refrigerator were separately inoculated on fresh PDA plates and then kept at 25 °C in an incubator for seven days. The endophytic fungal plug (5 mm diameter) of the mycelial inoculum was obtained by an autoclaved hole punch from the margin of an actively growing colony and then transferred into an Erlenmeyer flask (500 mL) containing 200 mL potato dextrose broth (three plugs per flask). All flasks were shaken in an incubator at 125 rpm at 25 °C for 14 days. The 14-day fermented broth cultures were extracted with 200 mL EtOAc three times. Each resulting EtOAc crude extract was collected and concentrated to dryness in a vacuum rotary evaporator at 40–45 °C and then washed with 3 mL EtOAc and transferred into a clean vial. Each empty vial was weighed before and after the EtOAc volatilized completely. The weight of EtOAc extract of each endophytic fungus was calculated.
4.7. Antifungal Assay for Endophytic Fungi
Antifungal activities of EtOAc extracts of 93 endophytic fungal isolates were carried out by colony radial mycelia growth method against pathogenic fungi
F. sambucinum and
P. zanthoxyli [
54]. EtOAc extracts of each endophytic fungus were dissolved in 1 mL EtOAc and then filtered through a 0.22-μm Millipore filter. Each filtration was added into an Erlenmeyer flask (250 mL) containing 100 mL PDA before the PDA solidified and then mixed; the PDA medium was poured into sterile petri dishes (9 cm diameter). Each petri dish contained 10 mL PDA medium. The final concentrations of endophytic fungi EtOAc extracts in media are summarized in
Table 3. The blank control was carried out without adding anything to the PDA media. The negative control was carried out with EtOAc addition into the PDA media (1%,
v/
v).
The pathogenic fungi
Fusarium sambucinum and
Pseudocercospora zanthoxyli were activated from dormant states by cultivation on PDA plates. After that, the pathogenic fungus plugs (5-mm diameter) from the margin of actively growing colonies were placed on the center of the PDA medium plates. Each treatment was carried out with five triplicates. All of the plates were then kept at 25 °C in the dark in an incubator for seven days, when the colony diameter was measured twice in perpendicular. The inhibitory rate (
IR) was calculated by Equation (9):
where
D0 is the average diameter of blank control,
Ds is the average diameter of the treated sample, and
Dn is the average diameter of the negative control.