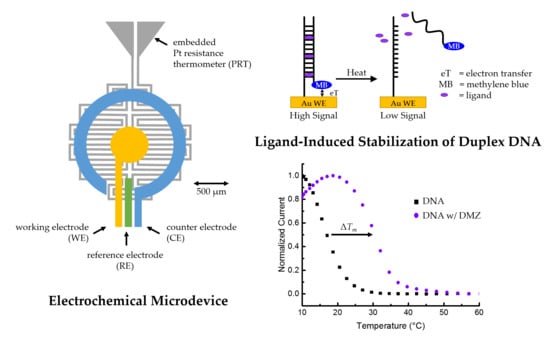
Ligand-Based Stability Changes in Duplex DNA Measured with a Microscale Electrochemical Platform
Abstract
:1. Introduction
2. Materials and Methods
2.1. Design of Microscale Platform
2.2. Materials
2.3. Microfabrication of Three-Electrode Device with Embedded Microheater
2.4. PRT Calibration
2.5. Platform Assembly: Resistive Heating
2.6. Platform Assembly: Thermoelectric Heating
2.7. Electrode Cleaning and Preparation of Duplex Self-Assembly on Au Electrode
2.8. Melting-Curve Analysis
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Neidle, S. DNA minor-groove recognition by small molecules. Nat. Prod. Rep. 2001, 18, 291–309. [Google Scholar] [CrossRef] [PubMed]
- Emami, S.; Dadashpour, S. Current developments of coumarin-based anti-cancer agents in medicinal chemistry. Eur. J. Med. Chem. 2015, 102, 611–630. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef]
- Kohn, K.W. Beyond DNA cross-linking: History and prospects of DNA-targeted cancer treatment--fifteenth Bruce F. Cain Memorial Award Lecture. Cancer Res. 1996, 56, 5533–5546. [Google Scholar] [PubMed]
- Pommier, Y.; Leo, E.; Zhang, H.; Marchand, C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010, 17, 421–433. [Google Scholar] [CrossRef]
- Rescifina, A.; Zagni, C.; Varrica, M.G.; Pistarà, V.; Corsaro, A. Recent advances in small organic molecules as DNA intercalating agents: Synthesis, activity, and modeling. Eur. J. Med. Chem. 2014, 74, 95–115. [Google Scholar] [CrossRef]
- Patel, A.G.; Kaufmann, S.H. How does doxorubicin work? Elife 2012, 1, e00387. [Google Scholar] [CrossRef] [PubMed]
- Gewirtz, D.A. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem. Pharmacol. 1999, 57, 727–741. [Google Scholar] [CrossRef]
- Turner, P.R.; Denny, W.A. The mutagenic properties of DNA minor-groove binding ligands. Mutat. Res. 1996, 355, 141–169. [Google Scholar] [CrossRef]
- Wilson, W.D.; Tanious, F.A.; Fernandez-Saiz, M.; Rigl, C.T. Evaluation of drug-nucleic acid interactions by thermal melting curves. Methods Mol. Biol. 1997, 90, 219–240. [Google Scholar] [CrossRef]
- Kelly, J.M.; Tossi, A.B.; Mcconnell, D.J.; Ohuigin, C. A Study of the Interactions of Some Polypyridylruthenium(II) Complexes with DNA Using Fluorescence Spectroscopy, Topoisomerization and Thermal-Denaturation. Nucleic Acids Res. 1985, 13, 6017–6034. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.L.; Lacroix, L. Analysis of thermal melting curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef] [PubMed]
- Nasef, H.; Ozalp, V.C.; Beni, V.; O’Sullivan, C.K. Melting temperature of surface-tethered DNA. Anal. Biochem. 2010, 406, 34–40. [Google Scholar] [CrossRef]
- Herne, T.M.; Tarlov, M.J. Characterization of DNA Probes Immobilized on Gold Surfaces. J. Am. Chem. Soc. 1997, 119, 8916–8920. [Google Scholar] [CrossRef]
- Steel, A.B.; Herne, T.M.; Tarlov, M.J. Electrochemical quantitation of DNA immobilized on gold. Anal. Chem. 1998, 70, 4670–4677. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Lai, R.Y.; Plaxco, K.W. Preparation of electrode-immobilized, redox-modified oligonucleotides for electrochemical DNA and aptamer-based sensing. Nat. Protoc. 2007, 2, 2875–2880. [Google Scholar] [CrossRef]
- Kelley, S.O.; Barton, J.K.; Jackson, N.M.; Hill, M.G. Electrochemistry of Methylene Blue Bound to a DNA-Modified Electrode. Bioconj. Chem. 1997, 8, 31–37. [Google Scholar] [CrossRef] [Green Version]
- Pei, H.; Lu, N.; Wen, Y.L.; Song, S.P.; Liu, Y.; Yan, H.; Fan, C.H. A DNA Nanostructure-based Biomolecular Probe Carrier Platform for Electrochemical Biosensing. Adv. Mater. 2010, 22, 4754–4758. [Google Scholar] [CrossRef] [Green Version]
- Boon, E.M.; Ceres, D.M.; Drummond, T.G.; Hill, M.G.; Barton, J.K. Mutation detection by electrocatalysis at DNA-modified electrodes. Nat. Biotechnol. 2000, 18, 1096–1100. [Google Scholar] [CrossRef]
- Lucarelli, F.; Marrazza, G.; Turner, A.P.F.; Mascini, M. Carbon and gold electrodes as electrochemical transducers for DNA hybridisation sensors. Biosens. Bioelectron. 2004, 19, 515–530. [Google Scholar] [CrossRef] [Green Version]
- Somasundaram, S.; Holtan, M.D.; Easley, C.J. Understanding Signal and Background in a Thermally Resolved, Single-Branched DNA Assay Using Square Wave Voltammetry. Anal. Chem. 2018, 90, 3584–3591. [Google Scholar] [CrossRef]
- Meunier-Prest, R.; Raveau, S.; Finot, E.; Legay, G.; Cherkaoui-Malki, M.; Latruffe, N. Direct measurement of the melting temperature of supported DNA by electrochemical method. Nucleic Acids Res. 2003, 31, e150. [Google Scholar] [CrossRef]
- Belozerova, I.; Levicky, R. Melting thermodynamics of reversible DNA/ligand complexes at interfaces. J. Am. Chem. Soc. 2012, 134, 18667–18676. [Google Scholar] [CrossRef]
- He, Y.K.; Zhang, J.; Ruffin, S.; Ji, L.N.; Wang, K.; Levicky, R.; Xia, X.H. An Electrochemical Study of the Surface Hybridization Process of Morpholino-DNA: Thermodynamics and Kinetics. Electroanal 2016, 28, 1647–1653. [Google Scholar] [CrossRef]
- Flechsig, G.U.; Peter, J.; Hartwich, G.; Wang, J.; Gründler, P. DNA hybridization detection at heated electrodes. Langmuir 2005, 21, 7848–7853. [Google Scholar] [CrossRef]
- Surkus, A.E.; Flechsig, G.U. Electrochemical Detection of DNA Melting Curves by Means of Heated Biosensors. Electroanal 2009, 21, 1119–1123. [Google Scholar] [CrossRef]
- Tseng, H.-Y.; Adamik, V.; Parsons, J.; Lan, S.-S.; Malfesi, S.; Lum, J.; Shannon, L.; Gray, B. Development of an electrochemical biosensor array for quantitative polymerase chain reaction utilizing three-metal printed circuit board technology. Sens. Actuators B Chem. 2014, 204, 459–466. [Google Scholar] [CrossRef]
- Fiche, J.B.; Fuchs, J.; Buhot, A.; Calemczuk, R.; Livache, T. Point mutation detection by surface plasmon resonance imaging coupled with a temperature scan method in a model system. Anal. Chem. 2008, 80, 1049–1057. [Google Scholar] [CrossRef]
- Melaine, F.; Coilhac, C.; Roupioz, Y.; Buhot, A. A nanoparticle-based thermo-dynamic aptasensor for small molecule detection. Nanoscale 2016, 8, 16947–16954. [Google Scholar] [CrossRef]
- Johnson, R.P.; Gale, N.; Richardson, J.A.; Brown, T.; Bartlett, P.N. Denaturation of dsDNA immobilised at a negatively charged gold electrode is not caused by electrostatic repulsion. Chem. Sci. 2013, 4, 1625–1632. [Google Scholar] [CrossRef]
- Papadopoulou, E.; Gale, N.; Thompson, J.F.; Fleming, T.A.; Brown, T.; Bartlett, P.N. Specifically horizontally tethered DNA probes on Au surfaces allow labelled and label-free DNA detection using SERS and electrochemically driven melting. Chem. Sci. 2016, 7, 386–393. [Google Scholar] [CrossRef]
- Yang, A.H.; Hsieh, K.; Patterson, A.S.; Ferguson, B.S.; Eisenstein, M.; Plaxco, K.W.; Soh, H.T. Accurate zygote-specific discrimination of single-nucleotide polymorphisms using microfluidic electrochemical DNA melting curves. Angew. Chem. Int. Ed. Engl. 2014, 53, 3163–3167. [Google Scholar] [CrossRef]
- Halperin, A.; Buhot, A.; Zhulina, E.B. Sensitivity, specificity, and the hybridization isotherms of DNA chips. Biophys. J. 2004, 86, 718–730. [Google Scholar] [CrossRef]
- Halperin, A.; Buhot, A.; Zhulina, E.B. On the hybridization isotherms of DNA microarrays: The Langmuir model and its extensions. J. Phys. Condens. Matter 2006, 18, S463–S490. [Google Scholar] [CrossRef]
- Shen, Z.; Sintim, H.O.; Semancik, S. Rapid nucleic acid melting analyses using a microfabricated electrochemical platform. Anal. Chim. Acta 2015, 853, 265–270. [Google Scholar] [CrossRef]
- Contento, N.M.; Semancik, S. Thermal characteristics of temperature-controlled electrochemical microdevices. Sens. Actuators B Chem. 2016, 225, 279–287. [Google Scholar] [CrossRef] [Green Version]
- Nicholson, B.H.; Peacocke, A.R. The inhibition of ribonucleic acid polymerase by acridines. Biochem. J. 1966, 100, 50–58. [Google Scholar] [CrossRef] [Green Version]
- Li, H.J.; Crothers, D.M. Relaxation studies of the proflavine-DNA complex: The kinetics of an intercalation reaction. J. Mol. Biol. 1969, 39, 461–477. [Google Scholar] [CrossRef]
- Kuriakose, S.; Muleme, H.M.; Onyilagha, C.; Singh, R.; Jia, P.; Uzonna, J.E. Diminazene aceturate (Berenil) modulates the host cellular and inflammatory responses to Trypanosoma congolense infection. PLoS ONE 2012, 7, e48696. [Google Scholar] [CrossRef]
- Zhou, J.; Le, V.; Kalia, D.; Nakayama, S.; Mikek, C.; Lewis, E.A.; Sintim, H.O. Diminazene or berenil, a classic duplex minor groove binder, binds to G-quadruplexes with low nanomolar dissociation constants and the amidine groups are also critical for G-quadruplex binding. Mol. Biosyst. 2014, 10, 2724–2734. [Google Scholar] [CrossRef] [Green Version]
- Mikek, C.G.; West, S.J.; Gwin, J.C.; Dayal, N.; Sintim, H.O.; Lewis, E.A. Berenil Binds Tightly to Parallel and Mixed Parallel/Antiparallel G-Quadruplex Motifs with Varied Thermodynamic Signatures. ACS Omega 2018, 3, 11582–11591. [Google Scholar] [CrossRef]
- Nakayama, S.; Kelsey, I.; Wang, J.; Sintim, H.O. c-di-GMP can form remarkably stable G-quadruplexes at physiological conditions in the presence of some planar intercalators. Chem. Commun. 2011, 47, 4766–4768. [Google Scholar] [CrossRef]






| Thermoelectric Cooling/Heating | Refrigerated w/Resistive Heating | |||||
|---|---|---|---|---|---|---|
| No ligand | Proflavine | DMZ | No Ligand | Proflavine | DMZ | |
| Tm,baseline (°C) | 17.0 ± 1.0 | 18.5 ± 0.1 | 16.8 ± 1.0 | 18.1 ± 1.8 | 11.8 ± 1.2 1 | 15.8 ± 0.8 |
| Tm, control (°C) | 17.0 ± 1.0 | - | - | 18.1 ± 2.2 | - | - |
| Tm, ligand (°C) | - | 30.3 ± 0.4 | 31.5 ± 2.0 | - | 21.9 ± 0.2 1 | 28.1 ± 1.6 |
| ΔTm (°C) | −0.3 ± 0.4 | 11.8 ± 0.3 | 14.6 ± 0.6 | 0.0 ± 1.1 | 10.1 ± 1.0 1 | 12.3 ± 1.7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robinson, S.M.; Shen, Z.; Askim, J.R.; Montgomery, C.B.; Sintim, H.O.; Semancik, S. Ligand-Based Stability Changes in Duplex DNA Measured with a Microscale Electrochemical Platform. Biosensors 2019, 9, 54. https://doi.org/10.3390/bios9020054
Robinson SM, Shen Z, Askim JR, Montgomery CB, Sintim HO, Semancik S. Ligand-Based Stability Changes in Duplex DNA Measured with a Microscale Electrochemical Platform. Biosensors. 2019; 9(2):54. https://doi.org/10.3390/bios9020054
Chicago/Turabian StyleRobinson, Sarah M., Zuliang Shen, Jon R. Askim, Christopher B. Montgomery, Herman O. Sintim, and Steve Semancik. 2019. "Ligand-Based Stability Changes in Duplex DNA Measured with a Microscale Electrochemical Platform" Biosensors 9, no. 2: 54. https://doi.org/10.3390/bios9020054