Renewed global threat by the novel SARS-CoV-2 variants ‘XBB, BF.7, BQ.1, BA.2.75, BA.4.6’: A discussion
- 1Department of Chemistry, Government College of Engineering, Keonjhar, Odisha, India
- 2Department of Medical Biochemical Analysis, College of Health Technology, Cihan University—Erbil, Erbil, Kurdistan Region, Iraq
- 3Guangzhou HC Pharmaceutical Co., Ltd., Guangzhou, China
- 4Department of General Surgery, Dr NTR University of Health Sciences, Vijayawada, Andhra Pradesh, India
- 5Department of Electrical Engineering, Government College of Engineering, Keonjhar, Odisha, India
- 6Department of Microbiology, Prathima Institute of Medical Sciences, Karimnagar, Telangana, India
- 7Department of Chemistry, School of Applied Sciences, Centurion University of Technology and Management, Odisha, India
- 8Department of Pharmaceutical Chemistry, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
- 9School of Biotechnology, KIIT Deemed-to-be-University, Bhubaneswar, Odisha, India
Introduction
Despite the numerous preventive and curative measures in vogue post the onset of the recent pandemic, the novel SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) continues evolving to more critical and infectious variants (1, 2). The reasons attributed to facilitating the transmission include, among others, low vaccination rate, overcrowding defying social distancing, poor compliance to personal hygiene like masking and hand sanitisation, and increased immunodeficient and susceptible population. Such situations favour viral replication and increased mutation risk leading to the emergence of novel variants (3). Omicron mutants allegedly primarily had altered spike protein that facilitated evasion of the innate immune capabilities n individuals including in the vaccinated ones thereby rendering Omicron and its sub-lineages more infectious (4, 5). The currently used vaccines may not necessarily restrict infections and thus might not restrict the transmission, which could invariably contribute to the emergence of variants (6). Omicron variant is currently the most predominant SARS-CoV-2 variant completely replacing all preceding variants like, among others, alpha, beta, gamma and delta. The unique mutations of the Omicron variant at the receptor binding domain (RBD) enhanced the transmissibility of the virus as well as its immune escape mechanisms. The mutated spike protein could provide resistance against vaccination and monoclonal antibody (mAb) therapy (7). Most available mAb therapies target the spike protein of the earlier SARS-CoV-2 variant. As the latest Omicron variants and the numerous sub-lineages demonstrably have significantly altered spike protein attributed to mutations, their efficacy of vaccines and mAbs is questionable. The USFDA recommended the use of Bebtelovimab/Etesevimab that has demonstrated efficacy against most of the recent Omicron sub-lineages including BA.1.1, BA.2, BA.4 and BA.5. However, Bebtelovimab is unavailable outside the USA, and also its efficacy against the XBB, BQ.1 and their descendants is yet to be confirmed (8).
Novel SARS-CoV-2 variants
The XBB (BA.2.10 strain), an Omicron subvariant, was detected in August simultaneously in nations like Singapore, Australia, Denmark, Bangladesh, India, the US and Japan. Despite a rise in cases in Singapore, currently there is no proof of the subvariant leading to serious effects. With the WHO identifying Omicron as a ‘variant of concern’, the second generation Omicron variants and their descendants are treated similar as in the earlier cases. The percentage of the Omicron subvariant (XBB) strain cases has grown during the past month in Singapore. Up to 54% of local XBB cases between October 3 and 9, 2022, it is currently the most prevalent subvariant in Singapore. However, it is not more fatal than the earlier variants although it is extremely contagious. As per the Singaporean health authority, the current COVID-19 wave would peak by mid-November there amid the growing concern about the ‘immunity-evasive’ XBB strain (https://www.bloomberg.com/news/articles/2022-10-13/xbb-bf-7-ba-5-1-7-new-covid-variants-renew-threats-to-the-world?leadSource=uverify%20wall). As of now, the XBB variant has been reported in nine Indian states with more than 500 cases. The numbers may not represent the real picture since the access to COVID-19 diagnosis and sequencing of SARS-CoV-2 in most health centres in India is limited. As per the data of the Indian SARS-CoV-2 Consortium on Genomics (INSACOG), the XBB sub-lineage accounts for more than 50% of the recently sequenced Indian samples (https://weather.com/en-IN/india/coronavirus/news/2022-11-01-covid-19-faqs-xbb-variant-infiltrates-9-indian-states). In the ongoing Indian festivities, maintaining strict vigilance especially towards variants like BF.7, BQ.1 in India is suggested. BQ.1 is an offshoot of Omicron variant, related to BA.5. Both BF.7 and BQ.1 sub-variants are mutated making them contagious by evading immunity. Further, clinicians may misdiagnose it due to the prevailing influenza in the region with the onset of winter. Recent available data hint at the dominance of XBB and BQ.1 variants in India and Singapore. The BQ.1 being currently responsible for most cases in the UK, South Africa, Germany, the US, Australia and South Korea, XBB variants may have spread to more than 15 countries and might have contributed to the increasing cases in the US, Australia and South Korea. Analysing the recent trends, soon the BQ.1 variant replacing by XBB in most countries throughout the world is predicted.
Unique mutations
The recent rise in infections in Singapore demonstrate that most cases were due to XBB (BA.2.10) strain (more than 50%) followed by BA.2.75 (24%) and BA.5 (21%) (https://health.economictimes.indiatimes.com/news/industry/singapore-monitoring-xbb-covid-strain-very-closely-health-minister/94804128). As per the WHO (9), XBB is a hybrid/recombinant variant that potentially emerged from BA.2 (BA.2.10.1 and BA.2.75) sub-lineages, i.e., BJ1 and BM.1.1.1, with a break in S1 and unique mutations of BA.2+ (S:V83A, S:Y144, S:Q183E, S:H146Q, S:V213E, S:G252V, S:R346T, S:G339H, S:L368I, S:G446S, S:V445P, S:N460K, S:F486S, S:F490S), and it could apparently become the next dominant variant. The currently circulating Omicron lineages that include BA.2.3.20, BA.2.75.2, BM.1.1.1, BR.2, CA.1, BN.1, BQ.1.1, BU.1 and XBB demonstrate unique mutations (R346, K444, N450, L452, V445, G446, N460, F490, F486 and R493) on the RBD that reinforce their immunity-escape mechanism and increase transmissibility. The XBB and BQ.1.1 exhibited the strongest resistance to mAbs that target RBD and increased ACE2-binding affinity (10).
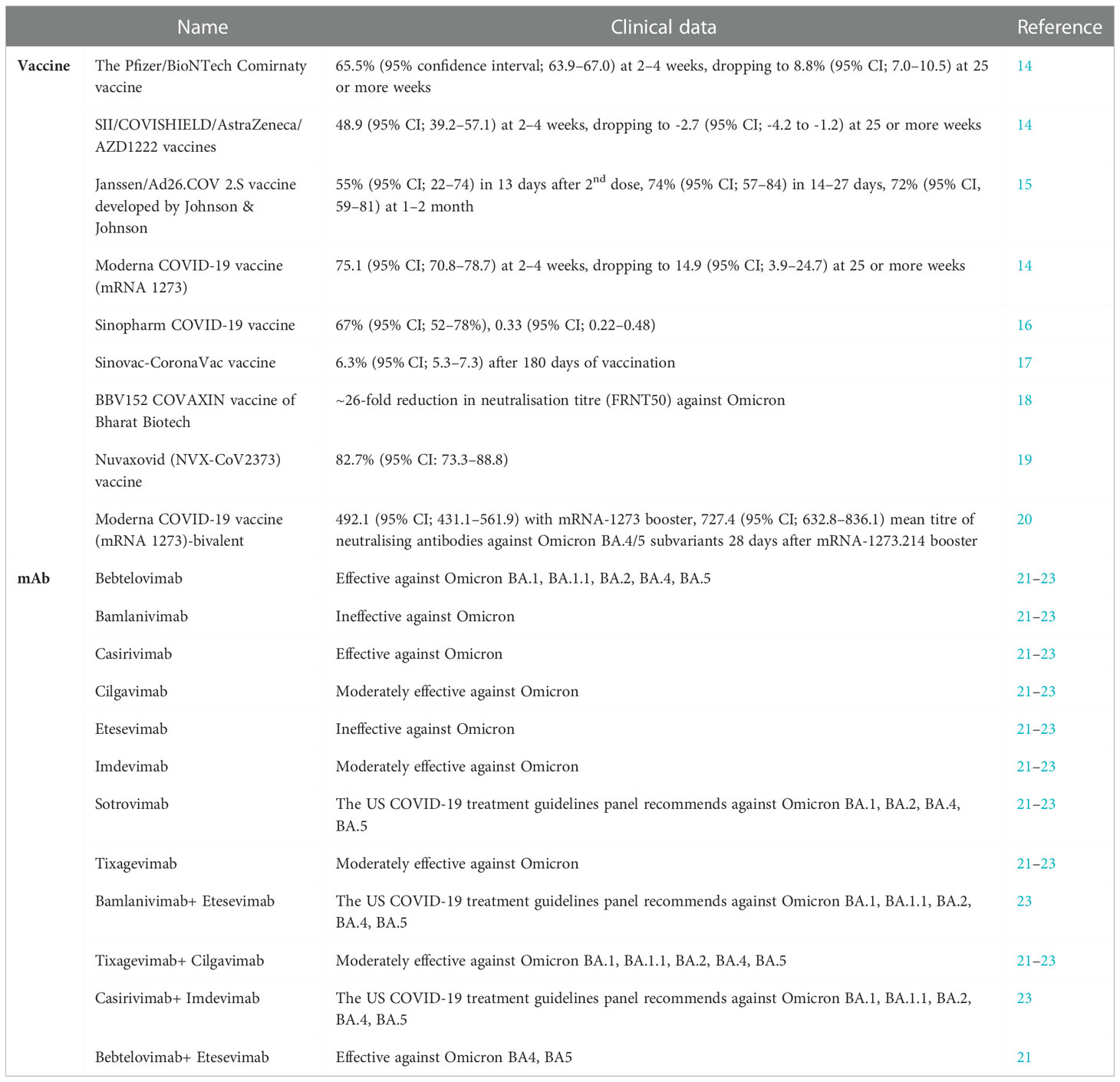
Vaccines and mAb therapy
The S gene analysis revealed that there is a significant difference in the representative codons among different variants of concern (11). The S gene of SARS-CoV-2 variants like alpha, beta, gamma and delta differ from the Omicron variant. Omicron variant emerging during the early stages of the pandemic and the remaining ones that stayed undetected until discovered later in South Africa is also hypothesised (11). Given the occurrence of unique mutations in the spike and RBD of the SARS-CoV-2 Omicron and its lineages, the efficacy of the current vaccines and the mAb therapies is uncertain (12). A bivalent vaccine that consisted of the spike proteins of the original SARS-CoV-2, and the Omicron BA.4 and BA.5 manufactured by the Moderna and Pfizer-BioNTech was recently approved by the USFDA (13). The currently available vaccines and mAb therapies for SARS-CoV-2 variants are listed in Table 1. Thus, the spread of such variants could contribute to increased infection that in turn could affect nations and their economies. It is prudent to reevaluate the current vaccines and improve the vaccination strategies to counter the variant and its lineages. An IgG antibody was synthesised that could block cell-virus fusion region, the COVID19-SF5 (24). The IgG antibody cross-reacted with six cell-adhesion facilitating spike protein sites. It is imperative that such antibodies are used in therapeutics to block infections, restrict viral replication and minimise the mutations.
It is evident that most of the recent variants have demonstrated significant mutations in the S region attributed to selective pressure on the survival of the virus against effective immune responses and vaccines. Future interventions must necessarily target such S variations that increase the virus’s resistance and infectivity (25). Understanding its evolution and predicting future viral variants is very important in public health perspective. Thermodynamics to increase the understanding of virus evolution was suggested (26, 27). The Gibbs energy of binding correlates well with the fusion and entry of the virus in to the host cells, and the Gibbs energy of growth correlates with the viral replication in the host. Thermodynamics implicate that the current SARS-CoV-2 virus could continue to evolve and develop as more pathogenic and more infective variants compared to native counterparts that are more infectious although less pathogenic. An urgent need to collaborate to develop broad-spectrum SARS-CoV-2 vaccines and mAb drugs is sensed.
Public health measures
Health experts in India advised against letting the guard down on the fight against the pandemic in face of the new variant that is anticipated to spread faster through the market places and the festival times. People taking preventative measures including following the government-recommended vaccination schedules and the COVID-appropriate behaviour in the public is suggested. The governments and administrations should ensure infection prevention measures like use of masks, and restricting public gatherings, among others to minimize transmission. Special attention may be extended to the Comorbid population with compromised immunity and flu-like symptoms. As both XBB and BQ.1 variants possess mutations that enable them to evade vaccine-related immunity and expedite transmission, people may further be prone to reinfection (10, 28).
With regard to the surveillance and readiness to counter the Omicron variants, it seems the Artificial Intelligence approach in line with the IoT (Internet of Things) could provide teeth to the drive. Artificial intelligence based machine learning approach shall pave an effective way to find solutions to such a burgeoning global menace as it is dependent on numerous dependent and interdependent factors and systemic issues that could be beyond human comprehension (29). An increasing need to apply machine learning techniques to accurately predict is felt. Without expressly programmed, machine learning could be elevated to artificial intelligence wherein the machine gains knowledge with experience. The data related to the pandemic could be analysed through various machine learning classifiers (Figure 1). Dataset is trained using machine learning classifiers and analysed based on the training. Healthcare system will appreciate this strategy and adopt it sooner or later for the common good.
Figure 1 Architecture of Artificial Intelligence (AI) system (reproduced from 29).
Author contributions
RM: conceptualised, updated and edited the draft. LK, AO, AM: Teamed up during the first draft. VK, AS: Updated the manuscript. SM: Teamed up during drafting and edited. All authors have critically reviewed and approved the final draft.
Acknowledgments
The authors are thankful to their respective institutions to provide necessary institutional facilities and encouragement.
Conflict of interest
Author AM was employed by Guangzhou HC Pharmaceutical Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Mohapatra RK, Kandi V, Verma S, Dhama K. Challenges of the omicron (B.1.1.529) variant and its lineages: A global perspective. ChemBioChem. (2022) 23:e202200059. doi: 10.1002/cbic.202200059
2. Mohapatra RK, Kandi V, Sarangi AK, Verma S, Tuli HS, Chakraborty S, et al. The recently emerged BA.4 and BA.5 lineages of omicron and their global health concerns amid the ongoing wave of COVID-19 pandemic. Int J Surg (2022) 103:106698. doi: 10.1016/j.ijsu.2022.106698
3. Dhawan M, Priyanka, Choudhary OP. Omicron SARS-CoV-2 variant: Reasons of emergence and lessons learnt. Int J Surg (2022) 97:106198. doi: 10.1016/j.ijsu.2021.106198
4. Mohapatra RK, Kandi V, Mishra S, Sarangi AK, Pradhan MK, Mohapatra PK, et al. Emerging novel sub-lineage BA.2.75: The next dominant omicron variant? Int J Surg (2022) 104:106835. doi: 10.1016/j.ijsu.2022.106835
5. Mohapatra RK, Tiwari R R, Sarangi AK, Islam MR, Chakraborty C, Dhama K. Omicron (B.1.1.529) variant of SARS-CoV-2: Concerns, challenges, and recent updates. J Med Virol (2022) 94(6):2336–42. doi: 10.1002/jmv.27633
6. Kannan SR, Spratt AN, Sharma K, Chand HS, Byrareddy SN, Singh K. Omicron SARS-CoV-2 variant: Unique features and their impact on pre-existing antibodies. J Autoimmun (2022) 126:102779. doi: 10.1016/j.jaut.2021.102779
7. Alkhatib M, Salpini R, Carioti L, Ambrosio FA, D'Anna S, Duca L, et al. Update on SARS-CoV-2 omicron variant of concern and its peculiar mutational profile. Microbiol Spectr. (2022) 10(2):e0273221. doi: 10.1128/spectrum.02732-21
8. Nichols RM, Deveau C, Upadhyaya H. Bebtelovimab: Considerations for global access to treatments during a rapidly evolving pandemic. Lancet Infect Dis (2022) 22(11):1531. doi: 10.1016/S1473-3099(22)00592-8
9. WHO. Tracking SARS-CoV-2 variants (2022). Available at: https://www.who.int/activities/tracking-SARS-CoV-2-variants (Accessed 20-10-22).
10. Cao Y, Jian F, Wang J, Yu Y, Song W, Yisimayi A, et al. Imprinted SARS-CoV-2 humoral immunity induces convergent omicron RBD evolution. bioRxiv (2022). doi: 10.1101/2022.09.15.507787
11. Paz M, Aldunate F, Arce R, Ferreiro I, Cristina J. An evolutionary insight into severe acute respiratory syndrome coronavirus 2 omicron variant of concern. Virus Res (2022) 314:198753. doi: 10.1016/j.virusres.2022.198753
12. Saxena SK, Kumar S, Ansari S, Paweska JT, Maurya VK, Tripathi AK, et al. Characterization of the novel SARS-CoV-2 omicron (B.1.1.529) variant of concern and its global perspective. J Med Virol (2022) 94(4):1738–44. doi: 10.1002/jmv.27524
13. FDA US. Coronavirus (COVID-19) update: FDA authorizes moderna and pfizer-BioNTech bivalent COVID-19 vaccines for use as a booster dose in younger age groups (2022). Available at: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-moderna-and-pfizer-biontech-bivalent-covid-19-vaccines.
14. Andrews N, Stowe J, Kirsebom F, Toffa S, Rickeard T, Gallagher E, et al. Covid-19 vaccine effectiveness against the omicron (B.1.1.529) variant. N Engl J Med (2022) 386(16):1532–46. doi: 10.1056/NEJMoa2119451
15. Gray G, Collie S, Goga A, Garrett N, Champion J, Seocharan I, et al. Effectiveness of Ad26.COV2.S and BNT162b2 vaccines against omicron variant in south Africa. N Engl J Med (2022) 386(23):2243–5. doi: 10.1056/NEJMc2202061
16. Al-Momani H, Aldajah K, Alda'ajah E, ALjafar Y, Abushawer Z. Effectiveness of Pfizer/BioNTech and sinopharm COVID-19 vaccines in reducing hospital admissions in prince hamza hospital, Jordan. Front Public Health (2022) 10:1008521. doi: 10.3389/fpubh.2022.1008521
17. Ranzani OT, Hitchings MDT, de Melo RL, de Franca GVA, Fernandes CFR, Lind ML, et al. Effectiveness of an inactivated covid-19 vaccine with homologous and heterologous boosters against omicron in Brazil. Nat Commun (2022) 13:5536. doi: 10.1038/s41467-022-33169-0
18. Ahmed TI, Rishi S, Irshad S, Aggarwal J, Happa K, Mansoor S. Inactivated vaccine Covaxin/BBV152: A systematic review. Front Immunol (2022) 13:863162. doi: 10.3389/fimmu.2022.863162
19. Heath PT, Galiza EP, Baxter DN, Boffito M, Browne D, Burns F, et al. Safety and efficacy of the NVX-CoV2373 COVID-19 vaccine at completion of the placebo-controlled phase of a randomized controlled trial. Clin Infect Dis (2022). doi: 10.1093/cid/ciac803
20. Chalkias S, Harper C, Vrbicky K, Walsh SR, Essink B, Brosz A, et al. A bivalent omicron-containing booster vaccine against covid-19. N Engl J Med (2022) 387(14):1279–91. doi: 10.1056/NEJMoa2208343
21. IDSA. Anti-SARS-CoV-2 monoclonal antibodies (2022). Available at: https://www.idsociety.org/covid-19-real-time-learning-network/therapeutics-and-interventions/monoclonal-antibodies/#BamlanivimabEtesevimab.
22. Takashita E, Kinoshita N, Yamayoshi S, Sakai-Tagawa Y, Fujisaki S, Ito M, et al. Efficacy of antibodies and antiviral drugs against covid-19 omicron variant. N Engl J Med (2022) 386(10):995–8. doi: 10.1056/NEJMc2119407
23. NIH. Anti-SARS-CoV-2 monoclonal antibodies(2022). Available at: https://www.covid19treatmentguidelines.nih.gov/therapies/anti-sars-cov-2-antibody-products/anti-sars-cov-2-monoclonal-antibodies/.
24. Wang H, Yang T, Jiang W, Qin M, Sun Z, Dai W, et al. Identification and characterization of a novel cell binding and cross-reactive region on spike protein of SARS-CoV-2. Sci Rep (2022) 12(1):15668. doi: 10.1038/s41598-022-19886-y
25. Magazine N, Zhang T, Wu Y, McGee MC, Veggiani G, Huang W. Mutations and evolution of the SARS-CoV-2 spike protein. Viruses. (2022) 14(3):640. doi: 10.3390/v14030640
26. Popovic M. Beyond COVID-19: Do biothermodynamic properties allow predicting the future evolution of SARS-CoV-2 variants? Microb Risk Anal (2022) 22:100232. doi: 10.1016/j.mran.2022.100232
27. Popovic M. Strain wars 4 - Darwinian evolution through gibbs' glasses: Gibbs energies of binding and growth explain evolution of SARS-CoV-2 from hu-1 to BA.2. Virology (2022) 575:36–42. doi: 10.1016/j.virol.2022.08.009
28. Callaway E. COVID 'variant soup' is making winter surges hard to predict. Nature. (2022) 611(7935):213–4. doi: 10.1038/d41586-022-03445-6
Keywords: SARS-CoV-2, COVID-19, novel variants, omicron, XBB sub-variant
Citation: Mohapatra RK, Mahal A, Kutikuppala LVS, Pal M, Kandi V, Sarangi AK, Obaidullah AJ and Mishra S (2022) Renewed global threat by the novel SARS-CoV-2 variants ‘XBB, BF.7, BQ.1, BA.2.75, BA.4.6’: A discussion. Front. Virol. 2:1077155. doi: 10.3389/fviro.2022.1077155
Received: 24 October 2022; Accepted: 02 December 2022;
Published: 23 December 2022.
Edited by:
Manish Chandra Choudhary, Brigham and Women’s Hospital and Harvard Medical School, United StatesReviewed by:
Vijaya Pandey, University of California, Los Angeles, United StatesGagandeep Singh, Icahn School of Medicine at Mount Sinai, United States
Shipra Sharma, The Scripps Research Institute, United States
Copyright © 2022 Mohapatra, Mahal, Kutikuppala, Pal, Kandi, Sarangi, Obaidullah and Mishra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ranjan K. Mohapatra, rkmohapatra@gcekjr.ac.in; Ahmed Mahal, ahmed.mahal@cihanuniversity.edu.iq; Venkataramana Kandi, ramana20021@gmail.com
†ORCID: Ranjan K. Mohapatra, orcid.org/0000-0001-7623-3343
 Ranjan K. Mohapatra
Ranjan K. Mohapatra Ahmed Mahal
Ahmed Mahal LV Simhachalam Kutikuppala
LV Simhachalam Kutikuppala Madhumita Pal5
Madhumita Pal5  Venkataramana Kandi
Venkataramana Kandi Ashish K. Sarangi
Ashish K. Sarangi Ahmad J. Obaidullah
Ahmad J. Obaidullah Snehasish Mishra
Snehasish Mishra