Inherent Serum Inhibition of Influenza Virus Neuraminidases
- 1Center for Vaccines and Immunology, University of Georgia, Athens, GA, United States
- 2Department of Infectious Diseases, University of Georgia, Athens, GA, United States
Influenza virus vaccines have been designed for human and veterinary medicine. The development for broadly protective influenza virus vaccines has propelled the vaccine field to investigate and include neuraminidase (NA) components into new vaccine formulations. The antibody-mediated protection induced by NA vaccines is quantified by inhibition of sialic acid cleavage. Non-immune inhibitors against influenza viruses naturally occur in varying proportions in sera from different species. In this brief report, the inherent ability of raw animal sera to inhibit a panel of influenza virus NA was determined. Raw sera from the same species inhibited more than 50% of influenza viruses tested from four different subtypes, but the breadth of inhibiting NA activity depended on the source of sera. Furthermore, different influenza viruses were inhibited by different sources of sera. Overall, additional studies are needed to ensure that scientific methods are consistent across studies in order to compare NA inhibition results. Through future investigation into the differences between sera from different animal species and how they influence NA inhibition assays, there can be effective development of a broadly protective influenza virus vaccines for veterinary and human use.
Introduction
Influenza viruses are global zoonotic and human pathogens, and vaccination remains the main preventative measure against infection. The influenza virus is a member of the Orthomyxoviridae family. The genome is composed of eight negative single-sense RNA segments that determines the viral genus, alpha-, beta-, delta-, and gammainfluenzavirus that correspond to the species influenza A, B, D, and C viruses, respectively. Of the four influenza types, Types A and D are commonly isolated from animals, whereas influenza Types B and C are most commonly associated with human infection especially in children (1). The Type A influenza viruses are further classified into subtypes determined by the two major surface proteins, hemagglutinin (HA) and neuraminidase (NA). Currently, there are 18 HA subtypes and 11 NA subtypes that can be paired to create different influenza subtypes.
Influenza viruses are of international importance due to the widespread infection in different livestock, leading to vaccination being utilized across the veterinary field (2). Equine influenza viruses are important horse pathogens with policies in place that require horses be vaccinated for equine influenza viruses before participation in events or importation (3, 4). Furthermore, due to the transmission of influenza viruses from horses to dogs, as well as the endemic infection of influenza viruses in the canine population, canine vaccination is also recommended for dogs with high risk of exposure (5, 6). The swine industry uses primarily whole inactivated vaccines [WIVs—reviewed in reference (7)] that are developed using split-inactivated technologies (7, 8). The poultry industry utilizes the greatest variety of vaccine platforms, including split-inactivated virus, HA protein antigens, HA DNA vaccines, and recombinant technologies with other backbone viruses (9, 10).
However, during infection both HA and NA proteins are targets for neutralizing antibodies (11). The NA glycoprotein mediates viral egress and virion de-aggregation by cleaving sialic acids as well as contributing to motility through cleaving mucins in the upper respiratory tract (12, 13). Polyclonal NA-specific sera and NA inhibition (NAI) titers reduce, modulate, and protect against disease (14, 15). Further research has identified monoclonal NA-specific antibodies that neutralize viral growth (16). Although NA antibodies hinder viral replication, the induction of NAI antibody titers following vaccination is not as great as the induction of HAI titers, potentially due to either the split-inactivated vaccines lacking a standardized concentration of NA protein or the NA protein being destroyed during the split-inactivation process (15). Recently, research and vaccine development have focused on live-attenuated viruses that elicit NA antibodies, or protein vaccines that include the NA (17).
Currently, the enzyme-linked lectin assay (ELLA), MUNANA substrate, thiobarbituric acid (TBA) fluorescent-based assay, and NA-Star chemiluminescent assay are methods for measuring antibodies against the NA molecule (18–23). As the NA glycoprotein undergoes antigenic drift, the protein's ability to cleave sialic acid can be measured and quantified using these assays. All techniques assess the elicited antibody-specific inhibition of the NA after vaccination or infection. The ELLA measures the ability of the viral NA to cleave sialic acids from a large substrate (fetuin) similar to infection when sialic acids are expressed on the surface of the host cell, whereas the MUNANA and NA-Star techniques measure cleavage of small soluble chemical substrates (24). However, only the ELLA was proposed as the assay for measuring serum NA-inhibiting antibodies as a correlate for protection for humans (25).
Components in raw sera have non-specific inhibitory activity against NA activity (20). These initial findings were conducted with ferret sera that varied using different viruses from different influenza subtypes. However, treating sera with receptor-destroying enzyme (RDE) overnight and then heat-inactivating the sera for 8 h at 55°C mitigated the non-specific inhibition without loss of NA- or HA-specific inhibitory activity (20). The animal models used for influenza virus research are growing and now include more species. Not only is there a need to compare serological results between animal models that are used for human influenza viruses, but also endemic influenza virus infection in agricultural animal species requires a consistent method to quantify the NA-inhibiting antibodies as well. Therefore, it may be necessary to handle sera from different species differently when quantifying the NA inhibition responses, which may be key to determining overall vaccine effectiveness.
Therefore, animal sera from different species were characterized for their inherent inhibition of the ELLA with a panel of influenza viruses. Sera were compared for their ability to non-specifically inhibit the NA proteins of many influenza viruses representing different viral subtypes. Sera were collected and tested from varying animal serum sources against H1 and H3 human- and swine-isolated influenza strains as well as avian-isolated viruses with N2 and N3 proteins. Overall, there are many different variables that contribute to the interpretation of the ELLA assay, and understanding the innate characteristics of the host origin of the sera is critical to conducting the assay and interpreting the results. Therefore, it is important to standardize methodologies that will allow for consistent and reproducible results to assess anti-NA antibodies.
Materials and Methods
Viruses
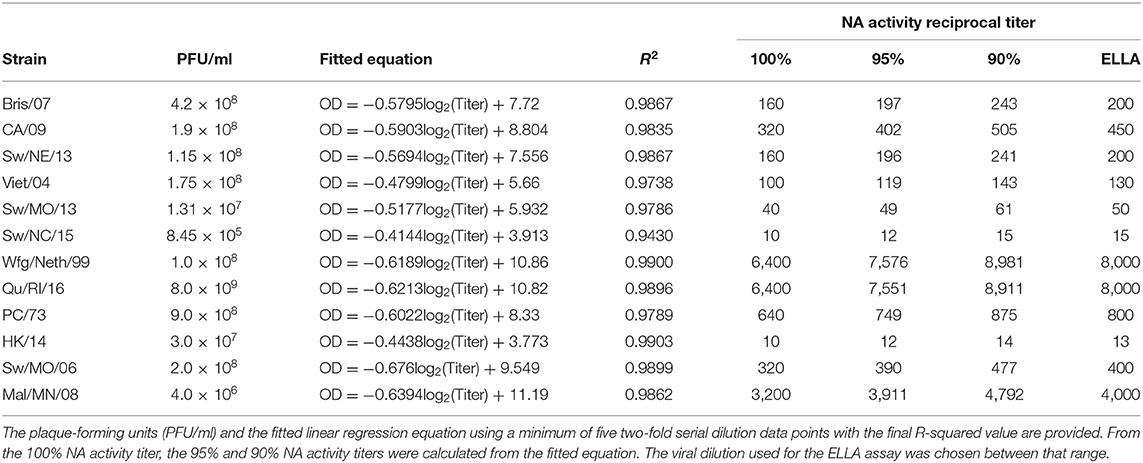
All swine viruses were passaged once in Madin–Darby canine kidney (MDCK) cell culture at 37°C, which was the same growth conditions as they were received in (26). The harvested virus was centrifuged at 2,500 rpm for 10 min to remove cell debris. Human and avian influenza viruses were propagated in 11-day-old embryonated chicken eggs. Virus lots were aliquoted for single-use applications and stored at −80°C. Viral titer of the frozen aliquots was determined with a plaque assay using MDCK cell culture in plaque-forming units per ml (PFU) (Table 1). The panel of viruses covered a range of N1 to N3 influenza NA subtypes, including A/Brisbane/59/2007 (H1N1) (Bris/07), A/California/07/2009xPR8 (6:2 viral reassortant with six internal genes from A/Puerto Rico/8/1934, and NA and HA external genes from virus indicated) (H1N1) (CA/09), A/swine/Nebraska/A10444614/2013 (H1N1) (Sw/NE/13), A/Vietnam/1203/2004xPR8 (H5N1) (Viet/04; HA gene contains mutation in multibasic cleavage site for BSL-2-level research), A/swine/Missouri/A10444664/2013 (H1N2) (Sw/MO/13), A/swine/North Carolina/152702/2015 (H1N2) (Sw/NC/15), A/white-fronted goose/Netherlands/22/1999 (H2N2) (Wfg/Neth/99), A/quail/Rhode Island/16-018622-1/2016 (H2N2) (Qu/RI/16), A/Port Chalmers/1/1973 (H3N2) (PC/73), A/Hong Kong/4801/2014 (H3N2) (HK/14), A/swine/Missouri/2124514/2006 (H2N3) (Sw/MO/06), and A/mallard/Minnesota/A108-3437/2008 (H2N3) (Mal/MN/08).
Animal Serum
Animal serum was either commercially sourced or generated in house. Sera were confirmed to be negative for preexisting antibodies to currently circulating human influenza viruses by HAI. Ferret sera originated from 6 to 8-months female finch ferrets (Mustela putorius furo, spayed, female, 6–8 months, descented) purchased from Triple F Farms (Sayre, PA); porcine sera originated from piglets at Auburn University; and rhesus macaque (Macaca mulatta) sera originated from previous dengue virus studies performed in the lab (27). The rat (cat #: 10710C), goat (cat #: 01-6201), horse (cat #: 31874), and mouse (cat #: 01-6501; NIH Swiss mouse) normal sera were harvested from non-immune animals (Invitrogen, Carlsbad, CA, USA) and rehydrated according to the manufacturer's specification; only one lot was tested for each commercial serum. Raw serum was not diluted any further before experimentation.
NA Activity and Inhibition Assay
High-affinity Immunoblot 4HBX 96-well flat-bottom plates (Thermo Fisher Scientific, Waltham, MA, USA) were coated overnight with 100 μl of 25 μg/ml fetuin (Sigma-Aldrich, St. Louis, MO, USA) in coating buffer (KPL coating solution concentrate; SeraCare Life Sciences Inc., Milford, MA, USA) and stored away from light for a maximum of 2 months at 4°C until use. Viruses were diluted to an initial dilution of 1:10 with Dulbecco's phosphate-buffered saline (DPBS) with Tween-20 and 1% BSA (DPBS-T-B), a PBS which contains 0.133 g/l CaCl2 and 0.1 g/l MgCl2 further supplemented with 1% BSA, and 0.5% Tween-20. Before virus addition, fetuin plates were washed three times in PBS-T (PBS + 0.05% Tween-20). Virus was diluted in two-fold serial dilutions within a range that allowed for linear regression analysis. After which, 50 μl of the viral dilutions was added to the fetuin-coated plate containing 50 μl of DPBS-T-B in duplicate. A negative control column was included containing 100 μl DPBS-T-B only. Plates were sealed and incubated for 16–18 h at 37°C and 5% CO2. After incubation, plates were washed six times in PBS-T. After washing, a diluted lectin was added to the plates to bind exposed galactose. Specifically, 100 μl of peanut agglutinin-HRPO (Sigma-Aldrich, St. Louis, MO, USA) diluted 1,000-fold in DPBS-B (DPBS, 1% BSA). Plates were incubated at RT for 2 h. Plates were washed three times in PBS-T, and 100 μl (500 μg/ml) of o-phenylenediamine dihydrochloride (OPD; Sigma-Aldrich, St. Louis, MO, USA) in 0.05 M phosphate-citrate buffer with 0.03% sodium perborate pH 5.0 (Sigma-Aldrich, St. Louis, MO, USA) was added to the plates. Plates were immediately incubated in the dark for 10 min at room temperature (20−22°C). The reaction was stopped with 100 μl of 1 N sulfuric acid. The absorbance was read at 490 nm. NA activity was determined after subtracting the mean background absorbance of the negative control wells. Linear regression analysis was used to determine the dilution of NA antigen necessary to achieve 90−95% NA activity and was used for subsequent NA inhibition ELLAs.
From each virus titration, at least five serial dilutions within the linear range were used to calculate the linear regression after transforming the dilutions by log2. The R-squared value above 0.9 was considered acceptable. The best-fit values for the slope (m) and y-intercept (b) were used to determine the 90−95% range. The lowest titer dilution used for regression was defined as the 100% NA activity dilution. Using the fitted linear regression equation, the optical density (OD100%) value for 100% NA activity was calculated. Then, the OD95% and OD90% were calculated by multiplying OD100% by 0.95 and 0.9, respectively. The range of viral dilution for 90−95% NA activity was then determined by using the OD95% and OD90% values in the linear regression equation to obtain lower and upper bounds for the virus dilution (Equation 1). Virus dilutions were then chosen between that range as indicated (Table 1).
The NI ELLA titers were determined from two-fold serially diluting sera in DPBS-T-B from 1:10 to 1:1,280. Duplicate dilutions were added to fetuin plates in 50 μl. The NA antigen was diluted to 90−95% NA activity in DPBS-T-B, and 50 μl was added to the plate. Controls were each a minimum of eight wells and included a positive NA antigen control (50 μl NA antigen + 50 μl DPBS-T-B) and a negative control (100 μl of DPBS-T-B) on each plate. Plates were incubated for 16–18 h at 37°C and 5% CO2 after which they were washed and processed, and absorbance was read as described above. Initially, the mean background absorbance from the negative control wells was subtracted from all wells. Then, NA percent activity was determined by dividing the serum absorbance by the mean virus-positive control wells multiplied by 100 (Equation 2).
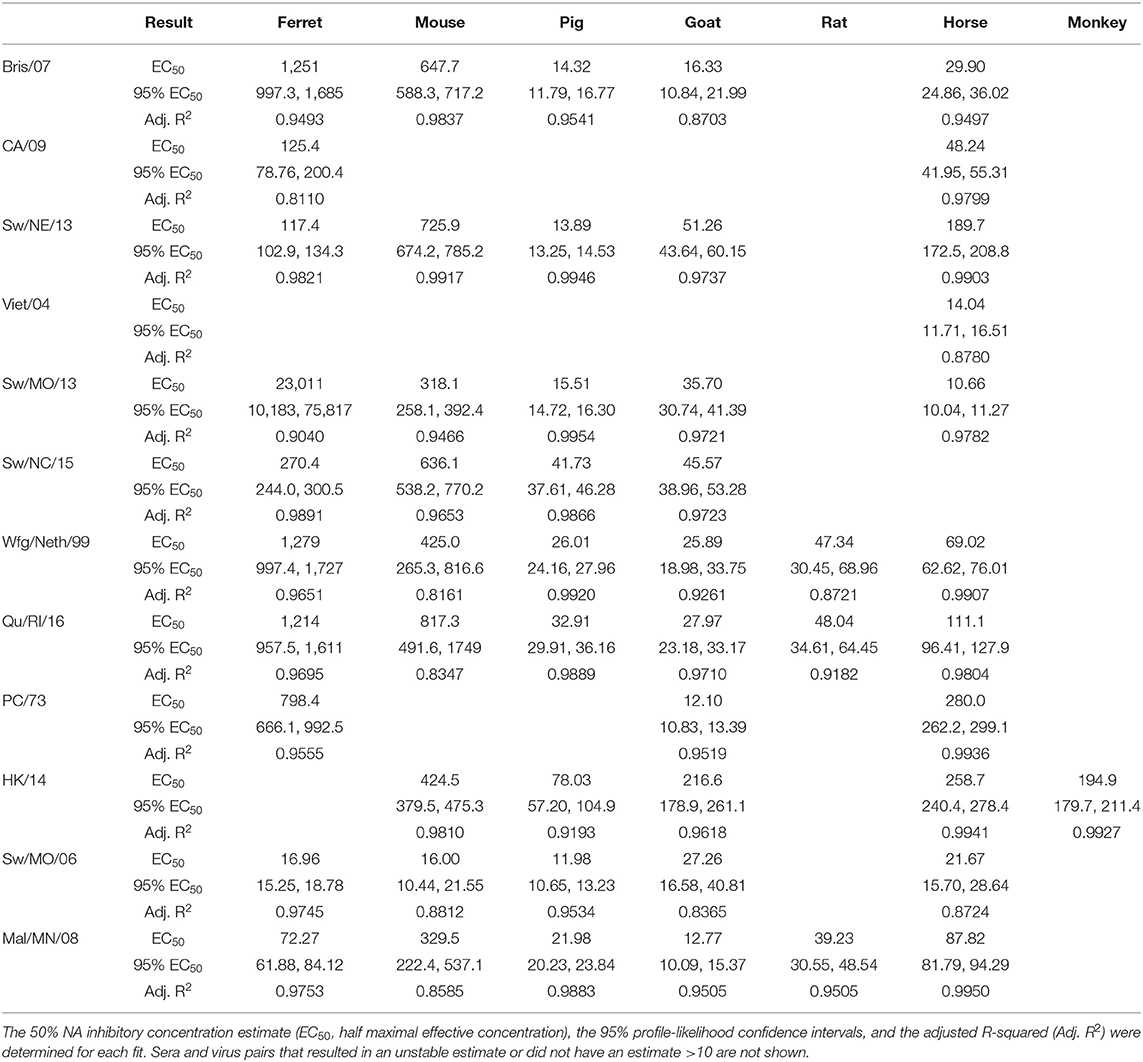
Non-linear regression fits were performed using GraphPad Prism version 9.1.1 (223) for MacOS (GraphPad Software, San Diego, CA, USA; www.graphpad.com), and the 50% NAI titer was estimated. Briefly, the “[Agonist] vs. normalized response—Variable slope” model was chosen which fits the model presented in Equation 3, which estimates the Hill slope and the half effective concentration (EC50). Outliers were not detected for or removed, and least-square regression with no weighting was used for the fitting. The model was constrained in that EC50 was >0. Asymmetrical profile-likelihood 95% confidence intervals of the EC50 were determined as well.
The lower limit of detection was 1:10, and the upper limit of detection was 1:1,280 due to the range of sera dilution tested.
Results
NA Titers of Influenza Viruses
The lowest dilution of virus needed to induce 100% NA activity varied between 1:10 and 1:6,400 for different influenza viruses (Table 1). Three HXN2 viruses had 100% NA titers below 100, 1:40 for Sw/MO/13 (H1N2), and 1:10 for both Sw/NC/15 (H1N2) and HK/14 (H3N2). Of these, the virus titer for only Sw/NC/15 was comparatively low at 8.45 × 105 PFU/ml, while the virus titers for Sw/MO/13 and HK/14 were 2.0 × 108 PFU/ml and 3.0 × 107 PFU/ml, respectively. The avian lineage H2N2 and H2N3 viruses had the highest 100% NA titers of 1:3,200 for Mal/MN/08 (H2N3) and 16,400 for both Wfg/Neth/99 (H2N2) and Qu/RI/16 (H2N2). The virus titer was not greater for these viruses than the others, therefore indicating that the increase in activity is not due to solely an increase in replicating virus.
Animal-Specific Raw Serum Inhibition of the Influenza NA
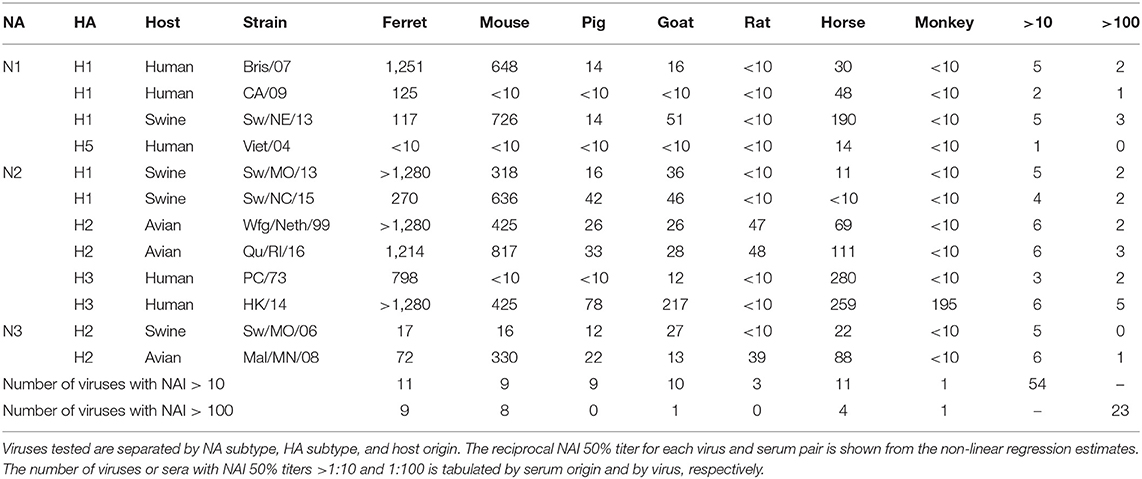
Sera collected from seven different sources were tested for the ability to inhibit the influenza virus NA activity as tested in the ELLA assay with fetuin substrate (Figure 1). Each serum sample was tested against 12 influenza viruses containing either NA type N1, N2, or N3. There were four swine origin viruses and three avian origin viruses. The 50% NAI titers were estimable for only some virus and serum pairs (Table 2).
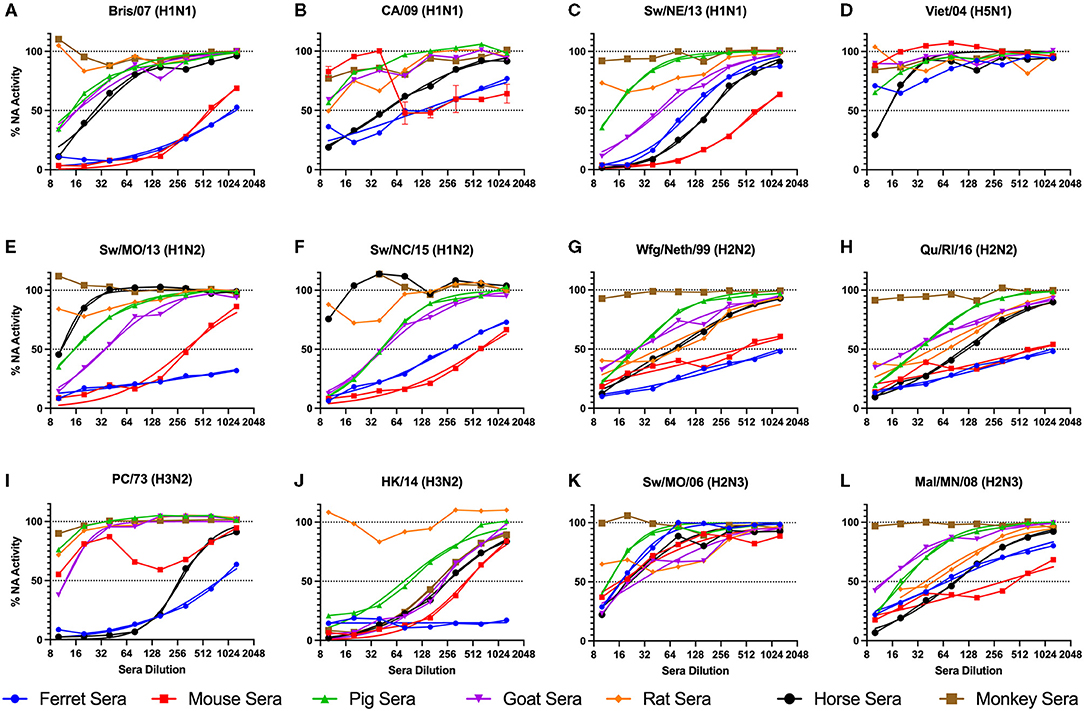
Figure 1. NA inhibition of influenza virus with addition of raw animal sera. A panel of influenza viruses were tested including N1 (A-D), N2 (E-J), and N3 (K,L) NA subtypes. The sera were two-fold serially diluted from the reciprocal dilutions of 10-1,280. Non-linear regression was conducted, and the regression that resulted in estimable parameters (as indicated in Table 2) are shown. The NA activity was normalized to 100% of a “virus only with no sera” control.
Ferret sera inhibited ELLA activity by 11 of the 12 viruses with a dilution titer >1:10 and 9 viruses with a titer >1:100 (Table 3). The rat sera inhibited the least number of viral NAs, inhibiting ELLA activity by three of the H2 viruses. Interestingly, not all animal sera inhibited all the same viruses (Table 3). For example, the Bris/07 (H1N1) virus was inhibited by ferret and mouse sera at a dilution >1:100, by pig, goat, and horse sera at a dilution >1:10, and was not inhibited by either rat or monkey sera. This variation was observed for other subtypes and host origin isolates. The Wfg/Neth/99 (H2N2) had a similar inhibition profile. The HK/14 (H3N2) virus was inhibited by the greatest number of sera. There was no distinguishable viral characteristic, such as host origin or HA or NA subtype, that was correlated with pattern of sera inhibition.
Discussion
Influenza vaccine formulations, including live-attenuated virus, whole-inactivated virus, and protein subunit minutes, use NA as a vaccine component to elicit NA-specific antibodies (28). However, components in raw sera have anti-NA properties that result in inhibition of NA activity. The ELLA is used to measure antibody-mediated NA inhibition for cleaving a large substrate, and has been used to assess the effectiveness of NA-containing vaccines and anti-NA antibodies (29–32).
In this study, seven raw animal sera were tested for inhibition of virus in the ELLA assay (Table 2). All sera, regardless of species, inhibited at least one influenza virus (50% inhibition) with a dilution of >1:10. Five of the seven samples inhibited 50% NAI activity at a titer of >1:100. Sera contain innate host influenza inhibitors, such as complement protein of the α-, β-, and γ-class serum inhibitors. In horse and pig sera, the α-2-macroglobulin (γ-class) is one of the major innate influenza virus-neutralizing factors (33, 34). The γ-class inhibitors express sialic acids that bind specifically to the HA protein on influenza viruses and may inhibit the NA through steric interactions. These γ-class inhibitors are inactivated through RDE treatment using Vibrio cholerae NA and are resistant to viral sialidase activity (34, 35). There appear to be minor innate factors that result in the ability of horse and pig sera to inhibit different viruses in the panel.
Not all sera inhibited NA activity of all viruses. There were distinct inhibition profiles against specific influenza viruses in the panel. Innate inhibitors interact with influenza viruses through competitive binding of sialic acids to the HA protein receptor-binding site (RBS) (α- and γ-class) and with mannose-binding lectins (β-class) (36, 37). Depending on the host origin of the virus, the HA RBS may have stronger affinity for α-2,3 or α-2,6 sialic acids. The glycosylation of HA proteins has been associated with mannose-binding lectins (37). Further research into the contributions of HA sialic acid binding specificity and the glycosylation of HA and NA surface proteins is needed to determine if it is significantly impacting the variation of NA inhibition observed here across the different viruses.
The innate NA inhibition of different species sera is useful for determining the appropriate treatment before conducting for ELLA assays. To account for the innate inhibitors observed here, sera may either be heat treated or RDE treated overnight at 37°C to cleave competing sialic acids from α- and γ-class inhibitors and heat inactivated at 56°C for a minimum of 30 min to inactivate the heat-labile β-class inhibitors and up to 8 h to fully inactivate the V. cholerae NA, when used with ferret sera (20). Immunoglobulins vary in their heat stability with IgG being more stable than IgA which is more stable than IgM (38). With researchers using different inactivation methods, it may be inappropriate to compare titers between sera heat inactivated for 30 min to RDE-treated sera that is heat inactivated for 8 h.
However, one of the major limitations of the study design was the inability to quantify within-species variability due to the limited sources of the sera. This variability can be further investigated to determine if age, sex, or husbandry practices, such as farm or laboratory origin animals, have any effect on the results. Furthermore, the serum inactivation procedure for conducting the ELLA may be different between species. To determine the appropriate method, positive control antiserum is necessary to confirm that no loss in NA-specific antibodies is observed during treatment. Given the wide panel of viruses and different animal models tested here, those samples were not available. Lastly, the wide variability in the NA activity titers observed between viruses (Table 1) may either be from increased enzymatic capacity, i.e., a virus' NA protein cleaves more sialic acid at a higher rate than another viral NA, or from having a higher NA content per PFU. Therefore, why different viruses had such variability in NA activity was undetermined.
In conclusion, with the increase in NA research, the RDE treatment, the inactivation time, and the temperature used to inactivate sialidases should be clearly described with the negative control data provided for each viral strain with serum species used for the assay in order to accurately interpret the results. This information will allow for comparison across species or if comparison of anti-NA serological results need to assessed within the same species.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by IACUC of University of Georgia.
Author Contributions
AS: concept, writing, analysis, and statistics. TR: editing and writing. All authors contributed to the article and approved the submitted version.
Funding
This work has been funded by the National Institute of Allergy and Infectious Diseases, a component of the NIH, Department of Health and Human Services, under the Collaborative Influenza Vaccine Innovation Centers (CIVICs) contract 75N93019C00052 and, in part, by the University of Georgia (UGA) (MRA-001). In addition, TR was supported by the Georgia Research Alliance as an Eminent Scholar.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Harrison Bergeron, Naoko Uno, and Z. Beau Reneer for assistance in obtaining sera. Furthermore, we would like to thank the following individuals for providing influenza viruses: Dr. Mark Tompkins at UGA for A/swine/North Carolina/152702/2015 (H1N2), Dr. Ralph Tripp at UGA for A/swine/Nebraska/A01444614/2013 (H1N1) and A/swine/Missouri/A01444664/2013 (H1N2), Dr. David Stallkecht at UGA for A/mallard/MN/2008 (H2N3), and Dr. Ron Fouchier at Erasmus MC for providing A/white-fronted goose/Netherlands/22/1999 (H2N2). Lastly, the remaining influenza A viruses were obtained through Virapur, BEI Resources, the International Reagent Resources (IRR), or from the Centers for Disease Control and Prevention, Atlanta, GA, USA.
References
1. Sederdahl BK, Williams JV. Epidemiology and clinical characteristics of influenza C virus. Viruses. (2020) 12:89. doi: 10.3390/v12010089
2. Tomley FM, Shirley MW. Livestock infectious diseases and zoonoses. Philos Trans R Soc Lond B Biol Sci. (2009) 364:2637–42. doi: 10.1098/rstb.2009.0133
3. Allkofer A, Garvey M, Ryan E, Lyons R, Ryan M, Lukaseviciute G, et al. Primary vaccination in foals: a comparison of the serological response to equine influenza and equine herpesvirus vaccines administered concurrently or 2 weeks apart. Arch Virol. (2021) 166:571–9. doi: 10.1007/s00705-020-04846-6
4. Paillot R, Hannant D, Kydd JH, Daly JM. Vaccination against equine influenza: quid novi? Vaccine. (2006) 24:4047–61. doi: 10.1016/j.vaccine.2006.02.030
5. Na W, Yeom M, Yuk H, Moon H, Kang B, Song D. Influenza virus vaccine for neglected hosts: horses and dogs. Clin Exp Vaccine Res. (2016) 5:117–24. doi: 10.7774/cevr.2016.5.2.117
6. Kim HH, Yang DK, Seo BH, Cho IS. Serosurvey of rabies virus, canine distemper virus, parvovirus, and influenza virus in military working dogs in Korea. J Vet Med Sci. (2018) 80:1424–30. doi: 10.1292/jvms.18-0012
7. Mancera Gracia JC, Pearce DS, Masic A, Balasch M. Influenza A virus in swine: epidemiology, challenges and vaccination strategies. Front Vet Sci. (2020) 7:647. doi: 10.3389/fvets.2020.00647
8. Everett HE, van Diemen PM, Aramouni M, Ramsay A, Coward VJ, Pavot V, et al. Vaccines that reduce viral shedding do not prevent transmission of H1N1 pandemic 2009 swine influenza a virus infection to unvaccinated pigs. J Virology. (2021) 95:e01787–20. doi: 10.1128/JVI.01787-20
9. Spackman E, Pantin-Jackwood MJ, Sitaras I, Stephens CB, Suarez DL. Identification of efficacious vaccines against contemporary North American H7 avian influenza viruses. Avian Dis. (2020) 65:113–21. doi: 10.1637/aviandiseases-D-20-00109
10. Wang T, Wei F, Liu J. Emerging role of mucosal vaccine in preventing infection with avian influenza A viruses. Viruses. (2020) 12:862. doi: 10.3390/v12080862
11. Chen YQ, Wohlbold TJ, Zheng NY, Huang M, Huang Y, Neu KE, et al. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell. (2018) 173:417–29 e10. doi: 10.1016/j.cell.2018.03.030
12. Cohen M, Zhang XQ, Senaati HP, Chen HW, Varki NM, Schooley RT, et al. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol J. (2013) 10:321. doi: 10.1186/1743-422X-10-321
13. Murti KG, Webster RG. Distribution of hemagglutinin and neuraminidase on influenza virions as revealed by immunoelectron microscopy. Virology. (1986) 149:36–43. doi: 10.1016/0042-6822(86)90084-X
14. Kilbourne ED. Comparative efficacy of neuraminidase-specific and conventional influenza virus vaccines in induction of antibody to neuraminidase in humans. J Infect Dis. (1976) 134:384–94. doi: 10.1093/infdis/134.4.384
15. Gilbert PB, Fong Y, Juraska M, Carpp LN, Monto AS, Martin ET, et al. HAI and NAI titer correlates of inactivated and live attenuated influenza vaccine efficacy. BMC Infect Dis. (2019) 19:453. doi: 10.1186/s12879-019-4049-5
16. Xiong FF, Liu XY, Gao FX, Luo J, Duan P, Tan WS, et al. Protective efficacy of anti-neuraminidase monoclonal antibodies against H7N9 influenza virus infection. Emerg Microbes Infect. (2020) 9:78–87. doi: 10.1080/22221751.2019.1708214
17. Krammer F, Fouchier RAM, Eichelberger MC, Webby RJ, Shaw-Saliba K, Wan H, et al. NAction! how can neuraminidase-based immunity contribute to better influenza virus vaccines? mBio. (2018) 9:e02332-17. doi: 10.1128/mBio.02332-17
18. Couzens L, Gao J, Westgeest K, Sandbulte M, Lugovtsev V, Fouchier R, et al. An optimized enzyme-linked lectin assay to measure influenza A virus neuraminidase inhibition antibody titers in human sera. J Virol Methods. (2014) 210:7–14. doi: 10.1016/j.jviromet.2014.09.003
19. Sandbulte MR, Eichelberger MC. Analyzing swine sera for functional antibody titers against influenza a neuraminidase proteins using an enzyme-linked lectin assay (ELLA). Anim Influenza Virus. (2014) 1161:337–45. doi: 10.1007/978-1-4939-0758-8_28
20. Westgeest KB, Bestebroer TM, Spronken MI, Gao J, Couzens L, Osterhaus AD, et al. Optimization of an enzyme-linked lectin assay suitable for rapid antigenic characterization of the neuraminidase of human influenza A(H3N2) viruses. J Virol Methods. (2015) 217:55–63. doi: 10.1016/j.jviromet.2015.02.014
21. Potier M, Mameli L, Belisle M, Dallaire L, Melancon SB. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate. Anal Biochem. (1979) 94:287–96. doi: 10.1016/0003-2697(79)90362-2
22. Buxton RC, Edwards B, Juo RR, Voyta JC, Tisdale M, Bethell RC. Development of a sensitive chemiluminescent neuraminidase assay for the determination of influenza virus susceptibility to zanamivir. Anal Biochem. (2000) 280:291–300. doi: 10.1006/abio.2000.4517
23. Wetherall NT, Trivedi T, Zeller J, Hodges-Savola C, McKimm-Breschkin JL, Zambon M, et al. Evaluation of neuraminidase enzyme assays using different substrates to measure susceptibility of influenza virus clinical isolates to neuraminidase inhibitors: Report of the neuraminidase inhibitor susceptibility network. J Clin Microb. (2003) 41:742–50. doi: 10.1128/JCM.41.2.742-750.2003
24. Shie JJ, Fang JM. Development of effective anti-influenza drugs: congeners and conjugates - a review. J Biomed Sci. (2019) 26:84. doi: 10.1186/s12929-019-0567-0
25. Krammer F, Weir JP, Engelhardt O, Katz JM, Cox RJ. Meeting report and review: Immunological assays and correlates of protection for next-generation influenza vaccines. Influenza Other Respir Viru. (2020) 14:237–43. doi: 10.1111/irv.12706
26. Organization WH Network WGIS. Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza. Geneva: World Health Organization (2011).
27. Uno N, Ross TM. A universal dengue vaccine elicits neutralizing antibodies against strains from all four dengue serotypes. J Virol. (2020) 95:e00658–20. doi: 10.1128/JVI.00658-20
28. Sandbulte MR, Gauger PC, Kitikoon P, Chen H, Perez DR, Roth JA, et al. Neuraminidase inhibiting antibody responses in pigs differ between influenza A virus N2 lineages and by vaccine type. Vaccine. (2016) 34:3773–9. doi: 10.1016/j.vaccine.2016.06.001
29. Desheva Y, Smolonogina T, Donina S, Rudenko L. Study of Neuraminidase-inhibiting antibodies in clinical trials of live influenza vaccines. Antibodies (Basel). (2020) 9:20. doi: 10.3390/antib9020020
30. Kaplan BS, Vincent AL. Detection and titration of influenza A virus neuraminidase inhibiting (NAI) antibodies using an enzyme-linked lectin assay (ELLA). Methods Mol Biol. (2020) 2123:335–44. doi: 10.1007/978-1-0716-0346-8_24
31. Paramsothy A, Lartey Jalloh S, Davies RA, Guttormsen AB, Cox RJ, Mohn KG. Humoral and cellular immune responses in critically Ill influenza A/H1N1 infected patients. Scand J Immunol. (2021) 2021:e13045. doi: 10.1111/sji.13045
32. Wang F, Wang Y, Wan Z, Shao H, Qian K, Ye J, et al. Generation of a recombinant chickenized monoclonal antibody against the neuraminidase of H9N2 avian influenza virus. AMB Express. (2020) 10:151. doi: 10.1186/s13568-020-01086-4
33. Ryan-Poirier KA, Kawaoka Y. Alpha 2-macroglobulin is the major neutralizing inhibitor of influenza A virus in pig serum. Virology. (1993) 193:974–6. doi: 10.1006/viro.1993.1208
34. Pritchett TJ, Paulson JC. Basis for the potent inhibition of influenza virus infection by equine and guinea pig alpha 2-macroglobulin. J Biol Chem. (1989) 264:9850–8. doi: 10.1016/S0021-9258(18)81736-7
35. Cwach KT, Sandbulte HR, Klonoski JM, Huber VC. Contribution of murine innate serum inhibitors toward interference within influenza virus immune assays. Influenza Other Respir Viruses. (2012) 6:127–35. doi: 10.1111/j.1750-2659.2011.00283.x
36. Krizanova O, Rathova V. Serum inhibitors of myxoviruses. Curr Top Microbiol Immunol. (1969) 47:125–51. doi: 10.1007/978-3-642-46160-6_6
37. Anders EM, Hartley CA, Jackson DC. Bovine and mouse serum beta inhibitors of influenza A viruses are mannose-binding lectins. Proc Natl Acad Sci USA. (1990) 87:4485–9. doi: 10.1073/pnas.87.12.4485
Keywords: neuraminidase, vaccine, ELLA, influenza, animal
Citation: Skarlupka AL and Ross TM (2021) Inherent Serum Inhibition of Influenza Virus Neuraminidases. Front. Vet. Sci. 8:677693. doi: 10.3389/fvets.2021.677693
Received: 08 March 2021; Accepted: 15 June 2021;
Published: 02 August 2021.
Edited by:
George C. Fthenakis, University of Thessaly, GreeceReviewed by:
Marie Rene Culhane, University of Minnesota, United StatesHongquan Wan, United States Food and Drug Administration, United States
Copyright © 2021 Skarlupka and Ross. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ted M. Ross, tedross@uga.edu
 Amanda L. Skarlupka
Amanda L. Skarlupka Ted M. Ross
Ted M. Ross