The FGFR4 Homolog KIN-9 Regulates Lifespan and Stress Responses in Caenorhabditis elegans
- Department of Biology, McMaster University, Hamilton, ON, Canada
Fibroblast growth factor receptors (FGFRs) regulate diverse biological processes in eukaryotes. The nematode Caenorhabditis elegans is a good animal model for studying the roles of FGFR signaling and its mechanism of regulation. In this study, we report that KIN-9 is an FGFR homolog in C. elegans that plays essential roles in aging and stress response maintenance. kin-9 was discovered as a target of miR-246, a microRNA that is positively regulated by the Axin family member pry-1. We found that animals lacking kin-9 function were long-lived and resistant to chemically induced stress. Furthermore, they showed a reduced expression of endoplasmic reticulum unfolded protein response (ER-UPR) pathway genes, suggesting that kin-9 is required to maintain a normal ER-UPR. The analysis of GFP reporter-based expression in transgenic animals revealed that KIN-9 is localized in the intestine. Overall, our findings demonstrate that kin-9 is regulated by miR-246 and may function downstream of pry-1. This study prompts future investigations to understand the mechanism of miRNA-mediated FGFR function in maintaining aging and stress response processes.
Introduction
Aging is the gradual deterioration of cellular and tissue functions that are regulated by both genetic and environmental factors (Kenyon, 2010; Lapierre and Hansen, 2012; Uno and Nishida, 2016). Genetic factors include components of conserved signaling pathways that regulate a host of cellular processes that are crucial for the development and proper functioning of healthy adults. Thus, it is essential to identify and understand the function of pathway components so that they can be targeted not only to promote normal growth, but also to develop potential treatments for associated diseases. To this end, our group is investigating the function of an Axin scaffolding protein homolog in the nematode Caenorhabditis elegans, PRY-1, which is necessary for the formation of many tissues and cell types, as well as for maintaining stress responses and aging (Mallick et al., 2019b, 2020, 2021a; Mallick and Gupta, 2022). To gain insights into the genetic network of pry-1, we performed mRNA and miRNA transcriptome profiling experiments (Ranawade et al., 2018; Mallick et al., 2019a). Together with biochemical and genetic studies, these data identified many factors that interact with pry-1, including aak-2/AMPK, daf-16/FOXO, and crtc-1/CRTCs (Mallick et al., 2020; 2021b). Our miRNA transcriptome analysis revealed six differentially expressed miRNAs in pry-1 mutants, five of which (lin-4, miR-237, miR-48, miR-84, and miR-241) were upregulated, and one (miR-246) was downregulated (Mallick et al., 2019a). miRNAs are non-coding RNAs that regulate target gene expression by binding to their conserved 3ʹ-untranslated region (UTR) or, less commonly, to the 5ʹ-UTR and coding sequences (Stefani and Slack, 2008; Ambros and Ruvkun, 2018; O’Brien et al., 2018). Studies have shown that miRNAs regulate diverse biological processes (Ambros and Ruvkun, 2018; O’Brien et al., 2018).
The present study focuses on miR-246 and one of its targets in mediating aging and stress resistance. miR-246 was reported to be the highest-fold upregulated gene during aging in both wild-type and long-lived daf-2 (insulin/insulin-like growth factor-1 signaling (IIS) receptor homolog) mutant (De Lencastre et al., 2010; Pincus et al., 2011). However, the mechanism underlying the action of miR-246 remains unclear. The results described here suggest that miR-246 acts genetically downstream of pry-1 to regulate the expression of a fibroblast growth factor receptor (FGFR) homolog to promote longevity and stress resistance in animals.
FGF signaling is conserved in eukaryotes and plays an important role in development and disease (DeVore et al., 1995; Ornitz and Itoh, 2015; Xie et al., 2020). Studies in mammalian models have revealed that the pathway is regulated by miRNAs (Yin et al., 2015; Copeland and Simoes-Costa, 2020); however, detailed mechanisms of interactions, specific processes, and pathway components are currently lacking. In this regard, the C. elegans system holds significant potential because it contains conserved miRNA families and FGF signaling that can be targeted by forward and reverse genetic approaches. While EGL-17 (FGF) and EGL-15 (FGFR) were previously shown to be necessary for sex specific muscle development (DeVore et al., 1995), they are not essential for the lifespan and stress responses in worms. Here, we provide evidence that KIN-9 is an FGFR4 family member that is repressed by mir-246. The receptor tyrosine kinase (RTK) domain of KIN-9 shows a high degree of sequence and structural similarity to those of FGFR4 mammalian family members. In agreement with this, we found that the KIN-9 overexpression phenotype resembled that of activated FGF signaling in C. elegans. A phenotypic analysis revealed that while kin-9 mutants are long-lived and stress-resistant, miR-246 mutants show the opposite phenotypes. We also found that kin-9 RNAi fully suppressed the lifespan and stress sensitivity of the miR-246 mutants. To further validate the regulatory relationship between miR-246 and kin-9, a chimeric GFP-kin-9-3ʹ UTR reporter was used, which showed increased fluorescence in miR-246 mutant animals. These data, together with the upregulation of KIN-9 in both the miR-246 and pry-1 mutants, support a model where pry-1 positively regulates miR-246, which in turn inhibits kin-9 expression. Analysis of kin-9::GFP transgenic animals revealed that the gene is expressed in the pharynx and intestine. Its presence in the intestine supports the role of kin-9 in lifespan maintenance, similar to that described for many other long-lived mutant strains (An and Blackwell, 2003; Libina et al., 2003; Taylor and Dillin, 2013). As kin-9 mutants are resistant to stress, we examined the expression of unfolded protein response (UPR) pathway components and chaperones. The results showed that the expression of the endoplasmic reticulum (ER) UPR components were downregulated, suggesting that lower kin-9 activity is beneficial for protein homeostasis. Overall, the results described in this study demonstrate the essential role of kin-9 in regulating mir-246-mediated lifespan and stress response in C. elegans.
Materials and Methods
Parts of the methods section are in the Supplementary Data S1.
Strains and Culture Conditions
Worms were cultured on standard NGM plates using established protocols and Escherichia coli strain OP50 as a food source (Brenner, 1974). The strains were maintained at 20°C unless otherwise mentioned.
RNAi
RNAi-mediated gene silencing was performed using a protocol previously published by our laboratory (Mallick et al., 2021a). Plates were seeded with Escherichia coli HT115 expressing either dsRNA specific to candidate genes or empty vector (L4440). Synchronized gravid adults were bleached, and eggs were plated. After becoming young adults, animals were analyzed for stress sensitivity and lifespan (Mallick et al., 2021a).
Computational Analysis
To identify the targets of miR-246, the computational algorithms TargetScan (Jan et al., 2011), PicTar (Lall et al., 2006), PITA (Kertesz et al., 2007) and STarMirDB (Rennie et al., 2016) were used. These tools predict miRNA targets based on 3ʹ UTR seed matches of genes. The searches generated miR-246 target candidates (Supplementary Figure S1; Supplementary Table S2).
Lifespan Analysis
Lifespan experiments were done following adult-specific RNAi treatment using a previously described protocol (Mallick et al., 2020). Animals were grown on NGM OP50 seeded plates till the late L4 stage after which they were transferred to RNAi plates. For lifespan analysis at different temperatures, animals were grown till the late L4 stage at 20°C following which they were shifted to either 15°C or 25°C. Plates were then screened daily for dead animals and surviving worms were transferred every other day till the progeny production ceased. Censoring was done for animals that either escaped, burrowed into the medium, showed a bursting of intestine from the vulva, or underwent bagging of worms (larvae hatch inside the worm and the mother dies).
U0126 Inhibitor Assay
NGM plates with U0126 inhibitor (U120-1 MG, Sigma-Aldrich) of 30 µM was adapted from described previously (Reiner et al., 2008; Sharanya et al., 2015). A stock concentration of 10 mM U0126 was prepared in DMSO and was added to plates right before pouring to achieve a plate concentration of 30 µM. Synchronized L1 animals were plated and allowed to grow till young adult stage following which stress sensitivity to 200 mM was examined.
Oil Red O Staining
Neutral lipid staining was done on synchronized day-1 adult animals using Oil Red O dye (Thermo Fisher Scientific, United States) following the previously published protocol. Quantification was then done using ImageJ software as described previously (Mallick and Gupta, 2020).
Results
We previously reported that PRY-1 regulates the expression of a set of heterochronic miRNAs, including the lin-4 and let-7 family members (Mallick et al., 2019a). miR-246, which affects the stress response and aging-related processes (De Lencastre et al., 2010), was also discovered in that study. It was found that miR-246 is not involved in the pry-1-mediated heterochronic pathway. Furthermore, its expression was downregulated in pry-1(mu38) larvae and adults (Mallick et al., 2019a).
kin-9 Expression is Regulated by miR-246 and its 3ʹ UTR Contains miRNA Consensus Binding Sites
To understand the biological role of miR-246, we focused on identifying its target genes. Because the phenotype caused by a miRNA deletion is expected to result from the increased activity of its target(s), we hypothesized that miR-246 loss-induced short lifespan and enhanced stress sensitivity would be suppressed by depletion of its target gene activity. The predicted targets of miR-246 were identified using computational algorithms (see Methods). We assessed the transcript levels of top three potential targets (cah-4, kin-9, and pbs-5) in an miR-246 deletion mutant that lacks the entire transcript and 5ʹ upstream sequence (n4636: 518 bp length) (Miska et al., 2007) (Figures 1A,B). It was expected that the transcript level of the target gene would be higher than normal in the miRNA mutant background. Our results revealed that, while kin-9 expression was significantly upregulated compared to the control, there was no change in cah-4 and pbs-5 expression (Figure 1B; Supplementary Figure S2).
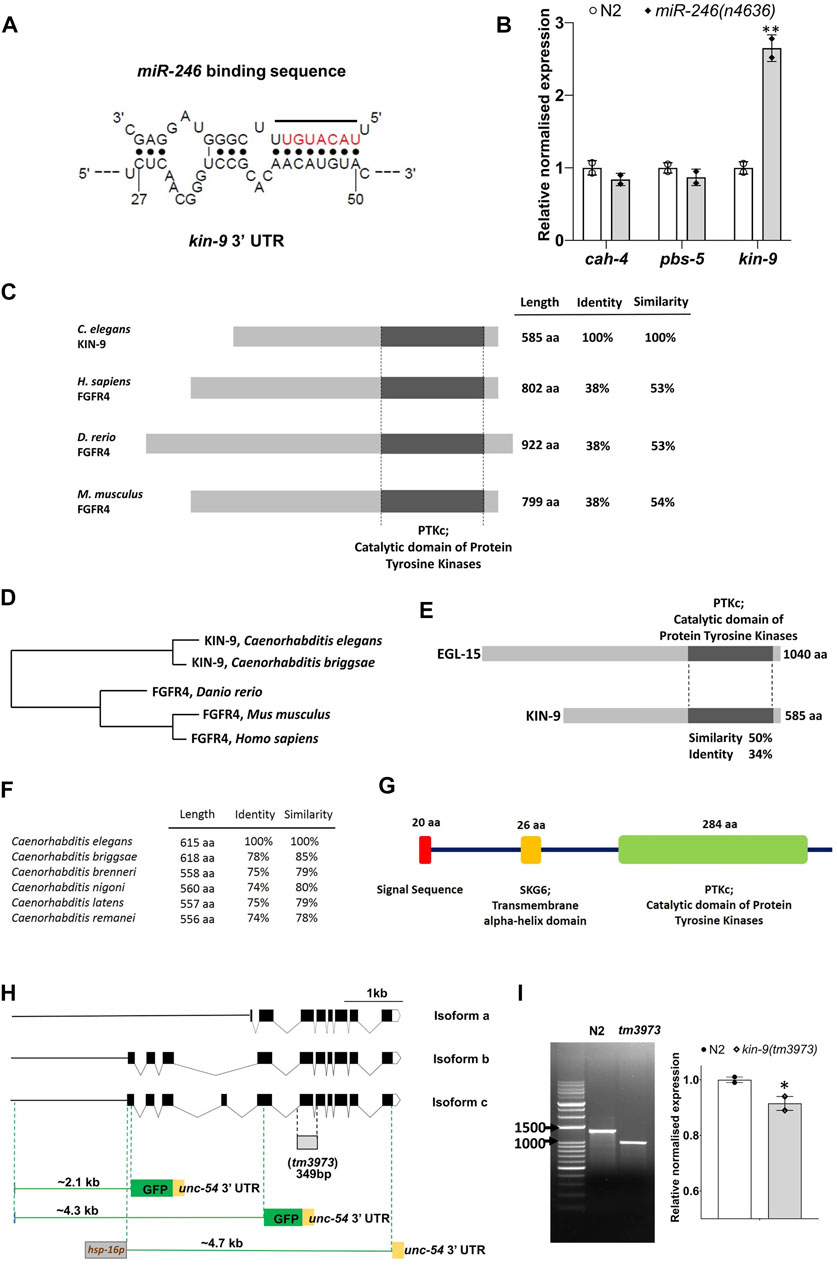
FIGURE 1. miR-246 mutants show overexpression of kin-9. (A) The predicted binding site of miR-246 at the 3′ UTR of kin-9 mRNA. (B) Expression analysis of three candidate genes cah-4, pbs-5 and kin-9 in the miR-246 mutants. (C) Schematic representation of KIN-9 and FGFR4 proteins from Caenorhabditis elegans, Homo sapiens, Danio rerio, and Mus musculus with percent identity and similarity indicated relative to C. elegans KIN-9. Conserved domains are aligned and are depicted with sizes, all presented to scale. (D) Phylogenetic tree of proteins shown in panel (C). (E) Schematic representation of C. elegans KIN-9 and EGL-15 proteins with percent identity and similarity indicated. (F) Similarities and identities between the KIN-9 proteins in the Caenorhabditis genus. (G) Protein domains and structure of KIN-9 protein. (H) Schematic dendrograms showing all the isoforms and tm3973 deletion allele of kin-9 with exons (black solid boxes), introns (bent lines) and upstream sequences (solid straight line). Regions used for creating transcriptional reporters and heat shock promoter-driven kin-9 overexpression are also shown. (I) PCR and qPCR analyses of the tm3973 allele. Gel image showing the shorter fragment of kin-9 transcript and bar graph showing kin-9 mRNA levels in the tm3973 mutants. (B, I) Each data represents the mean of two replicates and error bars the standard error of means. Significance was calculated using Bio-Rad software (one-way ANOVA) and significant differences are indicated by stars (*): * (p < 0.05), ** (p < 0.01).
Sequence analysis revealed that KIN-9 is a member of the FGFR4 family (http://www.wormbase.org). The RTK domain of KIN-9 is approximately 53% similar between the mouse and human FGFR4 proteins (Figures 1C,D). Outside this domain, no significant sequence similarity with FGFR4 was detected (Figure 1C). We also used the secondary structure prediction tool Jpred4 (http://www.compbio.dundee.ac.uk/jpred/) (Drozdetskiy et al., 2015), which confirmed KIN-9 homology with human FGFR4 (PDB ID: 6jpj, 6jpe, 5jkg, 4uxq, 4qrc, and 4qqt). Finally, to determine whether the KIN-9 RTK domain possesses conserved tyrosine kinase phosphorylation sites present in FGFR, the online program group-based prediction system (GPS 5) was utilized (http://gps.biocuckoo.cn/online.php) (Xue et al., 2011). The analysis revealed six such sites that, together with secondary structure prediction, established KIN-9 as a bona fide FGFR family member in C. elegans (Supplementary Figure S3). It is worth mentioning that a previously characterized FGFR in C. elegans, EGL-15, shares 50% sequence similarity with the RTK domain of KIN-9 (Figure 1E; Supplementary Figure S4) (Schutzman et al., 2001). KIN-9 orthologs were also found in other nematode species (http://www.wormbase.org) (Figures 1F). The kin-9 gene is predicted to produce three isoforms of varied sizes (478 aa (a), 585 aa (b), and 615 aa (c), all of which have a conserved RTK domain. The longest isoforms (b and c) also possess an N-terminal signal sequence and a transmembrane alpha-helix domain (Figures 1G,H).
kin-9 mutants are Stress Resistant and Long-Lived Whereas hsp::kin-9 Animals are Stress Sensitive and Die Prematurely
A deletion mutant of kin-9, tm3973, was used to investigate the gene function. The mutation was found to remove a portion of the RTK domain, leading to multiple in-frame stop codons (see Supplementary Data S1). These changes are expected to cause truncated proteins for all three isoforms (167 aa, 271 aa, and 304 aa) (Figure 1H). Interestingly, a cDNA analysis showed the presence of a roughly 990 bp transcript in kin-9 deletion mutant (tm3973) worms, indicating a readthrough transcription (Figure 1I). While it is unclear whether the tm3973 mutation allows translation, any products arising from this allele are expected to be non-functional.
kin-9 deletion mutant (tm3973) animals exhibited no obvious morphological defects but appeared to have a slight growth delay and lay significantly fewer eggs (Figures 2A,B); in addition, L1 larvae showed resistance to starvation (Figure 2C). Since brood size and L1 survival may be affected by lipid levels (Watts and Ristow, 2017; Ranawade et al., 2018), we performed Oil Red O staining and found no change in neutral lipid levels (Figure 2D). The tm3973 animals also showed increased resistance to paraquat and tunicamycin but were insensitive to heat stress (Figures 2E–G). These data suggest that kin-9 plays an important role in the stress response maintenance. Further support for this conclusion comes from the expression analysis of the endoplasmic reticulum unfolded protein response (ER-UPR) genes. We found that kin-9 mutants affected the ER-UPR pathway, as evidenced by the reduced expression of ire-1, pek-1, xbp-1, and the chaperone hsp-4 (Figures 2H,I). Consistent with these results, the mutants exhibited electrotaxis defects associated with chronic stress (Figure 2J) (Taylor et al., 2021). No change in the expression of mitochondrial UPR components was detected in the absence of kin-9 function (Figures 2H,I).
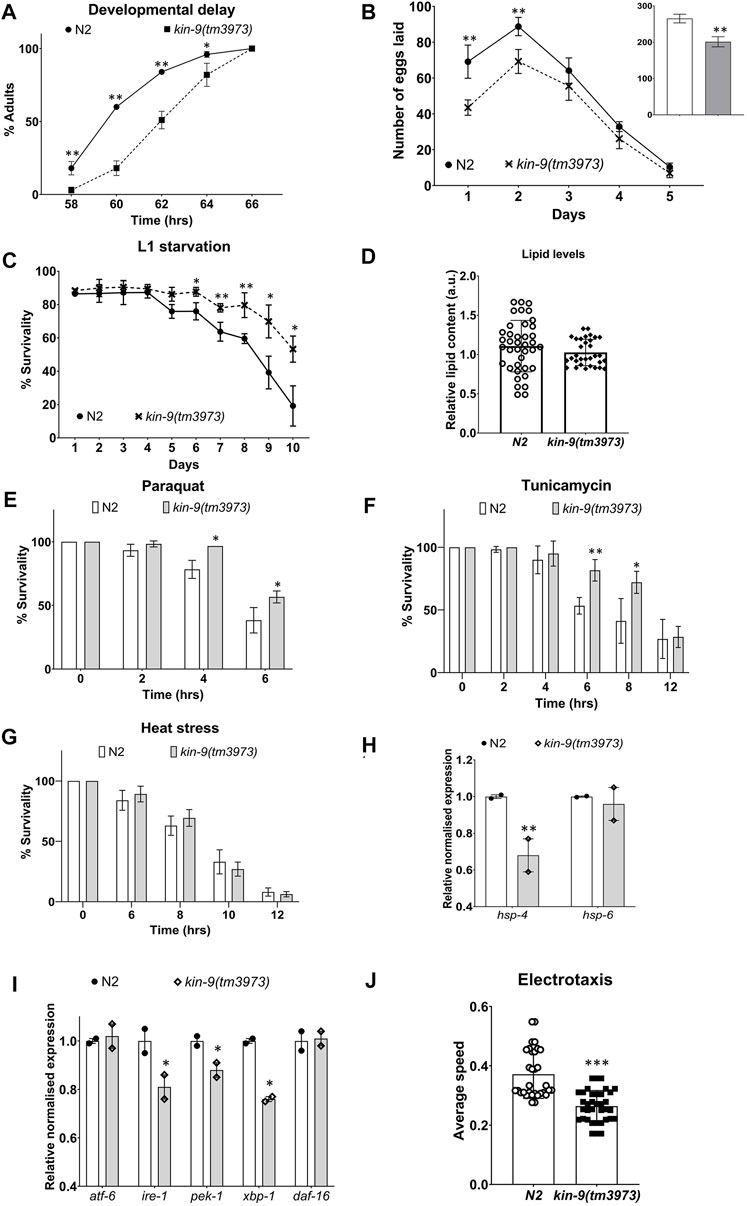
FIGURE 2. kin-9 mutants exhibit resistance to heat and chemical-induced stress. (A) Graph showing the time taken to reach adulthood. (B) Line graph showing the number of eggs laid by kin-9 mutants compared to N2. Smaller bar graph showing the total number of eggs laid (N2- clear bar; kin-9(tm3973) grey bar). (C) Survival graph of kin-9(tm3973) L1 worms upon starvation compared to N2. (D) Quantification of neutral lipids using Oil Red O staining. (E,F) Bar graph showing the survivability of kin-9 mutants compared to N2 following paraquat (200 mM) and tunicamycin (25 ng/μL) over a period of 6 and 12hrs respectively. (G) Survivability of kin-9 mutants compared to N2 at 35°C over a period of 12hrs. (H,I) qPCR analysis of daf-16 genes in kin-9(tm3973) adults compared to N2. Data in (H–I) represent the mean of two replicates and error bars the standard error of means. Significance was calculated using Bio-Rad software (one-way ANOVA). (J) Electrotaxis analysis of day-1 kin-9(tm3973) adults. (A–G,I) Data represent the mean of two replicates (n > 40 animals in each replicate) and error bars represent the standard deviation. Statistical analyses were done using multiple unpaired t-tests and significant differences are indicated by stars (*): * (p < 0.05), ** (p < 0.01), *** (p < 0.001).
Because miR-246 mutants are short-lived (De Lencastre et al., 2010), we examined the lifespan phenotype of animals lacking kin-9 function. Consistent with kin-9 being a downstream target, mutant animals showed an extended lifespan (Figure 3A; Supplementary Table S3). Similar lifespan changes were observed in the RNAi-treated animals (Figure 3B; Supplementary Table S3). The mutants exhibited slightly increased body bending rates but no change in pharyngeal pumping (Supplementary Figure S5).
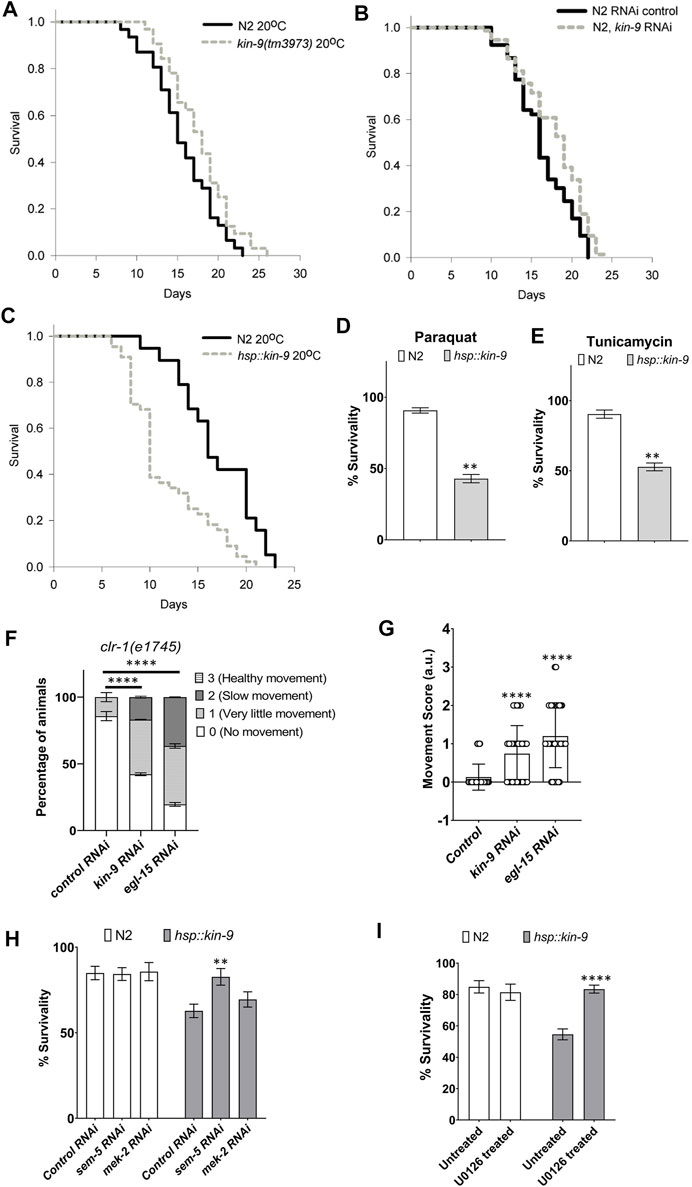
FIGURE 3. Loss of kin-9 function extends lifespan. (A) Lifespan graphs of kin-9 (tm3973) mutants compared to N2 (B) Lifespan analysis of animals following control and kin-9 RNAi knockdown during adulthood. (C) Lifespan graphs of hsp::kin-9 animals compared to N2. (A–C) See the Methods section and Supplementary Table S3 for statistics performed. (D,E) Bar graph showing the survivability of hsp::kin-9 adults compared to N2 following paraquat (200 mM) and tunicamycin (25 ng/μL) exposure for 2hrs. (F,G) Staggered bar graphs (F) and bar graphs (G) showing the analyses of movement defect in clr-1 mutants following control, kin-9 and egl-15 RNAi. In panel F movement is quantified on a scale of 0–3 and in panel G average score for movement has been plotted. (H) Bar graphs showing the survivability of N2 and hsp::kin-9 adults in 200 mM paraquat for 2hrs following control, sem-5 and mek-2 RNAi knockdown. (I) Bar graphs showing the survivability of N2 and hsp::kin-9 adults in 200 mM paraquat for 2hrs following U0126 inhibitor treatment. (D–I) Data represent the mean of two to three replicates (n > 50 animals in each replicate) and error bars represent the standard deviation. Statistical analyses were done using multiple unpaired t-tests with Welch correction (D,E,H,I), one-way ANOVA with multiple comparison test (G) and 2-way ANOVA with Sidak multiple comparison test (F). Significant differences are indicated by stars (*): ** (p < 0.01), *** (p < 0.001) and **** (p < 0.001).
If the absence of kin-9 results in a lifespan extension, then its increased levels should give rise to the opposite phenotype. To test this hypothesis, hsp::kin-9 transgenic lines were generated that overexpressed kin-9 following heat treatment (see Supplementary Data S1). High levels of kin-9 expression in these animals were confirmed using qPCR (Supplementary Figure S6). Consistent with the longer lifespan of kin-9 mutants, transgenic animals overexpressing kin-9 were small, short-lived, and exhibited slower pharyngeal pumping and body bending (Figure 3C; Supplementary Figures S5, S7, S8). The aging phenotype was similar at other growth temperatures (15 and 25°C, Supplementary Figure S7; Supplementary Table S3). Moreover, hsp::kin-9 animals exhibited an increased stress sensitivity to both paraquat and tunicamycin (Figures 3D,E). Altogether, these data demonstrate an important role for kin-9 in regulating the normal lifespan and stress responses of animals.
Interestingly, we found that hsp::kin-9 animals were unusually transparent when grown at 25°C (Supplementary Figure S8), an appearance that resembled ‘Clear (Clr)’ phenotype reported earlier in C. elegans that have activated FGF signaling (Supplementary Figure S8) (Borland et al., 2001). To investigate whether kin-9 genetically interacts with clr-1, we performed RNAi experiments. The results showed that while kin-9 RNAi had no effect on the Clr phenotype, it suppressed movement defects of animals (Figures 3F,G). In control experiments, egl-15 knockdown suppressed both the Clr and movement defects of clr-1 mutants (Figures 3F,G).
Additionally, we performed RNAi knockdowns of two of the FGF pathway components, MEK-2 (Erk Kinase) and SEM-5 (Grb2) (Borland et al., 2001). The results showed that sem-5 RNAi suppressed stress sensitivity of hsp::kin-9 animals (Figure 3H). No such response was not observed in the case of mek-2 (Figure 3H). Since C. elegans contains two Erk kinases (mek-1 and mek-2), we reasoned that there may be a genetic redundancy and, therefore, used a MEK inhibitor U0126 (Favata et al., 1998; Reiner et al., 2008; Sharanya et al., 2015). As expected, treating hsp::kin-9 animals with U0126 fully rescued their stress sensitivity to PQ (Figure 3I). Altogether, these data support our conclusion that KIN-9 function is mediated by the ERK kinase signaling, consistent with its role in regulating FGF signaling.
kin-9 is Expressed in the Pharynx, Intestine, and Tail Region
Considering the kin-9’s essential role in C. elegans, we wanted to identify the cells and tissues in which the gene is expressed. To this end, transgenic strains carrying kin-9::GFP reporters were generated. Two different constructs were utilized (see Supplementary Data S1), the longest of which (4.3 kb) contained a part of an exon that is common to all isoforms, whereas the shorter one (2.1 kb) is specific to isoforms b and c (Figure 1H). The analysis of transgenic lines showed GFP fluorescence throughout their lifespan, which agrees well with previously published transcriptomic data (Golden et al., 2008; Grün et al., 2014). We found that kin-9p(2.1 kb)::GFP adults exhibited fluorescence in the pharynx, intestine, and certain cells located in the tail (Figure 4A). While kin-9p(4.3 kb)::GFP animals exhibited a similar pattern, interestingly, no fluorescence was observed in the posterior region (Supplementary Figure S8). It is possible that the longer fragment has certain inhibitory sequences that contribute to differences in expression. More experiments involving the dissection of regulatory sequences are needed to investigate this possibility. The intestinal localization of KIN-9 and its persistence during adulthood agree well with the expression of other genes that promote lifespan and stress response maintenance (An and Blackwell, 2003; Libina et al., 2003; Taylor and Dillin, 2013). However, the specific role of KIN-9 in the pharynx remains to be determined.
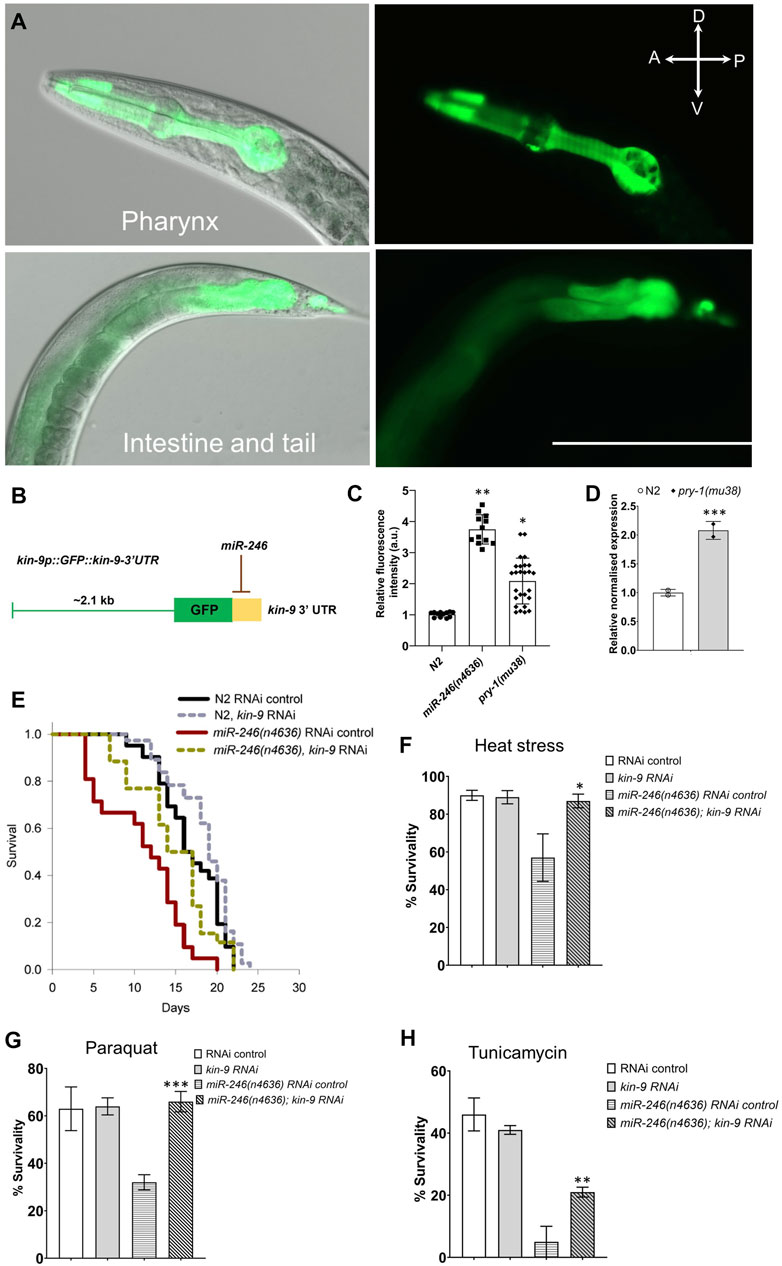
FIGURE 4. Lowering kin-9 activity suppresses miR-246 mutant defects. (A) kin-9p (2.1 kb):GFP analysis shows expression in the pharynx, intestine, and tail neurons. (B) Schematic diagram of a kin-9p::GFP::kin-9 3′ UTR construct showing potential miR-246 binding at the 3′ UTR of the kin-9 mRNA transcript. (C) Bar graph showing the GFP analysis using the array kin-9p::GFP::kin-9 3′ UTR in miR-246 (n4636) and pry-1(mu38) adults compared to control. Data represent the mean of two replicates (n > 25 animals in each replicate) and error bars represent the standard deviation. Statistical analyses were done using one-way ANOVA with Dunnett’s post hoc test and significant differences are indicated by stars (*): * (p < 0.05), ** (p < 0.01). (D) qPCR analysis of kin-9 gene in the pry-1(mu38) adults compared to control. Data represent the mean of two replicates and error bars the standard error of means. Significance was calculated using Bio-Rad software (one-way ANOVA) and significant differences are indicated by stars (*): *** (p < 0.001). (E) Lifespan analysis of N2 and miR-246 (n4636) animals following control and kin-9 RNAi knockdown. See the Supplementary Data S1 and Supplementary Table S3 for statistics performed. (F–H) Bar graphs showing survivability of N2 and miR-246 (n4636) animals following control and kin-9 RNAi when exposed to heat stress (35°C), paraquat (200 mM for 2 h) and tunicamycin (25 ng/μl for 2 h). Data represent the mean of two replicates (n > 50 animals in each replicate) and error bars represent the standard deviation. Statistical analyses were done using one-way ANOVA with Dunnett’s post hoc test and significant differences are indicated by stars (*): * (p < 0.05), ** (p < 0.01).
kin-9 3ʹ UTR is Targeted by miR-246 and kin-9 RNAi Rescues miR-246(n4636) Defects
Since miRNAs function mainly by binding to the 3ʹ UTR of target genes (Ambros and Ruvkun, 2018; O’Brien et al., 2018), we examined whether miR-246 affects the transcriptional regulation of kin-9. To this end, transgenic lines were generated containing a chimera of GFP and kin-9 3ʹ UTR under the control of the kin-9 promoter (kin-9p(2.1 kb)::GFP::kin-9 UTR) (Supplementary Data S1; Figure 4B). As expected, GFP fluorescence increased approximately four-fold when the construct was introduced into miR-246(n4636) animals (Figure 4C). Since miR-246 is positively regulated by pry-1 (Mallick et al., 2019a), we also examined kin-9p(2.1 kb)::GFP::kin-9UTR fluorescence in pry-1 mutants and found similar upregulation (Figure 4C). Consistent with this, qPCR analysis revealed that kin-9 transcript levels were high in animals lacking the pry-1 function (Figure 4D). Altogether, these data show that pry-1 and miR-246 negatively regulate kin-9 expression.
As kin-9 expression is upregulated in miR-246 mutants and miR-246 regulates kin-9 transcript levels, we tested whether miR-246(n4636) phenotypes could be rescued by reducing kin-9 function. The lifespan and stress sensitivity defects of miR-246 mutants were significantly rescued by kin-9 RNAi (Figures 4E–H; Supplementary Figure S9, Supplementary Table S3). Together with transcript analysis and data from previous sections, these results demonstrate that kin-9 is involved in lifespan regulation and acts downstream of miR-246. Interestingly, no phenotypic rescue was observed by knocking down kin-9 in the pry-1 mutant animals (Supplementary Figure S10, Supplementary Table S3), suggesting that kin-9 is not the sole effector of pry-1 function in aging-related processes. These findings are consistent with the interaction of pry-1 with multiple pathway components to regulate aging and the stress response in C. elegans.
Discussion
We identified kin-9 as a new target of the miRNA miR-246 in C. elegans and demonstrated its essential function in regulating stress responses and the lifespan of animals. Sequence analysis of kin-9 revealed that it is a member of the FGFR family, with the RTK domain being the most similar to FGFR4. Furthermore, its overexpression using a kin-9 transgenic system resulted in a Clr phenotype similar to that observed in activated FGF signaling conditions (Borland et al., 2001; Schutzman et al., 2001). The Clr phenotype is characterized by the accumulation of clear fluid within the pseudocoelomic cavity. Such animals appear to have floating intestines with fluid-filled body cavities and are short, immobile, and sterile (Borland et al., 2001; Schutzman et al., 2001). A similar trait was also observed in other FGF pathway component mutants (Schutzman et al., 2001). Interestingly, kin-9 RNAi did not affect the Clr phenotype of clr-1 mutants. However, movement defect of animals was rescued similar to egl-15 knockdown. Furthermore, we found that the MEK inhibitor U0126 fully suppressed the stress sensitivity of animals overexpressing kin-9. Overall, these data lead us to conclude that kin-9 regulates FGF signaling in C. elegans.
Studies in other animal models have reported the miRNA-mediated regulation of FGFs and FGFRs. For example, miR-140 regulates FGF9 during lung development, and miR-200a, miR-20a, and miR-217 regulate FGF4, FGF13, and FGFR12, respectively, during the establishment of the neural crest territory (Yin et al., 2015; Copeland and Simoes-Costa, 2020). The work described here represents the first such study in C. elegans and provides a unique opportunity to investigate the miRNA-mediated regulation of FGFR. It is worth mentioning that, while the mammalian systems contain twenty-three FGF family members (FGF1-23) and four FGFRs (FGFR1-4), C. elegans carries only two ligands (EGL-17 and LET-756) and two receptors (EGL-15 and KIN-9) (Borland et al., 2001; Ornitz and Itoh, 2015; Xie et al., 2020). The simplicity of the worm system, together with its short lifespan and powerful genetic approaches to manipulate the genes, make it possible to investigate the mechanism of kin-9 function and identify other pathway components that act genetically downstream of miR-246.
Previously, miR-246 was identified in a transcriptomic study in our lab as being positively regulated by pry-1 (Mallick et al., 2019a). Considering that miRNAs are necessary for lifespan maintenance (De Lencastre et al., 2010), we aimed to identify their targets to further understand the role of the pry-1-miR-246 genetic network. While our in-silico analysis revealed three genes with consensus miR-246 binding sites in their 3ʹ UTR, kin-9 was the only gene with increased expression in the miR-246 mutant background. Consistent with this, a GFP transgene containing the kin-9 3ʹ UTR was responsive to miR-246 activity, that is, in the absence of miR-246, fluorescence was significantly increased.
Analysis of kin-9 mutants has revealed the essential role of this gene in regulating stress responses and aging-related processes. While animals lacking kin-9 function showed resistance to stress treatments, reduced expression of heat shock chaperone and ER-UPR genes, and a longer lifespan, transgenic lines overexpressing kin-9 showed the opposite phenotypes. These data show that perturbations in kin-9 function have opposite effects on stress response and ER-UPR gene expression. Studies have shown that hsp-4 expression levels do not always correlate with stress sensitivity of animals. Thus, for example, hsp-4 is downregulated in both atf-6 (stress resistant) and xbp-1 (stress sensitive) mutants (Taylor and Dillin, 2013; Burkewitz et al., 2020). More work is needed to fully elucidate the cellular and molecular mechanisms of kin-9 in ER-UPR and stress response maintenance. It may be that a reduction in hsp-4 and other ER-UPR genes in kin-9 mutants is associated with beneficial effects such as improved proteostasis, thereby contributing to increased stress resistance and better health of animals. In this regard, experiments examining kin-9’s involvement in protein aggregation, various forms of stress, and degradation and clearing of intracellular factors should provide valuable information.
Consistent with the lifespan phenotype of kin-9 mutants, the gene is expressed in the intestine, a tissue known to be the primary player in nutrient uptake and metabolic activities (Libina et al., 2003; Rera et al., 2013). Studies on aging have shown that the intestine communicates with other parts of the body, such as neurons and muscles, leading to the activation of downstream effectors. For example, the IIS transcription factor DAF-16, which functions mainly in neurons and the intestine, affects muscle health and mitochondrial mass, suggesting cross-talk between these tissues (Libina et al., 2003; Uno and Nishida, 2016; Wang et al., 2019; Mallick et al., 2020; Mallick and Gupta 2022). Thus, it is conceivable that kin-9 regulates stress responses and lifespan by maintaining a healthy gut, which in turn signals to other tissues to promote their health.
Our data support a model in which pry-1 promotes miR-246 expression which in turn inhibits kin-9 expression. However, the precise mechanism underlying this genetic relationship remains unclear. Although kin-9 expression was inhibited by pry-1, knockdown of the gene did not suppress the pry-1 phenotype. It may be that kin-9 regulates only a small set of pry-1 downstream components and is unable to alter pry-1 signaling significantly in an independent manner. Previous studies on pry-1/Axin have revealed its genetic network, which includes multiple signaling components (Mallick et al., 2019b). For example, PRY-1 interacts with AAK-2/AMPK in the muscle in a cell-non-autonomous manner to regulate DAF-16 expression in the intestine to promote the lifespan and muscle health of animals (Mallick et al., 2020). PRY-1 also regulates the CABIN1 domain-containing protein, PICD-1, to affect calcineurin signaling and CRTC-1-dependent transcription (Mallick et al., 2021b). Recently, we identified several genes (cpz-1/CTSZ, cdk-1/CDK1, rnr-1/RRM1, his-7/H2AX, and ard-1/HSD17B10) that function downstream of pry-1 to regulate the lifespan and stress response of animals (Mallick et al., 2021a). Taken together, these findings demonstrate that pry-1 acts as a master regulator of aging-related processes and functions by coordinating the activities of diverse genes and pathways.
KIN-9 is the first identified FGFR family member in C. elegans that plays an essential role in aging and stress response maintenance. Previously, FGF signaling (EGL-15-EGL-17) was reported to be necessary to promote similar processes mediated by the homologs of the transmembrane proteins Klotho (KLO-1 and KLO-2), although EGL-15 and EGL-17 mutants independently do not have any such phenotypes (Château et al., 2010). Studies in higher eukaryotes have also shown that Klotho promotes lifespan and functions as a co-receptor for FGFRs (Kuro-o et al., 1999; Ornitz and Itoh, 2015). Moreover, there is evidence that mammalian FGFs cause age-related changes. For example, age-associated impairment of human mesenchyme-derived progenitor cells can be reversed by FGF2 treatment (Hurley et al., 2016). Additionally, the activated FGF2 pathway causes increased fat accumulation in aged human skeletal muscles (Mathes et al., 2021).
Our analysis of kin-9 as a target of miR-246 and its potential role genetically downstream of pry-1 prompt future studies to investigate the mechanism of its regulation and conservation across eukaryotes. Whether kin-9 utilizes the known components of FGF signaling remains to be determined. Interestingly, kin-9 RNAi was shown earlier to delay the development of let-60/Ras mutant animals (Byrne et al., 2007). The identification of kin-9 pathway components and their interactions with pry-1 holds significant promise in advancing our understanding of Axin signaling in stress maintenance and aging.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
Author Contributions
BPG and AM designed the study. AM, SKBT, LX, SM, and HH performed the experiments and analyzed data. A.M. produced the final figures and tables. BPG, AM, and SKBT wrote the manuscript.
Funding
This work was supported by funds from the National Sciences and Engineering Research Council of Canada (NSERC) Discovery grant to BG and NSERC Canada Graduate Doctoral scholarship to AM.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank M.H. Minhas for performing the electrotaxis experiment. The tm3973 allele was kindly provide by the National BioResource Project, Japan. Some of the strains were obtained from CGC, which is funded by the NIH Office of Research Infrastructure Programs (P400D01044).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fragi.2022.866861/full#supplementary-material
References
Ambros, V., and Ruvkun, G. (2018). Recent Molecular Genetic Explorations of caenorhabditis Elegans microRNAs. Genetics 209, 651–673. doi:10.1534/genetics.118.300291
An, J. H., and Blackwell, T. K. (2003). SKN-1 linksC. Elegansmesendodermal Specification to a Conserved Oxidative Stress Response. Genes Dev. 17, 1882–1893. doi:10.1101/gad.1107803
Borland, C. Z., Schutzman, J. L., and Stern, M. J. (2001). Fibroblast Growth Factor Signaling inCaenorhabditis Elegans. BioEssays 23, 1120–1130. doi:10.1002/bies.10007
Brenner, S. (1974). THE GENETICS OF CAENORHABDITIS ELEGANS. Genetics 77, 71–94. doi:10.1002/cbic.20030062510.1093/genetics/77.1.71
Burkewitz, K., Feng, G., Dutta, S., Kelley, C. A., Steinbaugh, M., Cram, E. J., et al. (2020). Atf-6 Regulates Lifespan through ER-Mitochondrial Calcium Homeostasis. Cell Rep. 32, 108125. doi:10.1016/j.celrep.2020.108125
Byrne, A. B., Weirauch, M. T., Wong, V., Koeva, M., Dixon, S. J., Stuart, J. M., et al. (2007). A Global Analysis of Genetic Interactions in Caenorhabditis elegans. J. Biol. 6, 8. doi:10.1186/jbiol58
Château, M.-T., Araiz, C., Descamps, S., and Galas, S. (2010). Klotho Interferes with a Novel FGF-Signalling Pathway and insulin/Igf-like Signalling to Improve Longevity and Stress Resistance in Caenorhabditis elegans. Aging 2, 567–581. doi:10.18632/aging.100195
Copeland, J., and Simoes-Costa, M. (2020). Post-transcriptional Tuning of FGF Signaling Mediates Neural Crest Induction. Proc. Natl. Acad. Sci. U.S.A. 117, 33305–33316. doi:10.1073/PNAS.2009997117
De Lencastre, A., Pincus, Z., Zhou, K., Kato, M., Lee, S. S., and Slack, F. J. (2010). MicroRNAs Both Promote and Antagonize Longevity in C. elegans. Curr. Biol. 20, 2159–2168. doi:10.1016/j.cub.2010.11.015
DeVore, D. L., Horvitz, H. R., and J.Stern, M. M. (1995). An FGF Receptor Signaling Pathway Is Required for the Normal Cell Migrations of the Sex Myoblasts in C. elegans Hermaphrodites. Cell 83, 611–620. doi:10.1016/0092-8674(95)90101-9
Drozdetskiy, A., Cole, C., Procter, J., and Barton, G. J. (2015). JPred4: A Protein Secondary Structure Prediction Server. Nucleic Acids Res. 43, W389–W394. doi:10.1093/nar/gkv332
Favata, M. F., Horiuchi, K. Y., Manos, E. J., Daulerio, A. J., Stradley, D. A., Feeser, W. S., et al. (1998). Identification of a Novel Inhibitor of Mitogen-Activated Protein Kinase Kinase. J. Biol. Chem. 273, 18623–18632. doi:10.1074/jbc.273.29.18623
Golden, T. R., Hubbard, A., Dando, C., Herren, M. A., and Melov, S. (2008). Age-related Behaviors Have Distinct Transcriptional Profiles inCaenorhabditis Elegans. Aging Cell 7, 850–865. doi:10.1111/j.1474-9726.2008.00433.x
Grün, D., Kirchner, M., Thierfelder, N., Stoeckius, M., Selbach, M., and Rajewsky, N. (2014). Conservation of mRNA and Protein Expression during Development of C. elegans. Cell Rep. 6, 565–577. doi:10.1016/j.celrep.2014.01.001
Hurley, M. M., Gronowicz, G., Zhu, L., Kuhn, L. T., Rodner, C., and Xiao, L. (2016). Age-Related Changes in FGF-2, Fibroblast Growth Factor Receptors and β-Catenin Expression in Human Mesenchyme-Derived Progenitor Cells. J. Cell. Biochem. 117, 721–729. doi:10.1002/jcb.25357
Jan, C. H., Friedman, R. C., Ruby, J. G., and Bartel, D. P. (2011). Formation, Regulation and Evolution of Caenorhabditis elegans 3′UTRs. Nature 469, 97–101. doi:10.1038/nature09616
Kertesz, M., Iovino, N., Unnerstall, U., Gaul, U., and Segal, E. (2007). The Role of Site Accessibility in microRNA Target Recognition. Nat. Genet. 39, 1278–1284. doi:10.1038/ng2135
Kuro-o, M., Matsumura, Y., Arawa, H., Kawaguchi, H., Suga, T., Utsugi, T., et al. (1999). Mutation of the Mouse Klotho Gene Leads to a Syndrome Resembling Ageing. Chemtracts 12, 703–707.
Lall, S., Grün, D., Krek, A., Chen, K., Wang, Y.-L., Dewey, C. N., et al. (2006). A Genome-wide Map of Conserved MicroRNA Targets in C. elegans. Curr. Biol. 16, 460–471. doi:10.1016/j.cub.2006.01.050
Lapierre, L. R., and Hansen, M. (2012). Lessons from C. elegans: Signaling Pathways for Longevity. Trends Endocrinol. Metabolism 23, 637–644. doi:10.1016/j.tem.2012.07.007
Libina, N., Berman, J. R., and Kenyon, C. (2003). Tissue-Specific Activities of C. elegans DAF-16 in the Regulation of Lifespan. Cell 115, 489–502. Available at: www.cell.com/cgi/content/full/115/4/489/DC1. doi:10.1016/s0092-8674(03)00889-4
Mallick, A., and Gupta, B. P. (2020). Vitellogenin-2 Acts Downstream of PRY-1/Axin to Regulate Lipids and Lifespan in C. elegans. Micropubl. Biol. 2020, 19–21. doi:10.17912/micropub.biology.000281
Mallick, A., and Gupta, B. P. (2022). AXIN-AMPK Signaling: Implications for Healthy Aging. F1000Res 10, 1259–1314. doi:10.12688/f1000research.74220.1
Mallick, A., Jhaveri, N., Jeon, J., Chang, Y., Shah, K., Hosein, H., et al. (2021a). Genetic Analysis of Caenorhabditis elegans pry-1/Axin Suppressors Identifies Genes Involved in Reproductive Structure Development, Stress Responses, and Aging. G3 Genes 12, jkab430. doi:10.1093/g3journal/jkab430
Mallick, A., Ranawade, A., and Gupta, B. P. (2019a). Role of PRY-1/Axin in Heterochronic miRNA-Mediated Seam Cell Development. BMC Dev. Biol. 19, 1–12. doi:10.1186/s12861-019-0197-5
Mallick, A., Ranawade, A., van den Berg, W., and Gupta, B. P. (2020). Axin-Mediated Regulation of Lifespan and Muscle Health in C. elegans Requires AMPK-FOXO Signaling. iScience 23, 101843. doi:10.1016/j.isci.2020.101843
Mallick, A., Taylor, S. K. B., Mehta, S., and Gupta, B. P. (2021b). picd-1, a Gene that Encodes CABIN1 Domain-Containing Protein, Interacts with pry-1/Axin to Regulate Multiple Processes in Caenorhabditis elegans. bioRxiv 1, 37. doi:10.1101/2021.09.27.462077
Mallick, A., Taylor, S. K. B., Ranawade, A., and Gupta, B. P. (2019b). Axin Family of Scaffolding Proteins in Development: Lessons from C. elegans. Jdb 7, 20–23. doi:10.3390/jdb7040020
Mathes, S., Fahrner, A., Ghoshdastider, U., Rüdiger, H. A., Leunig, M., Wolfrum, C., et al. (2021). FGF-2-dependent Signaling Activated in Aged Human Skeletal Muscle Promotes Intramuscular Adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 118, 1–12. doi:10.1073/pnas.2021013118
Miska, E. A., Alvarez-Saavedra, E., Abbott, A. L., Lau, N. C., Hellman, A. B., McGonagle, S. M., et al. (2007). Most Caenorhabditis elegans microRNAs Are Individually Not Essential for Development or Viability. PLoS Genet. 3, e215–2403. doi:10.1371/journal.pgen.0030215
O'Brien, J., Hayder, H., Zayed, Y., and Peng, C. (2018). Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 9, 1–12. doi:10.3389/fendo.2018.00402
Ornitz, D. M., and Itoh, N. (2015). The fibroblast growth factor signaling pathway. WIREs Developmental Biology 4, 215–266. doi:10.1002/wdev.176
Pincus, Z., Smith-Vikos, T., and Slack, F. J. (2011). MicroRNA predictors of longevity in caenorhabditis elegans. PLoS Genet 7, e1002306. doi:10.1371/journal.pgen.1002306
Ranawade, A., Mallick, A., and Gupta, B. P. (2018). PRY-1/Axin Signaling Regulates Lipid Metabolism in Caenorhabditis elegans. PLoS One 13, e0206540. doi:10.1371/journal.pone.0206540
Reiner, D. J., González‐Pérez, V., Der, C. J., and Cox, A. D. (2008). Use of Caenorhabditis elegans to Evaluate Inhibitors of Ras Function In Vivo. Methods Enzymol. 439, 425–449. doi:10.1016/S0076-6879(07)00430-2
Rennie, W., Kanoria, S., Liu, C., Mallick, B., Long, D., Wolenc, A., et al. (2016). STarMirDB: A Database of microRNA Binding Sites. RNA Biol. 13, 554–560. doi:10.1080/15476286.2016.1182279
Rera, M., Azizi, M. J., and Walker, D. W. (2013). Organ-specific Mediation of Lifespan Extension: More Than a Gut Feeling? Ageing Res. Rev. 12, 436–444. doi:10.1016/j.arr.2012.05.003
Schutzman, J. L., Borland, C. Z., Newman, J. C., Robinson, M. K., Kokel, M., and Stern, M. J. (2001). The Caenorhabditis elegans EGL-15 Signaling Pathway Implicates a DOS-like Multisubstrate Adaptor Protein in Fibroblast Growth Factor Signal Transduction. Mol. Cell. Biol. 21, 8104–8116. doi:10.1128/mcb.21.23.8104-8116.2001
Sharanya, D., Fillis, C. J., Kim, J., Zitnik, E. M., Ward, K. A., Gallagher, M. E., et al. (2015). Mutations inCaenorhabditis Briggsaeidentify New Genes Important for Limiting the Response to EGF Signaling during Vulval Development. Evol. Dev. 17, 34–48. doi:10.1111/ede.12105
Stefani, G., and Slack, F. J. (2008). Small Non-coding RNAs in Animal Development. Nat. Rev. Mol. Cell Biol. 9, 219–230. doi:10.1038/nrm2347
Taylor, R. C., and Dillin, A. (2013). XBP-1 Is a Cell-Nonautonomous Regulator of Stress Resistance and Longevity. Cell 153, 1435–1447. doi:10.1016/j.cell.2013.05.042
Taylor, S. K. B., Minhas, M. H., Tong, J., Selvaganapathy, P. R., Mishra, R. K., and Gupta, B. P. (2021). C. elegans Electrotaxis Behavior Is Modulated by Heat Shock Response and Unfolded Protein Response Signaling Pathways. Sci. Rep. 11, 1–17. doi:10.1038/s41598-021-82466-z
Uno, M., and Nishida, E. (2016). Lifespan-regulating Genes in c. Elegans. npj Aging Mech. Dis. 2, 16010. doi:10.1038/npjamd.2016.10
Wang, H., Webster, P., Chen, L., and Fisher, A. L. (2019). Cell-autonomous and Non-autonomous Roles of Daf-16 in Muscle Function and Mitochondrial Capacity in Aging C. elegans. Aging 11, 2295–2311. doi:10.18632/aging.101914
Watts, J. L., and Ristow, M. (2017). Lipid and Carbohydrate Metabolism in Caenorhabditis elegans. Genetics 207, 413–446. doi:10.1534/genetics.117.300106
Xie, Y., Su, N., Yang, J., Tan, Q., Huang, S., Jin, M., et al. (2020). FGF/FGFR Signaling in Health and Disease. Sig Transduct. Target Ther. 5, 181. doi:10.1038/s41392-020-00222-7
Xue, Y., Liu, Z., Cao, J., Ma, Q., Gao, X., Wang, Q., et al. (2011). GPS 2.1: Enhanced Prediction of Kinase-specific Phosphorylation Sites with an Algorithm of Motif Length Selection. Protein Eng. Des. Sel. 24, 255–260. doi:10.1093/protein/gzq094
Yin, Y., Castro, A. M., Hoekstra, M., Yan, T. J., Kanakamedala, A. C., Dehner, L. P., et al. (2015). Fibroblast Growth Factor 9 Regulation by MicroRNAs Controls Lung Development and Links DICER1 Loss to the Pathogenesis of Pleuropulmonary Blastoma. PLoS Genet. 11, e1005242–22. doi:10.1371/journal.pgen.1005242
Keywords: kin-9, FGFR4, pry-1, miR-246, aging, stress response, ER-UPR, Fgf signaling
Citation: Mallick A, Xu L, Mehta S, Taylor SKB, Hosein H and Gupta BP (2022) The FGFR4 Homolog KIN-9 Regulates Lifespan and Stress Responses in Caenorhabditis elegans. Front. Aging 3:866861. doi: 10.3389/fragi.2022.866861
Received: 31 January 2022; Accepted: 26 April 2022;
Published: 20 May 2022.
Edited by:
Carmen Nussbaum-Krammer, Heidelberg University, GermanyReviewed by:
Anders Olsen, Aalborg University, DenmarkKen Humphries, Oklahoma Medical Research Foundation, United States
Copyright © 2022 Mallick, Xu, Mehta, Taylor, Hosein and Gupta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bhagwati P. Gupta, guptab@mcmaster.ca
†Present Address: Leo Xu, Department of Cell and Systems Biology, University of Toronto, Toronto, CanadaHannah Hosein, Department of Biochemistry, McGill University, Montreal, Canada
‡These authors have contributed equally to this work
 Avijit Mallick
Avijit Mallick Leo Xu†‡
Leo Xu†‡  Bhagwati P. Gupta
Bhagwati P. Gupta