- 1Department of Microbiology, Yongchuan Hospital of Chongqing Medical University, Chongqing, China
- 2Department of Molecular Biology, Jiaxing Maternal and Child Health Hospital, Jiaxing, China
- 3Department of Microbiology, Affiliated Hangzhou Xixi Hospital, Zhengjiang University School of Medicine, Hangzhou, China
- 4Department of Microbiology, The First Affiliated Hospital of Jiamusi University, Jiamusi, China
Background: The sequence type 11 (ST11) carbapenem-resistant Klebsiella pneumoniae (CRKP) carrying blaKPC−2 has been widespread all over the world, and it has been reported frequently in China. The blaKPC−2 located on the mobile genetic element brings tremendous pressure to control the spread and outbreak of resistant bacteria. Whole-genome sequencing (WGS) technology can comprehensively and in-depth display the molecular characteristics of drug-resistant bacteria, providing a basis for evaluating the genetic diversity within the CRKP genome.
Methods: The ST11 CRKP in this study was collected in the intensive care unit of a major teaching hospital. PCR and Sanger sequencing confirmed the existence of blaKPC−2. The AST-GN card and the microbroth dilution test were used for antimicrobial susceptibility testing. The transferability of plasmid was verified by a conjugation test. The whole genome is sequenced using the Illumina HiSeq short-read and Oxford Nanopore long-read sequencing technology.
Results: The studied strain was named CRKP63, which is a multi-drug resistance bacteria, which carries blaKPC−2 and blaSHV−182. Its genome consists of a circular chromosome of 5,374,207 bp and an IncFII plasmid named pKPC-063001 of 359,625 bp. In the drug-resistant plasmid pKPC-063001, the key carbapenem resistance gene blaKPC−2 was located in the genetic context with insertion sequence ISKpn27 upstream and ISKpn6 downstream and bracketed by IS26. The three copies of the IS26–ISKpn27–blaKPC−2–ISKpn6–IS26 unit were present in tandem. blaKPC−2 can be transferred horizontally between other species by conjugation, the complete type IV secretion system (T4SS) structure helps to improve the adaptability of bacteria to the external environment, strengthen the existence of drug-resistant bacteria, and accelerate the spread of drug resistance.
Conclusion: High-throughput sequencing has discovered the different surrounding environments of blaKPC−2, which provides a new idea for further revealing the transmission and inheritance of blaKPC−2 at the molecular level. In order to control the further spread and prevalence of drug-resistant bacteria, we should pay close attention to the changes in the genetic environment of blaKPC−2 and further study the transcription and expression of T4SS.
Background
Carbapenem-resistant Enterobacteriaceae, especially Klebsiella pneumoniae, have emerged as important causes of morbidity and mortality among hospital-acquired and long-term care-associated infections (1). As of now, carbapenem-resistant K. pneumoniae (CRKP) strains have spread worldwide and posed a severe threat to public health. K. pneumoniae carbapenemases (KPC)-2, the most common variant of KPC enzymes, is a dominant factor mediating carbapenems resistance in CRKP (2, 3). The most predominant isolates of KPC-producing K. pneumoniae (KPC-kpn) belong to the clonal group 258 (CG258), two representative types of this CG: the ST258 and ST11 strains, have been identified worldwide – ST258 is mostly prevalent in America and Europe, while ST11 is the highly dominant clone in Asia (especially in China) (4).
The blaKPC−2 is a typical plasmid-mediated drug resistance gene and mainly carried on plasmids of different incompatibility (Inc) groups, such as IncFII, FIA, I2, A/C, N, X, P, and L/M (4). The blaKPC−2 on the plasmid can spread the resistance through different methods, such as gene duplication, transposon elements, or plasmid transfer. The horizontal transmission of drug-resistant plasmids can accelerate the spread of multidrug resistance genes and mediate the production of multidrug-resistant bacteria (MDR). An in-depth understanding of the plasmid structure and its genome characteristics will help to control and prevent the emergence and outbreak of drug-resistant bacteria.
The rapid development of whole-genome sequencing (WGS) technology has gradually matured its application in the field of clinical microbiology (5, 6). WGS has the characteristics of large data information and high resolution, and it plays an important value in the research and detection of MDR. In this study, a whole-genome sequence of a CRKP, which is ST11 type and carrying blaKPC−2 in Chongqing, China, was performed and further explored its microbiological and genomic characteristics.
Materials and Methods
Bacterial Collection
According to previous research, a total of 51 non-duplicated CRKP samples isolated from the ICU of Yongchuan Affiliated Hospital of Chongqing Medical University (a major teaching hospital in Chongqing, China) were collected from July 2018 to July 2020. Homology analysis based on the result of pulsed-field gel electrophoresis showed that 62.7% of the isolates belonged to the same cluster, indicating that there was a clonal transmission of ST11 carrying blaKPC−2 CRKP in the ICU of this hospital. In order to further study the molecular characteristics of CRKP in this ward, we selected one of the strains from the clone group, which carries multiple drug resistance genes and has the ability to conjugate—CRKP63—and conduct in-depth research on it. This blaKPC−2-positive isolate was collected after identification (VITEK-2 automated microbiology analyzer, bioMérieux, France) and routine antimicrobial susceptibility testing by the Microbiology Laboratory in April 2020. The strain was identified as blaKPC−2 producing carbapenem-resistant K. pneumoniae by PCR detection and drug sensitivity (carbapenems) review. The isolate was stored at −80°C for further study.
Antimicrobial Susceptibility Testing
The VITEK-2 Compact automatic microbiological analyzer Antimicrobial Susceptibility Testing-Gram-Negtive (AST-GN) card (bioMérieux, France) was used for routine antimicrobial susceptibility testing. Minimum inhibitory concentration (MIC) is defined as the lowest compound concentration (μg/ml) required to stop bacterial growth was determined by using the microbroth dilution method. Imipenem (IPM), meropenem (MEM), amikacin (AMK), levofloxacin (LEV), tigecycline (TIG), polymyxin B (PLB), and ceftazidime-avibactam (CAZ-AVI) were used to determine the MIC by the microbroth dilution method. ATCC 25922, ATCC 700603, and BAA-1705 were used as quality control strains. Three parallel assays were performed for each sample. The IPM, MEM, AMK, LEV, PLB, and CAZ-AVI results were interpreted based on the Clinical and Laboratory Standards Institute (CLSI) criteria (7), whereas the TIG results were interpreted based on the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (8) breakpoint recommendations.
Conjugation Experiment
The conjugation experiment was carried out using a membrane bonding experiment as previously described (9). Both the donor (CRKP) and the recipient strains (E. coli EC600) were mixed in Luria-Bertani broth at a ratio of 1:3, and the mixtures were placed on a membrane and incubated for 24 h at 35°C. Transconjugants were selected on Mueller-Hinton agar II (MHA) plates supplemented with rifampicin (600 μg/ml) and MEM (1 μg/ml). Colonies that grew on the selective medium were identified by the VITEK-2 Compact system and 16S rRNA sequence. A strain that harbored carbapenemase and exhibited higher MICs of resistance to carbapenems than EC600 was defined as the transconjugants and the presence of resistance determinants was confirmed by PCR.
WGS and Data Analysis
Genomic DNA was isolated using the MagAttract HMW DNA Kit (Qiagen, Hilden, Germany) and submitted to next-generation high-throughput sequencing (NGS) on a HiSeq 2000™ platform (Illumina Inc., San Diego, CA, USA) with 2 × 100-bp paired-end reads and to long-read high-throughput sequencing (LRS) on a MinION platform (Oxford Nanopore Technologies, Oxford, UK). The long reads generated by MinION were assembled using Canu v. 1.6 (10) and polished with the short reads generated by HiSeq using Pilon v1.22 (11) to obtain the whole genome and complete plasmid sequences. The chromosome and plasmid sequences were annotated using the prokaryotic gene prediction tool Prokka (12). The plasmid incompatibility type was searched using the online tool PlasmidFinder (https://cge.cbs.dtu.dk//services/PlasmidFinder/) (13). Antibiotic resistance genes were identified using both the Comprehensive Antibiotic Resistance Database (CARD) (14) and ResFinder database (https://cge.cbs.dtu.dk/services/ResFinder/) (15). Meanwhile, virulence-associated genes were identified using VirulenceFinder (https://cge.cbs.dtu.dk/services/VirulenceFinder/) (16). Transposon and insertion sequence (IS) elements were scanned using the ISfinder database (https://www-is.biotoul.fr/) (17). Comparative plasmid illustration was implemented by BRIG (http://brig.sourceforge.net) (18) or Easyfig tools (https://github.com/mjsull/Easyfig) (19). BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (20) was used for comparative analysis through the coverages and identities.
Nucleotide Sequence Accession Numbers
The complete sequences were submitted to GeneBank under accession numbers.
Result
Clinical Character
The blaKPC−2-positive CRKP, CRKP63, collected in this study was derived from a 90-year-old female patient in the ICU ward. Concurrently, this strain also carried blaSHV−182. The patient was admitted to the hospital due to acute exacerbation of chronic obstructive pulmonary disease on April 10, 2020, on the day of admission, the patient underwent mechanical ventilation. The patient underwent bronchoalveolar lavage 4 times during the hospitalization, and 10 days after admission, K. pneumoniae was detected in the bronchoalveolar lavage fluid, which was identified as a carbapenemase-producing CRKP. During the patient's hospitalization, infectious symptoms were repeatedly realized and anti-infective treatment continued. Twenty-eight days after admission, the condition of the patient did not improve, and finally, the patient and the patient's family gave up treatment and was discharged voluntarily. The main process of the patient during hospitalization is shown in Figure 1.

Figure 1. The main process of the patient during hospitalization. CMV, mechanical ventilation; BLA, bronchoalveolar lavage; SCF, cefoperazone/sulbactam; MXF, moxifloxacin; TZP, piperacillin tazobactam; SXT, sulfamethoxazole.
Result of Antimicrobial Susceptibility
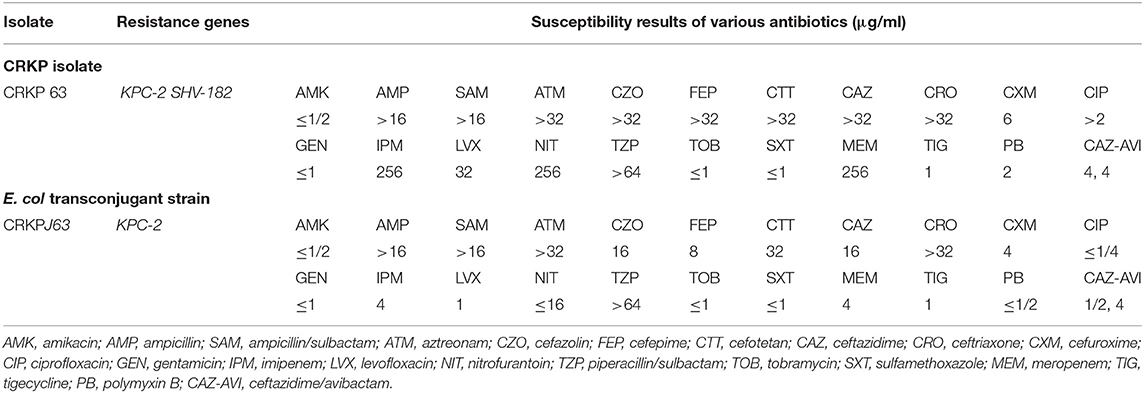
Carbapenem-resistant Klebsiella pneumoniae63 shows resistance to more than three antibiotics and can be defined as MDR (21). The MIC value of IPM and MEM was as high as 256 μg/ml. However, it was sensitive to tobramycin (TOB), sulfamethoxazole (SXT), TIG, and CAZ-AVI. The specific information of various antibiotics is shown in Table 1.
Conjugation Experiment
After the conjugation experiment was successful, the transconjugant CRKPJ63 was obtained. Compared with the original donor, CRKPJ63 still carried blaKPC−2, but not carried blaSHV−182. Obviously, the MIC value of transconjugant for carbapenems (IMP and MEM) was significantly decreased. The variation on donor and transconjugant susceptibility profiles is shown in Table 1.
Results of WGS
Carbapenem-resistant Klebsiella pneumoniae63 carries two drug resistance genes at the same time, blaKPC−2 and blaSHV−182. The extended-spectrum β-lactamase (ESBL) gene blaSHV−182 is located on the chromosome (position in contig 1,037,173–1,038,033). Yet, the key carbapenem resistance gene blaKPC−2 is located on the drug resistance plasmid pKPC-063001, which is of type IncFII. Therefore, the whole genome of CRKP63 consists of a circular chromosome of 5,374,207 bp and a drug-resistant plasmid (named pKPC-063001) of 359,625 bp. For chromosome, the final draft genome showed a G + C content of 60.4%, with a total of 5,165 annotated protein-coding sequences (CDSs). The visual circle map is shown in Figure 2. The Cluster of Orthologous Groups [(COGs) (of proteins)] database was used to annotate its genome, and it was found that genes related to metabolism and genes related to genetic information processing accounted for a relatively large proportion. In addition, there are also functional proteins related to gene processing and material conversion. The class of protein function and its number are shown in Figure 3.
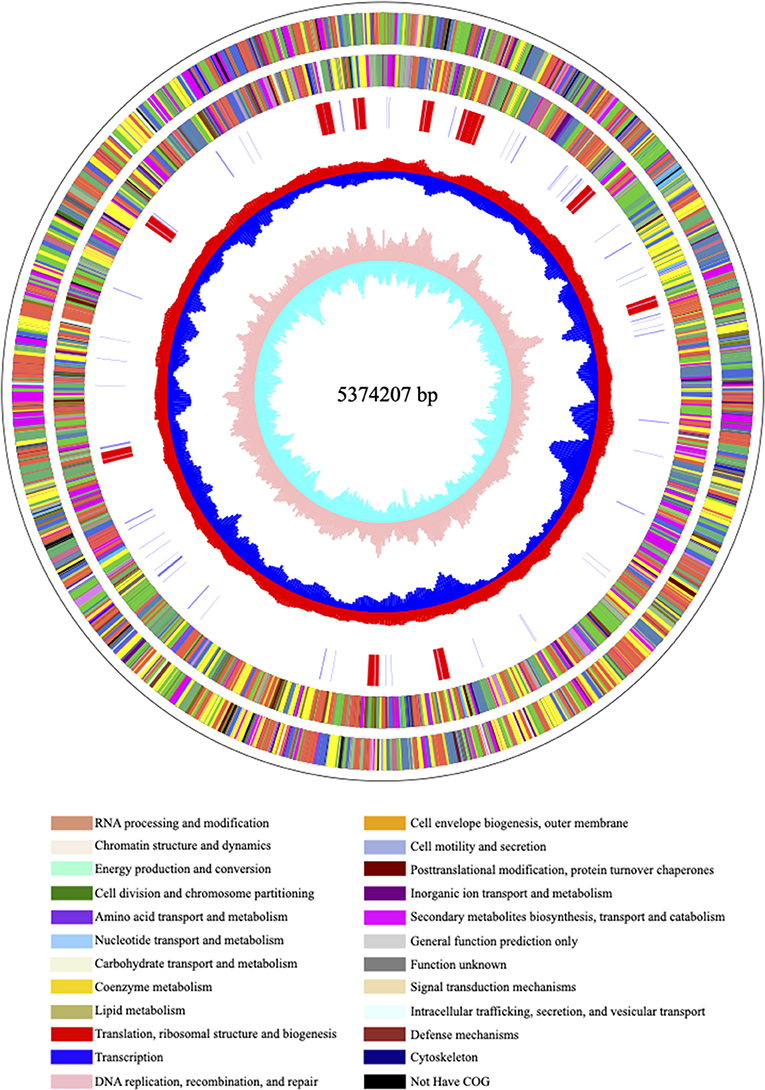
Figure 2. The visual circle map of the chromosome. From outside to inside, the first and second circles are CDS on the positive and negative strands, the third circle is rRNA and tRNA; the fourth circle is the GC content, and the outward red part indicates that the GC content of this region is higher than the average GC content of the whole genome. The higher the peak, the greater the difference from the average GC content, the inward blue part indicates that the GC content of the region is lower than the average GC content of the whole genome, the higher the peak, the greater the difference from the average GC content; the innermost circle (fifth) is the GC skew value, the specific algorithm is G – C/G + C, when the value is positive in the biological sense, the positive chain is more inclined to transcribe CDS, which is a negative value (22, 23).
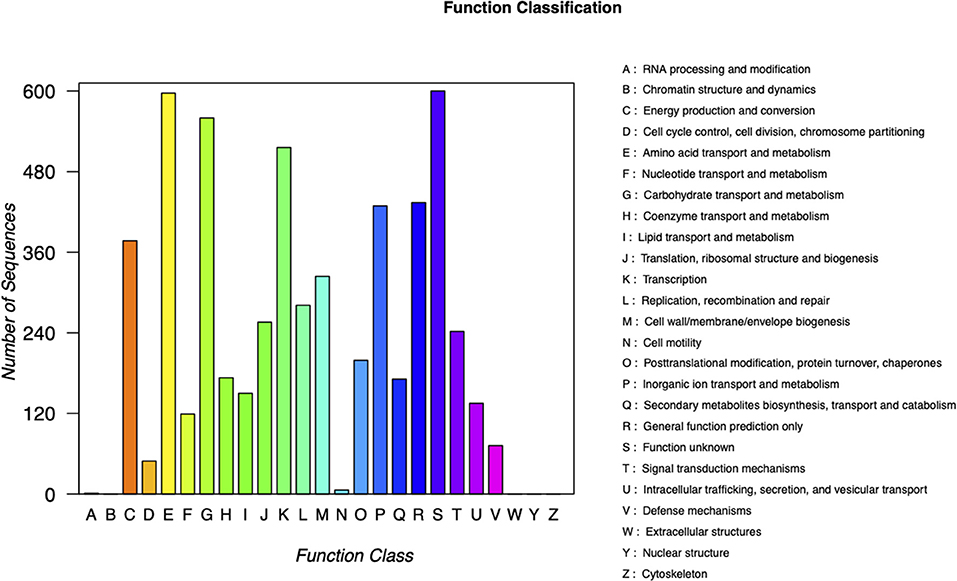
Figure 3. Classification of COG functions of chromosome in DNA libraries. COG, Cluster of Orthologous Groups.
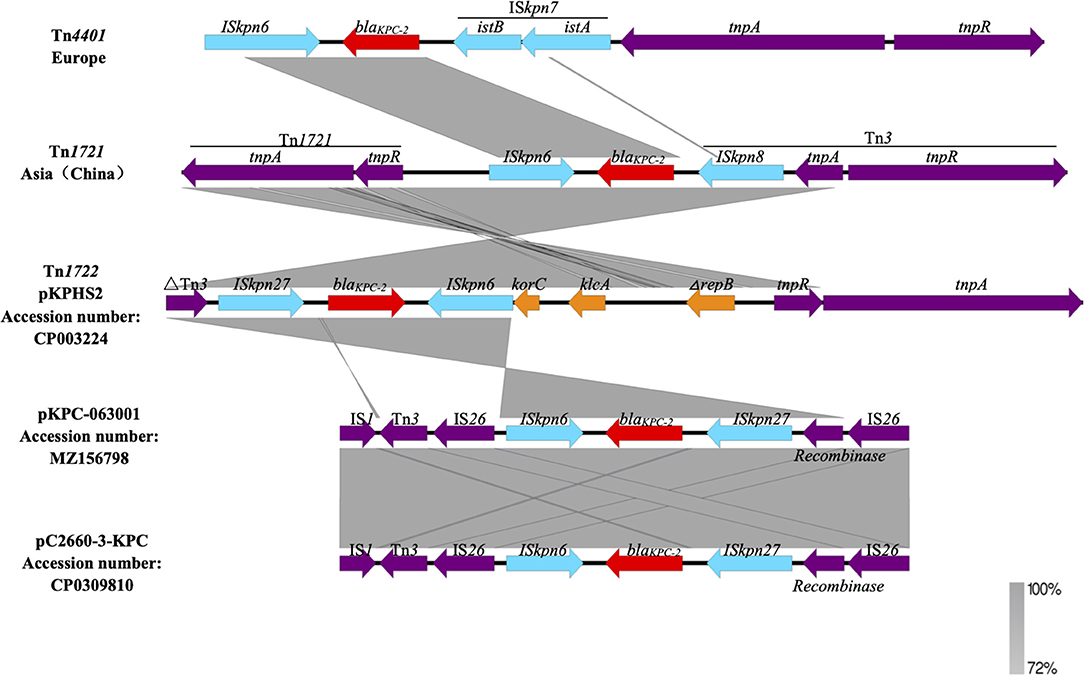
pKPC-063001 is a 359,625 bp circular molecule with an average G + C content of 58.19% and was predicted to encode a total of 409 CDSs. In addition to the blaKPC−2, it also contains virulence factors iucA, iucB, iucC, and iucD, plasmid replication protein (repA), plasmid stabilization protein (parA), and the type IV secretion system (T4SS) proteins traA, traB, traD, traM, and traK that mediate the conjugation and transfer of plasmids. The visual circle map is shown in Figure 4. After a detailed analysis of the surrounding structure of the key gene blaKPC−2, it was found that blaKPC−2 is located in a gene fragment with IS26 repeat inserts at both ends. This gene fragment has IS26 repeats at both ends, and ISkpn27 and ISkpn6 in the middle, and there is also a Tn3-tnpR structure between ISkpn27 and IS26 (Figure 5).
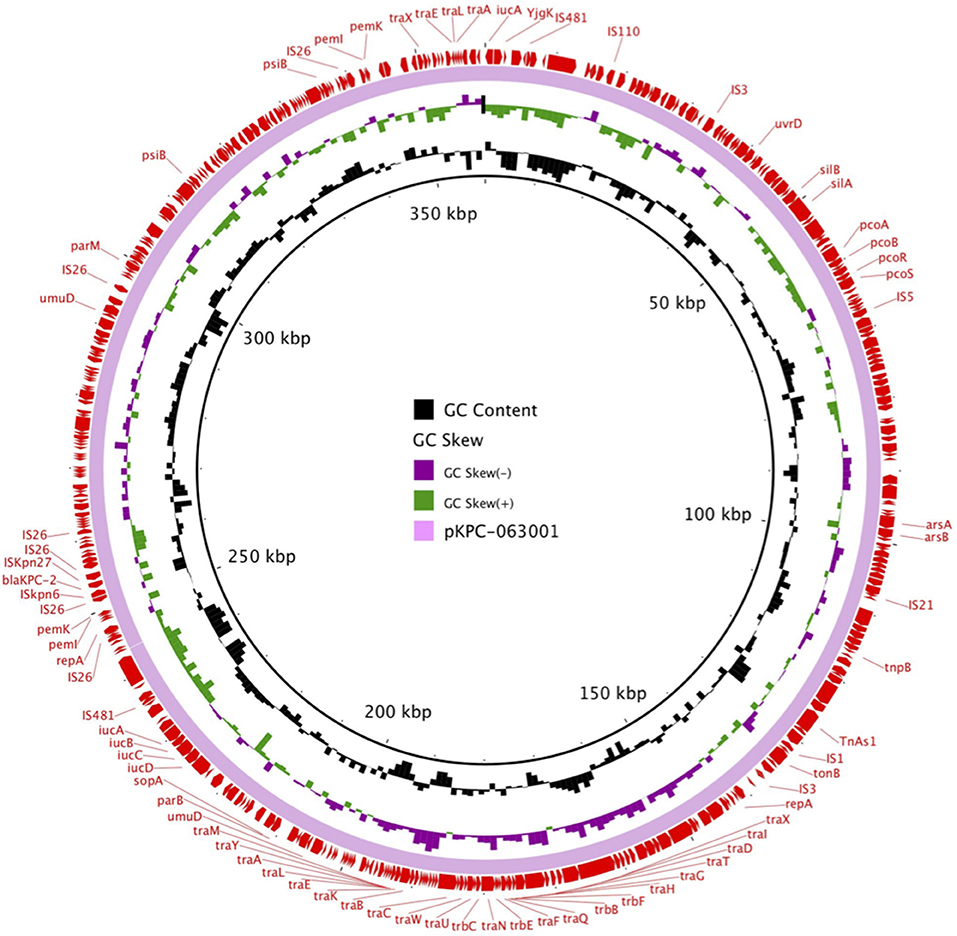
Figure 4. The visual circle map of plasmid pKPC-063001, where blaKPC−2 is located. GC content, GC skew+, and GC skew– are, respectively, indicated in black, green, and purple.
Accession Numbers
The result obtained by sequencing in this study has been uploaded to the Genebank website (www.ncbi.nlm.nih.gov). The accession number of the chromosome is CP077763 (GenBank: CP077763.1), and the accession number of the plasmid is MZ156798 (GenBank: MZ156798.1).
Discussion
The blaKPC gene still plays a key role in high-level carbapenem resistance (24). One of the key factors contributing to the rapid and wide dissemination of blaKPC−2 is its location on a transposable element (25, 26). In Europe, blaKPC−2 is mainly located on the conserved Tn3 family transposon Tn4401 (25) and is considered the origin of the acquisition and dissemination of this marker (27). Tn4401 is approximately 10 kb in size and delimited by two 39 bp imperfect inverted repeat sequences. It harbors two ISs flanking blaKPC, ISKpn6, and ISKpn7, in addition to transposase (tnpA) and resolvase (tnpR) genes. However, Tn1721-like transposons among ST11 K. pneumoniae are mainly responsible for the effective spread of the blaKPC−2 gene in China (28). There are three inverted repeats, IRR, IRL1, and IRL2, in the Tn1721a transposon. tnpA and tnpR are located between IRR and IRL1, while the 5′ end of blaKPC−2 has a complete Tn3 located between IRL1 and IRL2. When Tn3-tnp R is interrupted by IS26, Tn3-tnp A and IRL2 are lost, and a new structure, such as Tn1721-blaKPC−2-ΔTn3-IS26 (Tn1721b), is formed. The Tn1722-based unit transposons with the ISKpn27-blaKPC−2-ΔISKpn6 core structure (ISKpn27 is initially named in the ISfinder database) has been also reported in China, such as pKP048 (29), p628-KPC (30), and pHS102707.
In this study, the surrounding environment of the blaKPC−2 is different from the epidemic structure of domestic and international, we call it “composite transposon-based on IS26”. The original 1319bp ISKpn6 was truncated by IS26 and became 881bp ΔISKpn6. ISKpn27 is quite different from other inserted sequences, and there is a “recombinase” gene between ISKpn27 and IS26 repeats (Figure 5). Notably, different lengths of truncated ISKpn6 can be observed for these IS26-based transposons from different plasmids. This structure has also been reported by others (31), such as pC2660-3-KPC (32). This difference in transposons indicates that there is a certain variability and diversity surrounding the environment of the blaKPC−2 gene, which may also be one of the factors causing the rapid spread of blaKPC−2 strain and the different epidemic status in different regions. Loftie et al. (33) found that Tn6231 can significantly improve the persistence of resistant plasmid pMS0506 and broaden the host range of plasmid, which shows that the evolutionary behavior of plasmid (such as transposon) can affect the spread of resistance gene and improve the selection of plasmid host. Mutations in the genetic environment help to improve the durability of antibiotic resistance, so that the host adapts to changes in the external environment and then affects the spread of bacterial resistance. Therefore, the blaKPC−2 gene environment is constantly changing, and its horizontal transfer with transposable elements shows greater flexibility than plasmid transmission. In this way, the clinical harm will be greater.
As the key carbapenemase, blaKPC−2 can exist on many different types of plasmids. The blaKPC found in this study is located on the IncFII type plasmid. The success of the conjugation experiment also confirmed the conjugability of the plasmid. It can be seen that the pKPC-063001 has a complete functional skeleton structure, i.e., plasmid replication structure, plasmid stabilization structure, and plasmid conjugation transfer structure. These structures provide a structural basis for the widespread of blaKPC−2. Among them, theT4SS plays an important role in conjugative transfer (34, 35). The classic T4SS system was originated from Agrobacterium rhizogenes, it was generally composed of 12 proteins (i.e., 11 VirB proteins and 1 VirD coupling protein (VirD4)) (36, 37), coupling protein–relaxosome contact could lead to DNA unwinding, generating a single strand of DNA that is then transferred to the recipient in a 5′ to 3′ direction (38), so that the genetic information is transferred. Then, T4SS was also found in Gram-negative bacteria (39). Among the two major phylogenetic groups of gram-negative bacteria T4SSs, type IVA (the conjugation systems of the IncF and IncP plasmids) is more common (40, 41). Lawley et al. (34) reported that T4SSs, also known as the mating pair formation (Mpf) apparatus, are central to the dissemination of numerous genetic determinants between bacteria, as highlighted by the spread of antibiotic resistance among pathogens. After studying the gene deletion of the T4SS regulatory region of the pCTX-M3 plasmid of the IncM group, Dmowski et al. found that (42) the knockout of the conjunction structural gene will result in no transfer of the resistance gene or low conjunction rate. However, when orf35 and orf36 were knocked out, the plasmid conjugation rate could be improved, this is because two genes are involved in suppressing the transcriptional regulation of the T4SS gene according to transcription analysis. This shows that the existence of T4SS undoubtedly provides strong support for the global popularity of blaKPC−2. The complete T4SS structure helps to improve the adaptability of bacteria to the external environment, thereby enhancing the existence of drug-resistant bacteria and accelerating the spread of drug-resistant bacteria. An in-depth study of the function and transcriptional expression of T4SS will help prevent the further spread of blaKPC−2.
In addition, it should be noted that blaSHV−182 located on the chromosome did not transfer with blaKPC−2, simultaneously, the results of conjugation showed that the MIC value of conjugants for carbapenems decreased, indicating that other causes of drug resistance, such as membrane protein, or efflux pump, for example, did not transfer with the plasmid. This also suggests that the resistance of CRKP63 is caused by a variety of factors, not simply caused by pKPC-063001.
Combined with previous research (43), we know that CRKP63 was collected in the ICU ward of a hospital in Chongqing, China. There had been an outbreak of ST11 CRKP carrying blaKPC−2 in this ward. This outbreak was closely related to the horizontal transfer of blaKPC−2. Similarly, over the past period, IncFII plasmids carrying blaKPC−2 often have been reported in K. pneumoniae in China, especially the ST11 (44), the prevalence and dissemination of InFII plasmid carrying KPC-2 in China has become a fact. However, unlike the dominant Tn1721 in China, the presence of “IS26-based composite transposon” structure represented by pKPC-063001 implies the variability and complexity of blaKPC−2. In the future, “IS26-based composite transposon” is likely to become the dominant clone group in China and even the world with amazing speed and adaptability, thence, continuous monitoring will be necessary to prevent further dissemination of pKPC-063001 type plasmid.
In conclusion, this study reported the microbial and genomic characteristics of an ST11 CRKP carrying blaKPC−2 in Chongqing, China. Through WGS, we found the different surrounding environments of blaKPC−2, which provides a new research idea for further revealing the transmission and inheritance of blaKPC−2 at the molecular level. The differences in the surrounding environment of blaKPC−2 create convenience for its dissemination and popularity, and the complete T4SS structure provides a solid guarantee for it. Therefore, in order to control the further spread and prevalence of drug-resistant bacteria, we should also pay close attention to the genetic environment of blaKPC−2, and further study the transcription and expression of T4SS.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP077763 and https://www.ncbi.nlm.nih.gov/genbank/, MZ156798.
Ethics Statement
The studies involving human participants were reviewed and approved by The Ethics Committee of Yongchuan Hospital of ChongQing Medical University. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
XZ conceived and designed the study. LZ and JZ wrote this paper and contributed equally to this work. LZ, JW, CY, and WH performed the experiments. JL, KH, LY, and LZ analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the General Projects of Chongqing Natural Science Foundation (cstc2020jcyj-msxm0067), the Yongchuan Natural Science Foundation (2021yc-jckx20053), and the Talent introduction project of Yongchuan Hospital of Chongqing Medical University (YJYJ202005 and YJYJ202004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. Yu YunSong from Sir Run Run Shaw Hospital affiliated with Zhejiang University for the isolates of EC600.
References
1. Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. (2013) 13:785–96. doi: 10.1016/S1473-3099(13)70190-7
2. Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. (2009) 9:228–36. doi: 10.1016/S1473-3099(09)70054-4
3. Shi L, Feng J, Zhan Z, Zhao Y, Zhou H, Mao H, et al. Comparative analysis of bla KPC-2- and rmtB-carrying IncFII-family pKPC-LK30/pHN7A8 hybrid plasmids from Klebsiella pneumoniae CG258 strains disseminated among multiple Chinese hospitals. Infect Drug Resist. (2018) 11:1783–93. doi: 10.2147/IDR.S171953
4. Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. (2014) 22:686–96. doi: 10.1016/j.tim.2014.09.003
5. Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. (1995) 269:496–512. doi: 10.1126/science.7542800
6. Ferrari C, Corbella M, Gaiarsa S, Comandatore F, Scaltriti E, Bandi C, et al. Multiple Klebsiella pneumoniae KPC clones contribute to an extended hospital outbreak. Front Microbiol. (2019) 10:2767. doi: 10.3389/fmicb.2019.02767
7. CLSI. Performance Standards for Antimicrobial Suceptibility Testing. In: 30th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. (2020).
8. The European Committee on Antimicrobial Susceptibility Testing. In: Breakpoint tables for interpretation of MICs and zone diameters, version 10.0. (2020).
9. Gong X, Zhang J, Su S, Fu Y, Bao M, Wang Y, et al. Molecular characterization and epidemiology of carbapenem non-susceptible Enterobacteriaceae isolated from the Eastern region of Heilongjiang Province, China. BMC Infect Dis. (2018) 18:417. doi: 10.1186/s12879-018-3294-3
10. Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. (2017) 27:722–36. doi: 10.1101/gr.215087.116
11. Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. (2014) 9:e112963. doi: 10.1371/journal.pone.0112963
12. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. (2014) 30:2068–9. doi: 10.1093/bioinformatics/btu153
13. Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. (2014) 58:3895–903. doi: 10.1128/AAC.02412-14
14. McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. (2013) 57:3348–57. doi: 10.1128/AAC.00419-13
15. Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. (2020) 75:3491–500. doi: 10.1093/jac/dkaa345
16. Malberg Tetzschner AM, Johnson JR, Johnston BD, Lund O, Scheutz Scheutz F: In silico genotyping of Escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J Clin Microbiol. (2020) 58:e01269–20. doi: 10.1128/JCM.01269-20
17. Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M: ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. (2006) 34:D32–36. doi: 10.1093/nar/gkj014
18. Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. (2011) 12:402. doi: 10.1186/1471-2164-12-402
19. Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. (2011) 27:1009–10. doi: 10.1093/bioinformatics/btr039
20. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. (2009) 10:421. doi: 10.1186/1471-2105-10-421
21. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
22. Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. (1996) 13:660–5. doi: 10.1093/oxfordjournals.molbev.a025626
23. Necsulea A, Lobry JR. A new method for assessing the effect of replication on DNA base composition asymmetry. Mol Biol Evol. (2007) 24:2169–79. doi: 10.1093/molbev/msm148
24. Huang J, Hu X, Zhao Y, Shi Y, Ding H, Xv J, Ren J, Wu R, Zhao Z. Genetic Factors Associated with Enhanced bla KPC Expression in Tn3/Tn4401 Chimeras. Antimicrob Agents Chemother. (2020) 64:e01836–19. doi: 10.1128/AAC.01836-19
25. Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene. Antimicrob Agents Chemother. (2008) 52:1257–63. doi: 10.1128/AAC.01451-07
26. Cuzon G, Naas T, Nordmann P. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother. (2011) 55:5370–3. doi: 10.1128/AAC.05202-11
27. Tzouvelekis LS, Miriagou V, Kotsakis SD, Spyridopoulou K, Athanasiou E, Karagouni E, et al. KPC-producing, multidrug-resistant Klebsiella pneumoniae sequence type 258 as a typical opportunistic pathogen. Antimicrob Agents Chemother. (2013) 57:5144–6. doi: 10.1128/AAC.01052-13
28. Fu P, Tang Y, Li G, Yu L, Wang Y, Jiang X. Pandemic spread of blaKPC-2 among Klebsiella pneumoniae ST11 in China is associated with horizontal transfer mediated by IncFII-like plasmids. Int J Antimicrob Agents. (2019) 54:117–24. doi: 10.1016/j.ijantimicag.2019.03.014
29. Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, et al. Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob Agents Chemother. (2009) 53:4333–8. doi: 10.1128/AAC.00260-09
30. Wang L, Fang H, Feng J, Yin Z, Xie X, Zhu X, et al. Complete sequences of KPC-2-encoding plasmid p628-KPC and CTX-M-55-encoding p628-CTXM coexisted in Klebsiella pneumoniae. Front Microbiol. (2015) 6:838. doi: 10.3389/fmicb.2015.00838
31. Feng Y, Liu L, McNally A, Zong Z. Coexistence of three blaKPC-2 genes on an IncF/IncR plasmid in ST11 Klebsiella pneumoniae. J Glob Antimicrob Resist. (2019) 17:90–3. doi: 10.1016/j.jgar.2018.11.017
32. Gao H, Liu Y, Wang R, Wang Q, Jin L, Wang H. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. EBioMedicine. (2020) 51:102599. doi: 10.1016/j.ebiom.2019.102599
33. Loftie-Eaton W, Yano H, Burleigh S, Simmons RS, Hughes JM, Rogers LM, et al. Evolutionary paths that expand plasmid host-range: implications for spread of antibiotic resistance. Mol Biol Evol. (2016) 33:885–97. doi: 10.1093/molbev/msv339
34. Lawley TD, Klimke WA, Gubbins MJ, Frost LS. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. (2003) 224:1–15. doi: 10.1016/S0378-1097(03)00430-0
35. Cenens W, Andrade MO, Llontop E, Alvarez-Martinez CE, Sgro GG, Farah CS. Bactericidal type IV secretion system homeostasis in Xanthomonas citri. PLoS Pathog. (2020) 16:e1008561. doi: 10.1371/journal.ppat.1008561
36. Redzej A, Ukleja M, Connery S, Trokter M, Felisberto-Rodrigues C, Cryar A, et al. Structure of a VirD4 coupling protein bound to a VirB type IV secretion machinery. EMBO J. (2017) 36:3080–95. doi: 10.15252/embj.201796629
37. Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. (2009) 73:775–808. doi: 10.1128/MMBR.00023-09
38. Ohki M, Tomizawa J. Asymmetric transfer of DNA strands in bacterial conjugation. Cold Spring Harb Symp Quant Biol. (1968) 33:651–8. doi: 10.1101/SQB.1968.033.01.074
39. Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol. (2015) 13:343–59. doi: 10.1038/nrmicro3456
40. Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. (2005) 59:451–85. doi: 10.1146/annurev.micro.58.030603.123630
41. Christie PJ. The mosaic type IV secretion systems. EcoSal Plus. (2016) 7. doi: 10.1128/ecosalplus.ESP-0020-2015
42. Dmowski M, Golebiewski M, Kern-Zdanowicz I. Characteristics of the conjugative transfer system of the IncM plasmid pCTX-M3 and identification of its putative regulators. J Bacteriol. (2018) 200:e00234–18. doi: 10.1128/JB.00234-18
43. Zeng L, Yang C, Zhang J, Hu K, Zou J, Li J, et al. An outbreak of carbapenem-resistant Klebsiella pneumoniae in an intensive care unit of a major teaching hospital in Chongqing, China. Front Cell Infect Microbiol. (2021) 11:656070. doi: 10.3389/fcimb.2021.656070
Keywords: carbapenem-resistant Klebsiella pneumoniae, ST11, whole-genome sequencing, nanopore, KPC-2
Citation: Zeng L, Zhang J, Hu K, Li J, Wang J, Yang C, Huang W, Yin L and Zhang X (2022) Microbial Characteristics and Genomic Analysis of an ST11 Carbapenem-Resistant Klebsiella pneumoniae Strain Carrying blaKPC−2 Conjugative Drug-Resistant Plasmid. Front. Public Health 9:809753. doi: 10.3389/fpubh.2021.809753
Received: 05 November 2021; Accepted: 17 December 2021;
Published: 27 January 2022.
Edited by:
Xiaojiong Jia, Harvard Medical School, United StatesReviewed by:
Paul Kirchberger, University of Texas at Austin, United StatesFatima Bachir Halimeh, Lebanese University, Lebanon
Copyright © 2022 Zeng, Zhang, Hu, Li, Wang, Yang, Huang, Yin and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoli Zhang, jmszxl123@163.com
†These authors have contributed equally to this work
 Lingyi Zeng1,2†
Lingyi Zeng1,2† Jianmin Wang
Jianmin Wang Wan Huang
Wan Huang Xiaoli Zhang
Xiaoli Zhang