- 1Key Laboratory of Tropical Fruit Biology, Ministry of Agriculture and Rural Affairs of China, Zhanjiang, China
- 2Key Laboratory of Hainan Province for Postharvest Physiology and Technology of Tropical Horticultural Products, South Subtropical Crops Research Institute, Chinese Academy of Tropical Agricultural Sciences, Zhanjiang, China
- 3National Key Laboratory for Tropical Crop Breeding, Sanya Research Institute, Chinese Academy of Tropical Agricultural Sciences, Sanya, China
- 4College of Horticulture and Forestry Science, Huazhong Agricultural University, Wuhan, China
Backgrounds: Potato is sensitive to heat stress particularly during plant seedling growth. However, limited studies have characterized the expression pattern of the StGATA family genes under heat stress and lacked validation of its function in potato plants.
Methods: Potato plants were cultivated at 30°C and 35°C to induce heat stress responses. qRT-PCR was carried out to characterize the expression pattern of StGATA family genes in potato plants subjected to heat stress. StGATA2 loss-of-function and gain-of-function plants were established. Morphological phenotypes and growth were indicated by plant height and mass. Photosynthesis and transpiration were suggested by stomatal aperture, net photosynthetic rate, transpiration rate, and stomatal conductance. Biochemical and genetic responses were indicated by enzyme activity and mRNA expression of genes encoding CAT, SOD, and POD, and contents of H2O2, MDA, and proline.
Results: The expression patterns of StGATA family genes were altered in response to heat stress. StGATA2 protein located in the nucleus. StGATA2 is implicated in regulating plant height and weight of potato plants in response to heat stresses, especially acute heat stress. StGATA2 over-expression promoted photosynthesis while inhibited transpiration under heat stress. StGATA2 overexpression induced biochemical responses of potato plant against heat stress by regulating the contents of H2O2, MDA and proline and the activity of CAT, SOD and POD. StGATA2 overexpression caused genetic responses (CAT, SOD and POD) of potato plant against heat stress.
Conclusion: Our data indicated that StGATA2 could enhance the ability of potato plants to resist heat stress-induced damages, which may provide an effective strategy to engineer potato plants for better adaptability to adverse heat stress conditions.
Highlights
1. Expression patterns of StGATAs are altered in response to heat stresses;
2. StGATA2 affects plant morphological phenotypes and growth under heat stress conditions;
3. The photosynthesis and transpiration are regulated by StGATA2 in response to heat stress;
4. StGATA2 induces biochemical and genetic responses of potato plants to heat stress.
Introduction
The GATA-binding transcription factors comprise a protein family whose members contain either one or two highly conserved zinc finger DNA-binding domains (Teakle et al., 2002).GATA factors normally recognize the consensus sequence W-GATA-R (W, thymidine (T) or an adenosine (A); R, guanidine (G) or adenosine (A)) (Behringer and Schwechheimer, 2015). The DNA binding domain of GATA factors is constituted by a type IV zinc finger in the form CX2CX17-20CX2C followed by a highly basic region (Gronenborn, 2005). GATA proteins present two of zinc fingers, where only the C-terminal finger (C-finger) is involved in DNA binding, and the N-terminal zinc finger (N-finger) can modulate the binding of the C-finger to specific GATA sites, bind DNA with different specificity, or mediate the interaction with transcription cofactors of the friend of GATA family (Bates et al., 2008). The majority of the plant GATA factors contain a single zinc finger domain and mostly fall into two different categories: those with 17-residue loops (CX2CX17CX2C; also called zinger finger type IVa) and those with 18-residue loops (CX2CX18CX2C; also called zinger finger type IVb) (Scazzocchio, 2000).
Members of GATA family have been identified in plant organisms. Reyes et al. documented the existence of 29 different loci encoding putative GATA factors in Arabidopsis and 28 in rice (Reyes et al., 2004). In rapeseed, 96 GATAs have been identified and classified into 4 subfamilies based on the phylogenetic relationships, DNA binding domains and intron-exon structures (Zhu et al., 2020). Unevenly distributing on 21 chromosomes, 79 GATAs have been characterized in wheat (Feng et al., 2022a). In Ophiorrhiza pumila (Rubiaceae), genome-wide survey identified a total of 18 GATA genes classified into 4 subfamilies (Shi et al., 2022). In Brachypodium distachyon, Peng et al. systematically identified 28 GATA transcription factors distributing on 5 chromosomes (Peng et al., 2021). As for Solanum (Family: Solanaceae) plants, 30 GATAs were recognized in tomato (Solanum lycopersicum) by genome-wide analysis (Yuan et al., 2018). Recently, Yu et al. confirmed 49 GATAs randomly distributing on 12 chromosomes in potato (Solanum tuberosum) (Yu et al., 2022).
Generally speaking, there appears to be evolutionary conservation with regard to function of GATA factors, which has been validated to play roles In the control of normal physiological processes, such as chloroplast development, plant architecture (Hudson et al., 2013), chlorophyll synthesis, glucose metabolism (Bi et al., 2005), apical meristem and flower development (Zhao et al., 2004). Additionally, GATAs are light-regulated genes and respond to signals from the circadian clock (Manfield et al., 2007). Studies have revealed the transcript abundance of several GATAs is responsive to salinity, drought, exogenous ABA, and gibberellins (Gupta et al., 2017; Guo et al., 2021). However, there is limited studies on the responses of GATAs to heat stresses in potato plants. Herein, the current study described the expression patterns of StGATAs under heat stress conditions. Further, we analyzed the functional aspects of StGATA2 and stress responses-related genes. The biological functions of GATA2 gene have been reported in biological processes, such as photomorphogenesis (Luo et al., 2010). However, its responses to abiotic or biotic stresses are rarely reported.
Materials and methods
Plant material and heat stress
Potato (Solanum tuberosum L.) cultivar “Atlantic” was used in this study, which is mainly due to its high yields of tubers and its resistance to common scab, latent and mild mosaics. Potato plants were in vitro planted in Murashige and Skoog (MS) medium with pH adjusted among 5.8-6.0. The medium was supplemented with 3% sucrose for seedling growth. To induce tuber generation, 8% sucrose was utilized. The cultivation was conducted in a controlled biotron, with the following conditions: 22°C/15°C (day/night), 16-h/8-h light/dark photoperiod (2,800 Lux), and 50% air humidity. Potato tubers with a sprouted bud of 1 mm in height were transferred into soil and vermiculite (1:1, v/v) pots measuring 18 cm × 26 cm × 27 cm and maintained for 5 weeks. To induce heat stress, the plants were next fostered under mild (30°C) and acute (35°C) conditions. Potato leaves were collected for detecting transcriptional patterns of StGATA family genes, 0 h, 1 h, 2 h, 4 h, 8 h, 12 h, 24 h, and 48 h after heat treatment. For the quantitation of StGATAs genes expression, there were 144 seedlings (1 line × 2 heat treatments × 8 different time periods × 3 replications × 3 pots); For the quantitation of heat-responsive genes, there were 630 seedlings (7 lines × 2 heat treatments × 5 different time periods × 3 replications × 3 pots); For the examination of physiological and photosynthetic indexes, there were 630 seedlings (7 lines × 2 heat treatments × 5 different time periods × 3 replications × 3 pots).
Construction of transgenic plants
To generate potato plants highly expressing StGATA2 gene (OE for short), StGATA2 protein-encoding gene (GenBank Accession No. XM_006347854.2) was amplified using the specific primers (forward, 5’-CTCGAGATGGATGTCTACGGCGTGCACTCT-3’ and reverse 5’-GTCGACGCAGACCCGAAAGTGATGTCCGTACATTC-3’) (Bioeditas, Shaanxi, China). The PCR products were cloned into pBI121-EGFP plasmid according to a previous method (Li et al., 2020). The constructed plasmid was introduced into Agrobacterium tumefaciens strains LBA4404, followed by infecting the potato tuber slide (2 mm) according to Si’s method (Si et al., 2003). Potato plants with StGATA2 knocked down (Ri for short) were constructed with a previous method (Lu et al., 2019). The sense cDNA sequence was amplified using forward primer (Kpn I) and reverse primer (EcoR I) and inserted as an Kpn I-EcoR I fragment into pHANNIBAL (pHAN-StMAPK1-F); The anti-sense cDNA sequence was amplified using forward primer (Hind III) and reverse primer (BamH I), and inserted as a Hind III-BamH I fragment into pHANNIBAL (pHAN-StGATA2-R). The pHAN-StGATA2-RF were subcloned at Sac I and Spe I sites into pART vector (pART-StGATA2-RNAi). pART-StGATA2-RNAi was introduced into LBA4404, which was used to infect potato tuber slides.
After agrobacterium-mediated transformation, the tuber slides were cultivated on a solid medium in the dark at 28°C for 48 h. Then plants were continually cultivated on differentiation media in the dark at 22°C with 16-h/8-h light/dark photoperiod (2,400 Lux). The medium was changed every 12 days and the plants were cultured for about 3 to 4 weeks for shoot differentiation. Plant shoots were cut after growing to 1.5 cm and the shoots were transferred to the rooting medium for screening with 75% mg/mL of kanamycin. After 3 weeks of cultivation on MS medium, the leaves were collected for detecting StGATA2 by PCR using the specific primers for RNAi (forward: 5’-ATGGATGTCTACGG CGTGCACTCT-3’ and reverse: 5’- ATTAGCCGGAAAATTACTAAATGAAT-3’) and NPT II (forward: 5’-CTCACCTTGCTCCTGCCGAGA-3’ and reverse: 5’-CGCCTTGAGCCTGG CGAACAG-3’).
qRT-PCR
mRNA expression of StGATA family genes was examined 0 h, 1 h, 2 h, 4 h, 8 h, 12 h, 24 h, and 48 h after heat stress that has been described as abovementioned. For StGATA2 mRNA expression in transgenic plants, the leaves were collected 3 weeks after cultivation on MS medium. To examine the expression of heat stress genes (StSOD, StCAT, StPOD and StP5CS), plants leaves were collected 0 h, 8 h, 12 h, 24 h and 48 h after heat treatments. TRIzol RNA Extraction Kit (Invitrogen, Carlsbad, CA, USA). The first-strand cDNA was synthesized using the First-Strand cDNA Synthesis Kit (TransGen Biotech, Beijing, China). qPCR was performed using 100 ng of cDNA, 10 μL of SYBR Premix Ex Taq (2 ×) (Takara, Tokyo, Japan), and 0.8 μL of specific primers (0.5 μM) on ABI3000 System (Applied Biosystems, Foster City, CA, USA). The reaction procedures were: one cycle at 94°C for 3 min and 36 cycles of amplification at 94°C for 45 s, 59°C for 34 s, and 72°C for 1 min. The relative mRNA expression was calculated using the formula 2-△△Ct. Solanum tuberosum translation elongation factor 1α gene (StEF1α) served as an internal control. The specific primers were listed in Table 1.
Subcellular localization
Protein-coding sequence of StGATA2 was amplified and ligated to the expression vector pPBI121-EGFP, using the following primers: forward, 5’-CTCGAGATGGATGTCTACGGC GTGCACTCT-3’ and reverse, 5’-GTCGACGCAGACCCGAAAGTGATGTCCGTACATTC-3’. The constructed plasmid was transformed into Agrobacterium tumefaciens GV3101. The transformed strain infiltrated tobacco epidermal cells in accordance with a previous method reported by Sparkes et al. (2006). The green fluorescence was detected 48 h after infiltration under a Leica TCA confocal scanning laser microscopy (Leica, Weztlar, Germany).
Phylogenetic analysis and amino acid alignment
Phylogenetic tress for GATA domain-containing homologs with highly similarity was constructed using MEGA 5.05 Software. Multiple-sequence alignment of GATA domain-containing amino acid sequence was analyzed using DNAman tool (Lynnon Biosoft, San Ramon, CA, USA)
Measurements of plant growth
Potato plants were imaged 2 days after heat stress. Potato plants cultivated at 35°C were transferred to 22°C conditions and maintained for 7 days. CK plants were cultivated at 22°C for 2 days. The plants were measured for plant height, plant fresh weight (FW), plant dry weight (DW), root FW and root DW.
Stomatal apertures
Stomatal apertures were examined according to a previous method reported by Tricker et al. (2005). After 35 days of cultivation, transgenic or non-transgenic plants were subjected to heat stress treatment for 24 h or 48 h. The comparable fully-expanded leaves of transgenic or non-transgenic plants were selected between 9:30-11:00 am for measuring stomatal aperture. Clear nail polish was applied to the third fully expanded fully functional leaf located at the top of the plant, followed by covering with scotch tape. The leaves were mounted on glass slides and imaged using a Zeiss-Axiolmager M2 microscope (Zeiss, Germany). Three biological replicates were performed for each condition separately.
Net photosynthetic rate, transpiration rate, and stomatal conductance
The third leaf from the plant top was collected when it fully expands during 9:30-11:30. Net photosynthetic rate, transpiration rate and stomatal conductance were examined using a portable photosynthetic LI-6400XT system (Li-COR, Lincoln, NE, USA). The photon flux density was set as 1,500 μmol·m-2·s-1. The relative humidity in leaf chamber was 50%-70%. CO2 concentration was 400 μmol/mol.
Contents of H2O2, proline, and MDA, and activities of CAT, SOD, and POD
To assess physiological and biochemical changes in plants under heat stress, 5-week-old StGATA2 transgenic plants and non-transgenic control plants grown under greenhouse conditions were subjected to heat stress treatments at 30°C and 35°C. Contents of H2O2, proline, and malondialdehyde (MDA) and activities of catalase (CAT), superoxide dismutase (SOD), and peroxidase (POD) were determined according to our previous methods (Zhu et al., 2021).
Statistical analysis
All experiments were carried out with three biological replicates and three technological replicates. Data are shown as the mean ± standard deviation. Statistical analysis was done with IBM SPSS 19.0 Statistical Software (IBM, Chicago, IL, USA) and GraphPad Prism Software (GraphPad, San Diego, CA, USA). Multiple comparisons were analyzed by one-way ANOVA with Tukey test or Dunnett’s T3 for post-hoc analysis or two-way ANOVA corrected by Sidak’s multiple comparisons test. Histograms were computed with GraphPad Prism software, heatmap by the pheatmap function in R version 4.2.3.
Results
Expression signatures of GATA family genes in potato plants in response to heat stress
To provide an overview of expression patterns of GATA family genes, we cultivated potato plants under heat stresses. qRT-PCR results showed that mRNA expression of the 57 members from GATA family genes were distinctly altered during mild (30°C) or acute (35°C) heat stresses (0-48 h), which was presented in heatmaps (Figures 1A, B). Importantly, under heat stress conditions, StGATA2 expression was induced 1-48 h after heat stress (30°C and 35°C) treatment and peaked at 48 h with 7.5-fold and 8.4-fold increases in mRNA expression, respectively (Figure 1). Notably, StGATA2 transcript levels were maintained at higher levels (expression increased more than 7 folds) at 8 h, 12 h, 24 h and 48 h after heat stress treatment (P < 0.05). The results indicated that heat stress at 30°C and 35°C induced a sustained and stable high expression of StGATA2. Consequently, we speculated that StGATA2 gene may play molecular functions in response to heat stresses.

Figure 1 Heatmap suggesting mRNA expression of StGATA family genes in potato leaves in response to heat stress. Five-week-old normally grown plants were subjected to 0 h, 1 h, 2 h, 4 h, 8 h, 12 h, 24 h, or 48 h of (A) mild (30°C) or (B) acute (35 °C) heat stresses. Multiple comparisons were analyzed by one-way ANOVA with Tukey test or Dunnett’s T3 for post-hoc analysis. Difference letters indicates the difference between the two groups (p<0.05).
Potato plants encoding GATA2 gene with CTHC-X18-CNAC motif
Phylogenetic tree of GATA factors was generated, including Solanum tuberosum GATA2 (StGATA2), Solanum lycopersicum GATA2 (SlGATA2), Nicotiana tabacum GATA4 (NtGATA4), Capsicum annuum GATA4 (CaGATA4), Capsicum baccatum GATA2 (CbGATA2), Arabidopsis thaliana GATA2 (AtGATA2), Oryza sativa GATA12 (OsGATA12) (Figure 2A). Solanum tuberosum GATA2 contains CTHC-X18-CNAC motif (Figure 2B). To examine StGATA2 function, we obtained StGATA2 overexpression plants by expressing pBI121-EGFP-StGATA2 and generated a loss-of-function plants by introducing pART-StGATA2-RNAi. The relative mRNA expression of StGATA2 in transgenic plants were shown in Figures 2C, D, which significantly increased or knocked down when compared to the wild type (p<0.001). Amongst the overexpression lines, OE-1, OE-2 and OE-5 were selected as the significant over-expressors, and Ri-1, Ri-4 and Ri-6 as the significant under-expressors used for the functional analysis in response to heat stresses. StGATA2 protein obviously located in the nucleus (Figure 3). The subcellular localization of GATA2 to the nucleus is the structural premise for DNA binding and transcription activation.
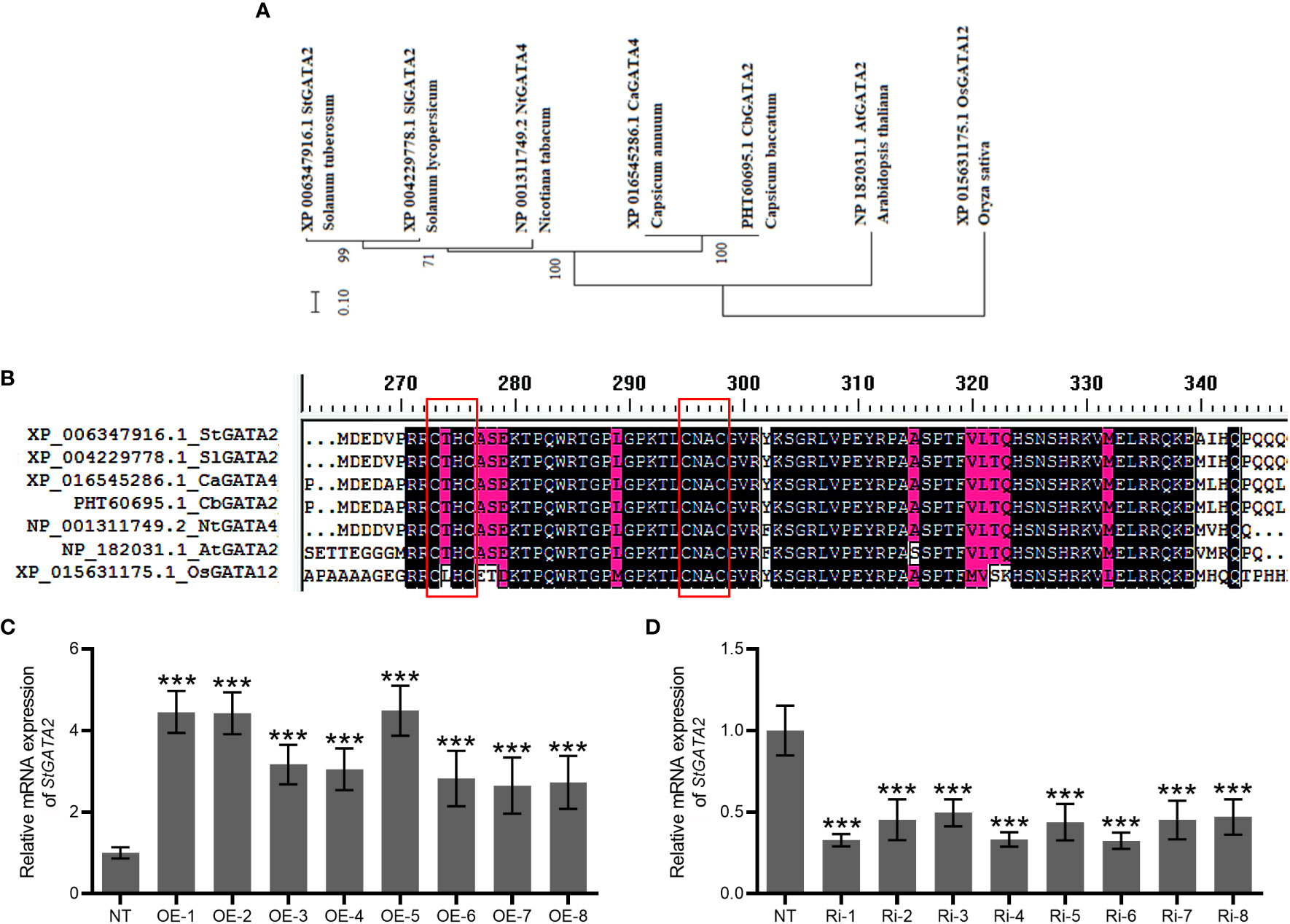
Figure 2 Bioinformatic analysis of StGATA2 protein and construction of StGATA2 overexpression or under-expression plants. (A) Phylogenetic tree and schematic representation of GATA factors from Solanum tuberosum (St), Solanum lycopersicum (Sl), Nicotiana tabacum (Nt), Capsicum annuum (Ca), Capsicum baccatum (Cb), Arabidopsis thaliana (At), Oryza sativa (Os). The scale bar represents the difference degree of amino acid sequences. (B) Multiple-sequence alignment of conserved single CTHC-X18-CNAC motif highlighted in red boxes. (C, D) The quantification of StGATA2 mRNA expression in potato plants; NT, non-transgenic plants; OE, pBI121-EGFP-StGATA2-transgenic lines; Ri, pART-StGATA2-RNAi-transgenic plants; Mean ± standard deviation; Student’s t-test, ***p<0.001 (n = 9).
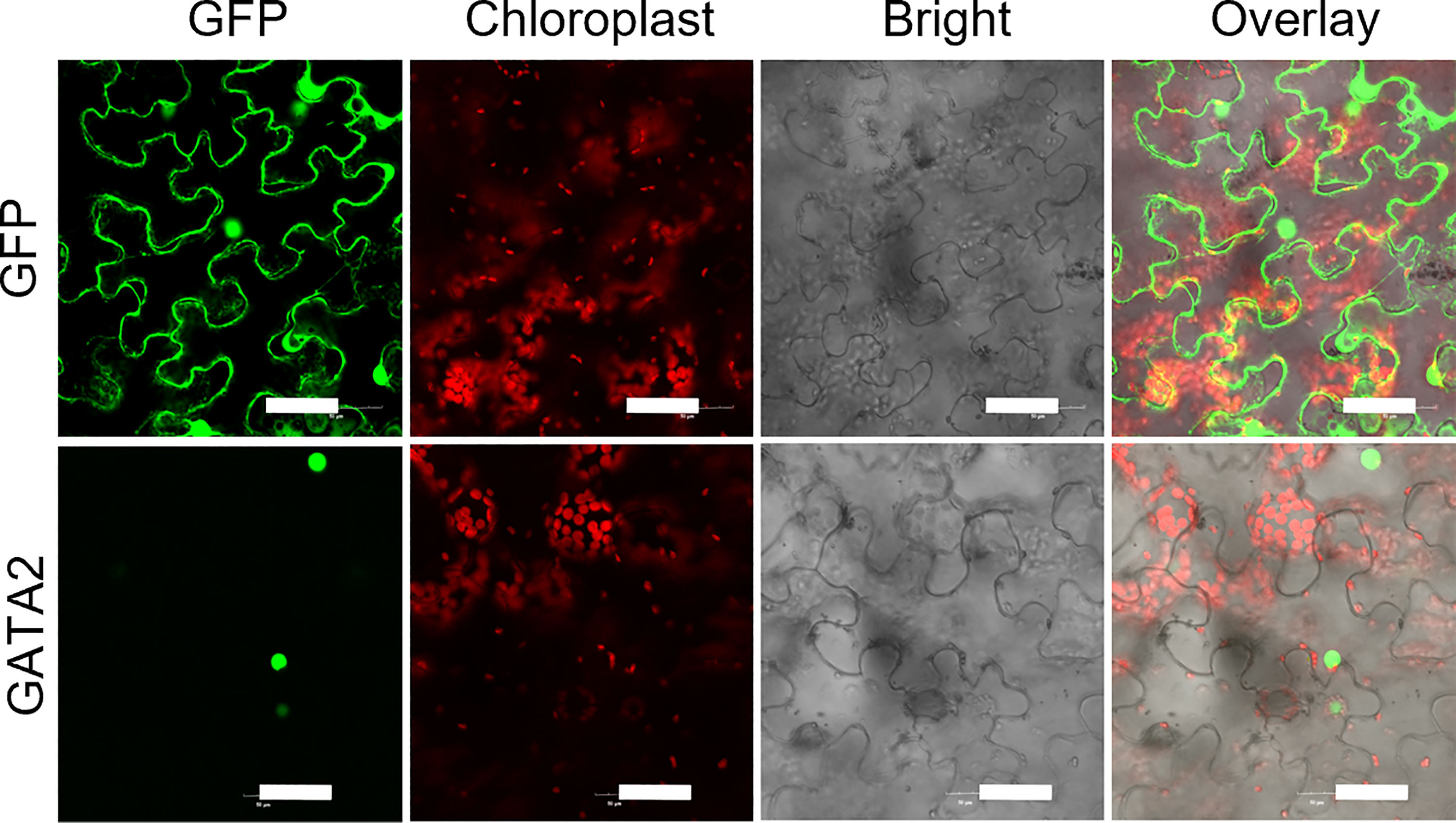
Figure 3 StGATA2 protein locates on the cellular nucleus of tobacco epidermal cells. GFP-StGATA2 fusion protein was transiently expressed in tobacco leaves and observed using a laser scanning confocal microscope. Bar = 50 μm.
Effects of StGATA2 on plant morphological phenotypes and growth in response to heat stresses
Next, we cultivated the 5-week-old transgenic or non-transgenic plants under heat stresses for 2 days, and investigated whether StGATA2 gene is involved in regulating plant morphology or growth such as plant height, plant weight and root weight. The morphological features of the plants were imaged 2 days after cultivation at 22°C (CK), 30°C, or 35°C. Figure 4A showed that neither StGATA2 over-expression nor under-expression affected plant morphological structure, compared to wild-type plants at 22°C. However, the height of OE plants in Figure 4B or Ri plants in Figure 4C was significantly different from the non-transgenic plants.
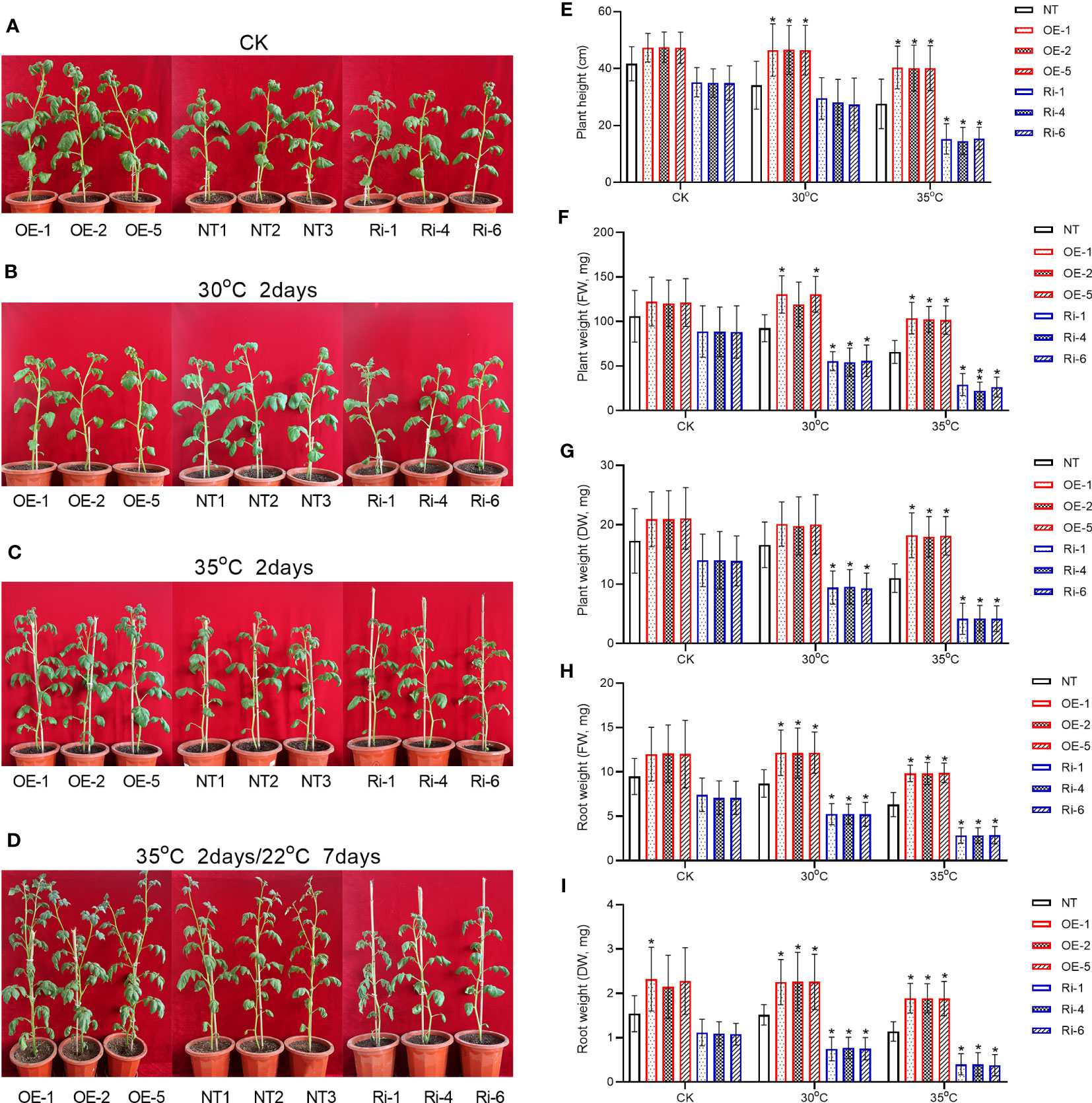
Figure 4 Growth characterizations of StGATA2 over-expression or under-expression plants in response to heat stresses. (A–D) Representative photographs of potato plants with the specified genotypes (NT, OE, and Ri); Five-week-old plants were cultivated under (A) 22°C (CK), (B) 30°C (30°C), (C) 35°C, or (D) 35°C followed by moving to 22°C condition; Plants in A, B, and C were imaged 2 days after heat stress; After heat stress, plants in D were imaged 7 days after normal cultivation. (E) Plant height, (F) plant fresh weight (FW), (G) plant dry weight (DW), (H) root FW, and (I) root DW of transgenic or non-transgenic plants were measured 2 days after cultivation under the specified conditions (22°C, 30°C, and 35°C). NT, non-transgenic plants; OE, pBI121-EGFP-StGATA2-transgenic lines; Ri, pART-StGATA2-RNAi-transgenic plants; Mean ± standard deviation; Ordinary two-way ANOVA with Tukey’s multiple comparisons test, *p<0.05 (n = 9).
The statistical analysis results revealed that under mild heat stress (30°C), OE plants were characterized by increased plant height (Figure 4E), while plant fresh weight (Figure 4F), plant dry weight (Figure 4G), root fresh weight (Figure 4H) or root dry weight (Figure 4I) were unchanged (p>0.05) relative to NT plants (p<0.05). In contrast, the height of Ri plants was not significantly different from NT plants (p>0.05) (Figure 4E), while StMAPK1 under-expression reduced plant fresh weight (Figure 4F), plant dry weight (Figure 4G), root fresh weight (Figure 4H) or root dry weight (Figure 4I) (p<0.05) 2 days after growth at 30°C. As for the acute heat stress (35°C), OE plants displayed significant increases in plant height, plant fresh weight, plant dry weight, root fresh weight, and root dry weight, which were significantly decreased in Ri plants compared to non-transgenic plants (p<0.05).
Further, we compared the morphological structures of transgenic plants to non-transgenic plants 7 days after the plants receiving heat stresses were cultivated at 22°C. Figure 4D showed that compared to the non-transgenic plants, OE plants showed an increased plant height and more apical branches. However, Ri plants grows slowly, with a shorter height compared to NT plants. These results indicated that StGATA2 is implicated in the growth of potato plants in response to heat stresses, especially acute heat stress.
StGATA2 over-expression enhanced photosynthetic rate and inhibited transpiration in response to heat stresses
Measurements revealed that average ratio of stomal width to length was significantly decreased in StGATA2 overexpressing plants compared with NT plants, 24 or 48 h after mild heat stresses (p<0.05, p<0.01) (Figures 5A, B). StGATA2 under-expression resulted in a significant increase in stomatal aperture (p<0.05), 24 h or 48 h after mild or acute heat stresses. Under heat stress conditions, the net photosynthetic rate was increasingly enhanced in OE plants, while decreased in Ri plants (p<0.05) (Figures 5C, D). Additionally, there were evidence of StGATA2 overexpression inhibiting transpiration rate (Figures 5E, F) and decreasing stomatal conductance (Figures 5G, H) (p<0.05, p<0.01). Comparing to the NT plants, 30°C or 35°C heat stresses resulted in significant increases in transpiration rate and stomatal conductance of StGATA2 under-expression plants (p<0.05, p<0.01, p<0.001). All of these results confirmed that StGATA2 is closely correlated with heat stress-induced modifications in photosynthesis and transpiration rate.
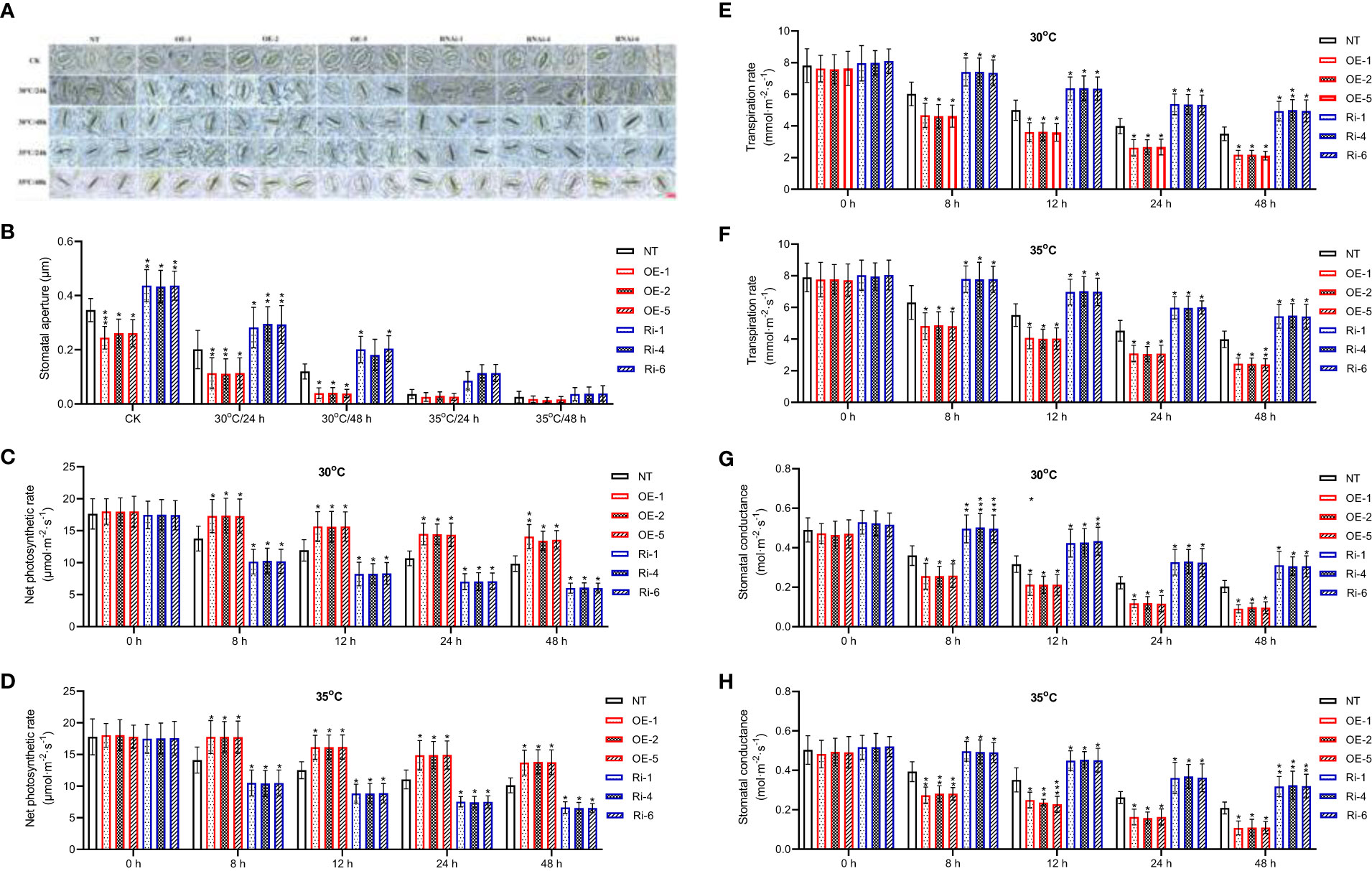
Figure 5 Stomatal apertures and photosynthesis of StGATA2 over-expression or under-expression plants in response to heat stresses. (A) Representative images of Solanum tuberosum stomata; The scale bar equates to 10 μm. (B) Stomatal apertures depicted by the ratio of pore width to length. (C, D) Net photosynthetic rate, (E, F) transpiration rate and (G, H) stomatal conductance of potato leaves. The plants were measured 2 days after cultivation under the specified conditions (22°C, 30°C, and 35°C). NT, non-transgenic plants; OE, pBI121-EGFP-StGATA2-transgenic lines; Ri, pART-StGATA2-RNAi-transgenic plants. Mean ± standard deviation. Ordinary two-way ANOVA with Tukey’s multiple comparisons test, *p<0.05, **p<0.01,***p<0.001 (n = 9).
Biochemical and genetic responses of StGATA2-transgenic potato plants to heat stress
Biochemical changes in potato plants were assayed to evaluate the capacity of StGATA2 to trigger protective mechanisms against heat stress. There is a significantly increase in contents of H2O2, MDA, and proline, as well as the activity of CAT, SOD and POD, following mild and acute heat stress treatment (p<0.05, p<0.01) (Figure 6). Compared to the NT plants, StGATA2 overexpression significantly restrained the accumulation of H2O2 and MDA, while further induced the generation of proline (Figures 6A–F). Under heat stress conditions, StGATA2 overexpression enhanced the activity of CAT, SOD and POD compared to the non-transgenic plants (Figures 6G–L). However, there was an opposite trend toward the contents of H2O2, MDA and proline, as well as the activity of CAT, SOD, and POD in the Ri plants following heat stress, compared with OE plants. These data suggested that StGATA2 overexpression induced biochemical responses of potato plant against heat stress.
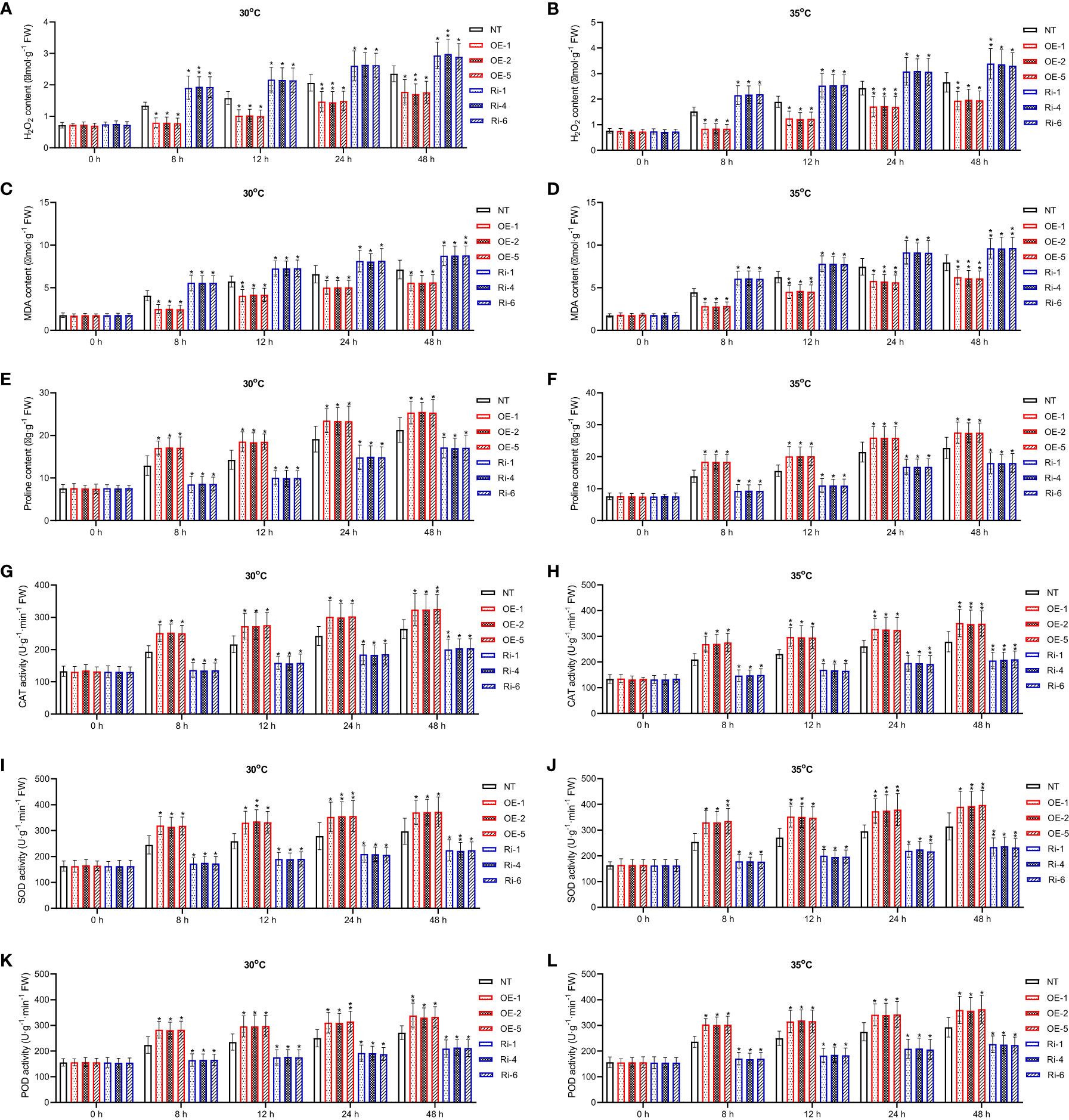
Figure 6 Effects of StGATA2 expression on biochemical indexes of potato plants in response to heat stresses. (A, B) H2O2 content, (C, D) MDA content, (E, F) proline content, (G, H) CAT activity, (I, J) SOD activity, and (K, L) POD activity in potato leaves were estimated 2 days after cultivation under the specified conditions (22°C, 30°C, and 35°C). NT, non-transgenic plants; OE, pBI121-EGFP-StGATA2-transgenic lines; Ri, pART-StGATA2-RNAi-transgenic plants. Mean ± standard deviation. Ordinary two-way ANOVA with Tukey’s multiple comparisons test, *p<0.05, **p<0.01, ***p<0.001 (n = 9).
Besides, under heat stress conditions, mRNA expression of heat stress responsive genes was increased in non-transgenic, including StSOD (Figures 7A, B), StCAT (Figures 7C, D), StPOD (Figures 7E, F), StP5CS (Figures 7G, H) (p<0.05, p<0.01, p<0.001). However, StGATA2 over-expression further elevated the transcription of these 4 heat stress-responsive genes following heat stress treatment, compared to non-transgenic plants. In contrast, Ri plants showed the opposite results. This analysis indicated that StGATA2 overexpression induced genetic responses of potato plant against heat stress.
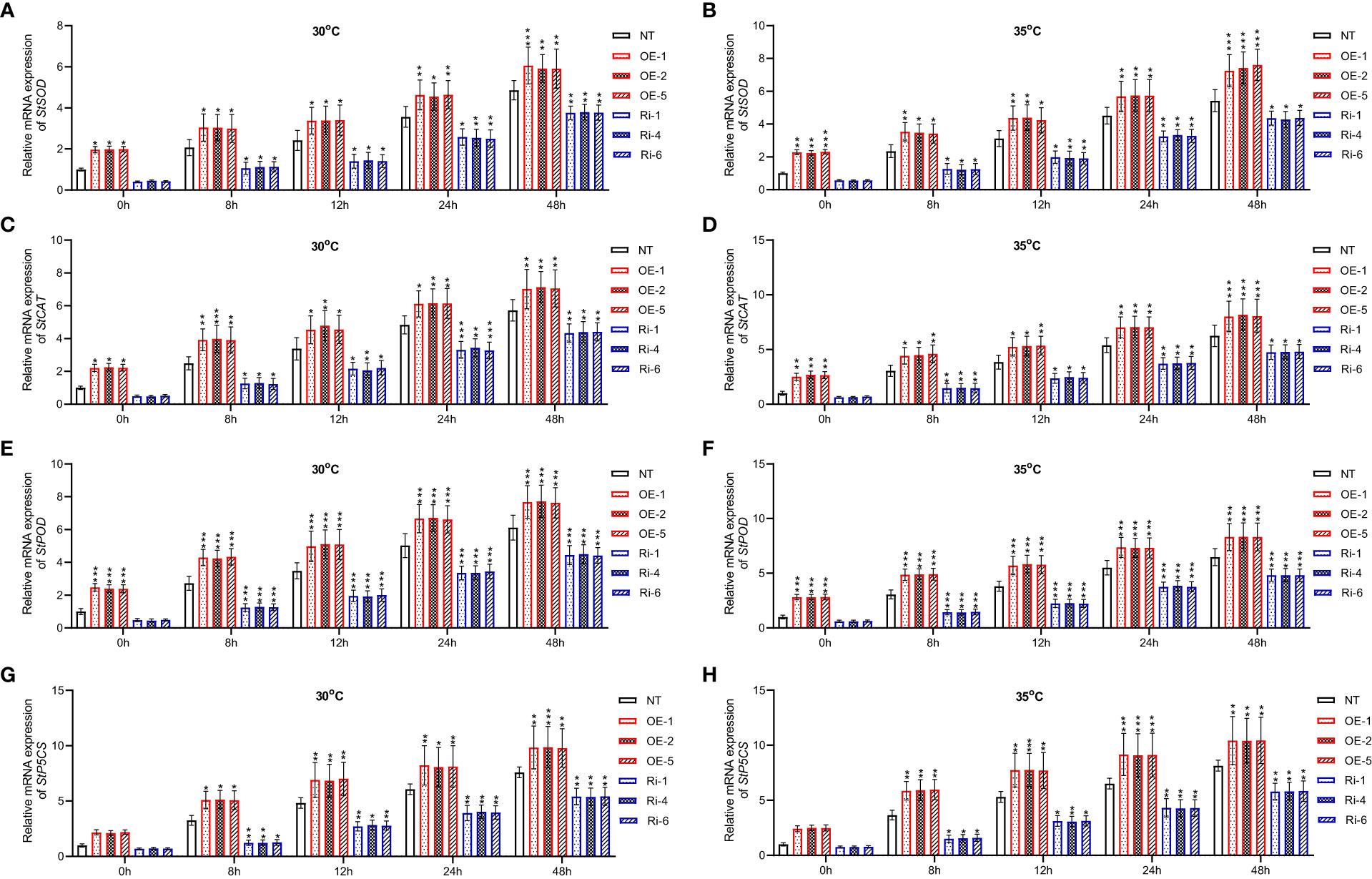
Figure 7 Changes in relative mRNA expression of heat-stress responsive genes in StGATA2 over-expression or under-expression plants in response to heat stresses. (A, B) mRNA expression of StSOD, (C, D) StCAT, (E, F) StPOD, and (G, H) StP5CS in potato leaves were estimated 2 days after cultivation under the specified conditions (22°C, 30°C, and 35°C). NT, non-transgenic plants; OE, pBI121-EGFP-StGATA2-transgenic lines; Ri, pART-StGATA2-RNAi-transgenic plants. Mean ± standard deviation. Ordinary two-way ANOVA with Tukey’s multiple comparisons test, *p<0.05, **p<0.01, ***p<0.001 (n = 9).
Discussion
High temperature limited plant morphophysiological growth, photosynthesis, dark respiration, carbohydrate metabolism (Lafta and Lorenzen, 1995; Fahad et al., 2016), root system architecture (Luo et al., 2020), stomal regulation and water use (Körner, 2006). GATA family genes are heat-responsive genes, which has been identified in cucumber (Zhang et al., 2021), wheat (Feng et al., 2022b), and pepper (Yu et al., 2021). However, these studies only characterized the expression pattern of the GATA family genes under heat stress and lacked validation of its function. In the present study, we present the first evidence that StGATA2 plays an important role in maintaining plant growth against heat stresses, through the regulation of heat-responsive gene expression in Solanum tuberosum.
Genome-wide analysis has identified the GATA family genes in wheat (Feng et al., 2022b), pepper (Yu et al., 2021), cucumber (Zhang et al., 2021), rice (Gupta et al., 2017), and foxtail millet (Lai et al., 2022). Based on the whole potato genome data, Yu et al. systematically identified 49 GATA proteins, of which mRNA expression is affected by Ralstonia solanacearum, abscisic acid and methyl jasmonate (Yu et al., 2022). Our previous study identified 57 GATA proteins randomly distributed on 12 chromosomes (unpublished data). The expression profile of GATA genes was significantly altered when plants are subjected to heat stress (Yu et al., 2021; Feng et al., 2022b), suggesting their potential roles involved in biological functions associated with heat tolerance. The expression pattern of GATA genes is still unexplored when potato plants are exposed to heat stresses. As a result, we cultivated potato plants under mild and acute heat stresses and determined mRNA expression of GATA genes. In mature potato plants, we noted an evident alteration in mRNA expression of GATA family genes under heat stress conditions during the observed intervals. We found a substantially and stably up-regulated gene StGATA2 after heat stress treatment. This suggested that StGATA2 is a positive heat stress-responsive gene in the wild-type potato plant. StGATA2 gene was then selected for constructing transgenic plants to identify its roles in response to heat stress.
StGATA2 belongs to 18-residue loops (i.e. CX2CX18CX2C; also called zinger finger type IVb). The zinc finger motifs are categorized into two groups: those with 17-residue loops (CX2CX17CX2C zinc finger type IVa) and those with 18-residue loops (CX2CX18CX2C zinc finger type IVb), which are based on the spacing between the cysteine pairs at the zinc finger loop (Park et al., 2006). GATA transcription factors play a crucial role in plant growth, biomass accumulation and plant height, of which PdGNC have been confirmed in poplar, for example (An et al., 2019). Studies have revealed the involvement of GATAs in responding to multiple stresses when the crops are exposed to salinity, drought, exogenous ABA, acid, alkali, dark, flooding, heat, and cold (Gupta et al., 2017; Yu et al., 2021; Zhang et al., 2021; Feng et al., 2022b; Lai et al., 2022). For instance, in tomato plants, SlGATA17 expression was induced by heat stress (Wang et al., 2023). Transgenic Arabidopsis plants overexpressing GATA factors ZIM show enhanced petiole and hypocotyl cell elongation (Shikata et al., 2004). However, limited studies have been conducted to demonstrate the roles of GATAs in plant growth in response to heat stress. Potato is characterized by specific temperature requirements and develops best at about 20°C. Subsequently we constructed the loss-of-function and gain-of-function variants to examine the role of StGATA2 in potato plant growth. Our results confirmed that potato plant growth was greatly reduced at 30 °C and 35°C higher than optimum, responding with decreases in plant height and weight. The transfer of plants from 35°C to 22°C had apparent effect on shoot growth. Besides, He et al. have validated that GATA is a crucial regulator of the flag leaf development in rice (He et al., 2018). These results suggested that StGATA2, a member of the StGATA family genes, is involved in the growth process of several organs such as root, shoot and leaf of the potato plants.
Stomata protects plants against immediate or long-term injuries caused by environmental alterations. It was recently shown that heat stress initiated a rapid local and systemic stomatal opening responses (Devireddy et al., 2020). In Arabidopsis, Klermund et al. reported that LLM-domain B-class GATA genes strongly promotes stomata formation, most strikingly in hypocotyls but also in cotyledons (Klermund et al., 2016). Whereas it is still unclear about how GATA genes affect the stomatal aperture. In our study, consistent heat stress decreases the stomatal aperture, and StGATA2 expression significantly further inhibited the stomatal aperture, which may play roles in leaf temperature, CO2 exchange rates and water vapor loss. In poplar, PdGNC affects chlorophyll content and photosynthetic rate (An et al., 2019). The conserved GATA transcription factor mediates plant architecture and chloroplast development by altering chlorophyll, chloroplast number, photosynthesis, and amnio acid and starch biosynthesis, which may be associate with a semidwarf phenotype of the transgenic rice (Hudson et al., 2013). Transcriptomic analysis found that PdGNC may potentially mediate nitrogen uptake in the root, cell division and carbohydrate utilization in the stem, and photosynthetic electron transfer and carbon assimilation in the leaf (An et al., 2019). However, the biological function of GATAs has not been studied under environmental stresses. Our data clearly demonstrated that StGATA2 was involved in maintaining photosynthetic rate and mitigating respiratory rate.
Catalase functions as antioxidant enzyme, dissolving hydrogen peroxide into water and oxygen peroxisomes, of which encoding genes were found with cis-elements related to stress responses and plant hormones signaling in potato plants (Jbir Koubaa et al., 2023). Superoxide dismutase encoded by StCuZnSODs, StFeSOD3 and StMnSOD play a significant role in catalyzing the conversion or dismutation of toxic superoxide anion radical into H2O2 and O2, representing the first line of antioxidant defense against ROS induced by heat stress (Rudić et al., 2022). Peroxidase also serves as ROS-scavenging enzyme, catalyzing the conversion of H2O2 into H2O with ascorbate as a specific electron donor (Caverzan et al., 2012). The expression of peroxidase-encoding gene is regulated in response to abiotic stresses such as heat stress during plant development (Caverzan et al., 2012). Our data suggested that the changes in enzyme activities of SOD, CAT, and POD at high temperature were coordinated with alterations in the transcription of the protein-encoding genes. The increases in activities of antioxidant enzymes at high temperature is more likely due to an increase in StGATA2 expression. In the present experiments, increased proline content was observed at high temperature, which has been related to elevated transcripts of StP5CS. A study has confirmed the efficacy of proline in countering the damages in plant growth and enzymes of carbon and antioxidative metabolism (Kaushal et al., 2011). We found that StGATA2 gene was associated with the maintained expression of StP5CS and proline level.
Plant GATAs containing GATA-type zinc fingers with two CX2C motifs interspaced by a 17-20 amino acid long loop recognize the DNA sequence W-G-A-T-A-R through a single type IV zinc finger (Patient and McGhee, 2002; Reyes et al., 2004). Sugimoto et al. reported that the GATA-type zinc finger region is required for binding activity and activate expression from NtMyb2 promoter (Sugimoto et al., 2003). In reverse, Gupta et al. presented that the alternative splice variant of OsGATA23 (OsGATA23b) was unable to respond to abiotic tresses, which throws a novel light on the tight regulation of the spliced variants of OsGATA genes in response to salinity, drought and exogenous ABA (Gupta et al., 2017). Wang et al. performed yeast one assay and dual-luc assay and confirmed that the GATA-box of SlGATA17 promoter was bound by SlHY5 and its expression was then regulated, which causes the hypersensitivity of Arabidopsis to NaCl at the post-germination stage (Wang et al., 2023). However, it is not yet possible to explain how the StGATA2 gene regulates the expression of heat responsive genes based on the available results of this study. GATA-encoded proteins contain higher conserved DNA binding domain that consist of two zinc finger motifs and two adjacent stretches of basic amino acids (Molkentin, 2000). The N-terminal finger can interact with adjacent GATA DNA sequence elements or with protein cofactors (Trainor et al., 1996; Weiss et al., 1997). The nuclear localization and transcriptional activation domains of GATA2-encoding protein may promote or inhibit the transcriptional activation of StSOD, StCAT, StPOD, and StP5CS. However, the identification of the protein domain needs protein domain deletion analysis.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors. The GATA protein sequences described in this article are available at the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) with the following number: SlGATA2 (XP_004229778.1), StGATA2 (XP_006347916.1), CaGATA4 (XP 016545286.1), CbGATA2 (PHT60695.1), NtGATA4 (NP_001311749.2), AtGATA2 (NP_182031.1), OsGATA12 (XP_015631175.1).
Author contributions
XZ, HD, and YZ planned and designed the research. XZ, HD, HJ, SC, ZC, SS, JT, and YZ collected the data. XZ, HD, HJ, SC, ZC, SS, JT, and YZ analyzed the data. XZ, HD, and YZ drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This Research Program was financially supported by the Hainan Provincial Natural Science Foundation of China (No. 322MS116, 323MS095), and Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (No. 1630062023001, 1630062023002).
Acknowledgments
We thank Rongkai Wang (Bioediates, Shaanxi, China) for providing the plasmids pBI121-EGFP, pHANNIBAL, and pART.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
An, Y., Zhou, Y., Han, Y., Shen, C., Wang, S., Liu, C., et al. (2019). The GATA transcription factor GNC plays an important role in photosynthesis and growth in poplar. J. Exp. Bot. 71 (6), 1969–1984. doi: 10.1093/jxb/erz564
Bates, D. L., Chen, Y., Kim, G., Guo, L., Chen, L. (2008). Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA. J. Mol. Biol. 381 (5), 1292–1306. doi: 10.1016/j.jmb.2008.06.072
Behringer, C., Schwechheimer, C. (2015). B-GATA transcription factors – insights into their structure, regulation, and role in plant development. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00090
Bi, Y-M., Zhang, Y., Signorelli, T., Zhao, R., Zhu, T., Rothstein, S.. (2005). Genetic analysis of arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 44 (4), 680–692. doi: 10.1111/j.1365-313X.2005.02568.x
Caverzan, A., Passaia, G., Rosa, S. B., Ribeiro, C. W., Lazzarotto, F., Margis-Pinheiro, M., et al. (2012). Plant responses to stresses: role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 35, 1011–1019. doi: 10.1590/S1415-47572012000600016
Devireddy, A. R., Arbogast, J., Mittler, R. (2020). Coordinated and rapid whole-plant systemic stomatal responses. New Phytol. 225 (1), 21–25. doi: 10.1111/nph.16143
Fahad, S., Hussain, S., Saud, S., Hassan, S., Ihsan, Z., Shah, A. N., et al. (2016). Exogenously applied plant growth regulators enhance the morpho-physiological growth and yield of rice under high temperature. Front. Plant Sci. 7, 1250. doi: 10.3389/fpls.2016.01250
Feng, X., Yu, Q., Zeng, J., He, X., Liu, W. (2022a). Genome-wide identification and characterization of GATA family genes in wheat. BMC Plant Biol. 22 (1), 372. doi: 10.1186/s12870-022-03733-3
Feng, X., Yu, Q., Zeng, J., He, X., Liu, W. (2022b). Genome-wide identification and characterization of GATA family genes in wheat. BMC Plant Biol. 22 (1), 1–15. doi: 10.1186/s12870-022-03733-3
Gronenborn, A. M. (2005). The DNA-binding domain of GATA transcription factors–a prototypical type IV Cys2-Cys2 zinc finger (Springer Boston, MA: Molecular Biology Intelligence Unit).
Guo, J., Bai, X., Dai, K., Yuan, X., Guo, P., Zhou, M., et al. (2021). Identification of GATA transcription factors in brachypodium distachyon and functional characterization of BdGATA13 in drought tolerance and response to gibberellins. Front. Plant Sci. p, 2386. doi: 10.3389/fpls.2021.763665
Gupta, P., Nutan, K. K., Singla-Pareek, S. L., Pareek, A. (2017). Abiotic stresses cause differential regulation of alternative splice forms of GATA transcription factor in rice. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01944
He, P., Wang, X., Zhang, X., Jiang, Y., Tian, W., Zhang, X., et al. (2018). Short and narrow flag leaf1, a GATA zinc finger domain-containing protein, regulates flag leaf size in rice (Oryza sativa). BMC Plant Biol. 18 (1), 273. doi: 10.1186/s12870-018-1452-9
Hudson, P., Guevara, D. R., Hand, A. J., Xu, Z., Hao, L. (2013). Rice cytokinin GATA transcription Factor1 regulates chloroplast development and plant architecture. Plant Physiol. 162 (1), 132–144. doi: 10.1104/pp.113.217265
Jbir Koubaa, R., Ayadi, M., Saidi, M N., Charfeddine, S., Bouzid, R. G, Nouri-Ellouz, O. (2023). Comprehensive genome-wide analysis of the catalase enzyme toolbox in potato (Solanum tuberosum l.). Potato Res. 66 (1), 23–49. doi: 10.1007/s11540-022-09554-z
Kaushal, N., Gupta, K., Bhandhari, K., Kumar, S., Thakur, P., Nayyar, H. (2011). Proline induces heat tolerance in chickpea (Cicer arietinum l.) plants by protecting vital enzymes of carbon and antioxidative metabolism. Physiol. Mol. Biol. Plants 17, 203–213. doi: 10.1007/s12298-011-0078-2
Klermund, C., Ranftl, Q. L., Diener, J., Bastakis, E., Richter, R., Schwechheimer, C. (2016). LLM-domain b-GATA transcription factors promote stomatal development downstream of light signaling pathways in arabidopsis thaliana hypocotyls. Plant Cell 28 (3), 646–660. doi: 10.1105/tpc.15.00783
Körner, C. (2006). Significance of temperature in plant life. Plant Growth Climate Change 100, 48–66. doi: 10.1002/9780470988695.ch3
Lafta, A. M., Lorenzen, J. H. (1995). Effect of high temperature on plant growth and carbohydrate metabolism in potato. Plant Physiol. 109 (2), 637–643. doi: 10.1104/pp.109.2.637
Lai, D., Yao, X., Yan, J., Gao, A., Yang, H., Xiang, D., et al. (2022). Genome−wide identification, phylogenetic and expression pattern analysis of GATA family genes in foxtail millet (Setaria italica). BMC Genomics 23 (1), 549. doi: 10.1186/s12864-022-08786-0
Li, G., Cao, C., Yang, H., Wang, J., Wei, W., Zhu, D., et al. (2020). Molecular cloning and potential role of DiSOC1s in flowering regulation in davidia involucrata baill. Plant Physiol. Biochem. 157, 453–459. doi: 10.1016/j.plaphy.2020.11.003
Lu, H., Klocko, A. L., Brunner, A. M., Ma, C., Magnuson, A. C., Howe, G. T., et al. (2019). RNA Interference suppression of AGAMOUS and SEEDSTICK alters floral organ identity and impairs floral organ determinacy, ovule differentiation, and seed-hair development in populus. New Phytol. 222 (2), 923–937. doi: 10.1111/nph.15648
Luo, X.-M., Lin, W.-H., Zhu, S., Zhu, J.-Y., Sun, Y., Fan, X.-F., et al. (2010). Integration of light-and brassinosteroid-signaling pathways by a GATA transcription factor in arabidopsis. Dev. Cell 19 (6), 872–883. doi: 10.1016/j.devcel.2010.10.023
Luo, H., Xu, H., Chu, C., He, F., Fang, S. (2020). High temperature can change root system architecture and intensify root interactions of plant seedlings. Front. Plant Sci. 11, 160. doi: 10.3389/fpls.2020.00160
Manfield, I. W., Devlin, P. F., Jen, C.-H., Westhead, D. R., Gilmartin, P. M. (2007). Conservation, convergence, and divergence of light-responsive, circadian-regulated, and tissue-specific expression patterns during evolution of the arabidopsis GATA gene family. Plant Physiol. 143 (2), 941–958. doi: 10.1104/pp.106.090761
Molkentin, J. D. (2000). The zinc finger-containing transcription factors GATA-4,-5, and-6: ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 275 (50), 38949–38952. doi: 10.1074/jbc.R000029200
Park, J., Kim, H., Kim, S., Kong, S., Park, J., Kim, S., et al. (2006). A comparative genome-wide analysis of GATA transcription factors in fungi. Genomics Inf. 4 (4), 147–160.
Patient, R. K., McGhee, J. D. (2002). The GATA family (vertebrates and invertebrates). Curr. Opin. Genet. Dev. 12 (4), 416–422. doi: 10.1016/S0959-437X(02)00319-2
Peng, W., Li, W., Song, N., Tang, Z., Liu, J., Wang, Y., et al. (2021). Genome-wide characterization, evolution, and expression profile analysis of GATA transcription factors in brachypodium distachyon. Int. J. Mol. Sci. 22 (4), 2026. doi: 10.3390/ijms22042026
Reyes, J. C., Muro-Pastor, M. I., Florencio, F. J. (2004). The GATA family of transcription factors in arabidopsis and rice. Plant Physiol. 134 (4), 1718–1732. doi: 10.1104/pp.103.037788
Rudić, J., Dragićević, M. B., Momčilović, I., Simonović, A. D., Pantelić, D. (2022). In silico study of superoxide dismutase gene family in potato and effects of elevated temperature and salicylic acid on gene expression. Antioxidants 11 (3), 488. doi: 10.3390/antiox11030488
Scazzocchio, C. (2000). The fungal GATA factors. Curr. Opin. Microbiol. 3 (2), 126–131. doi: 10.1016/S1369-5274(00)00063-1
Shi, M., Huang, Q., Wang, Y., Wang, C., Zhu, R., Zhang, S., et al. (2022). Genome-wide survey of the GATA gene family in camptothecin-producing plant ophiorrhiza pumila. BMC Genomics 23 (1), 256. doi: 10.1186/s12864-022-08484-x
Shikata, M., Matsuda, Y., Ando, K., Nishii, A., Takemura, M., Yokota, A., et al. (2004). Characterization of arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J. Exp. Bot. 55 (397), 631–639. doi: 10.1093/jxb/erh078
Si, H.-J., Xie, C.-H., Liu, J. (2003). An efficient protocol for agrobacterium-mediated transformation with microtuber and the introduction of an antisense class I patatin gene into potato. Acta Agronomica Sin. 29 (6), 801–805.
Sparkes, I. A., Runions, J., Kearns, A., Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1 (4), 2019–2025. doi: 10.1038/nprot.2006.286
Sugimoto, K., Takeda, S., Hirochika, H. (2003). Transcriptional activation mediated by binding of a plant GATA-type zinc finger protein AGP1 to the AG-motif (AGATCCAA) of the wound-inducible myb gene NtMyb2. Plant J. 36 (4), 550–564. doi: 10.1046/j.1365-313X.2003.01899.x
Teakle, G. R., Manfield, I. W., Graham, J. F., Gilmartin, P. M. (2002). Arabidopsis thaliana GATA factors: organisation, expression and DNA-binding characteristics. Plant Mol. Biol. 50, 43–56. doi: 10.1023/A:1016062325584
Trainor, C. D., Omichinski, J. G., Vandergon, T. L., Gronenborn, A. M., Clore, G. M., Felsenfeld, G. (1996). A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high-affinity interaction. Mol. Cell Biol. 16 (5), 2238–2247. doi: 10.1128/MCB.16.5.2238
Tricker, P. J., Trewin, H., Kull, O., Clarkson, G. J. J., Eensalu, E., Tallis, M. J., et al. (2005). Stomatal conductance and not stomatal density determines the long-term reduction in leaf transpiration of poplar in elevated CO 2. Oecologia 143, 652–660. doi: 10.1007/s00442-005-0025-4
Wang, Y., Cao, X., Zhang, D., Li, Y., Wang, Q., Ma, F., et al. (2023). SlGATA17, a tomato GATA protein, interacts with SlHY5 to modulate salinity tolerance and germination. Environ. Exp. Bot. 206, 105191. doi: 10.1016/j.envexpbot.2022.105191
Weiss, M. J., Yu, C., Orkin, S. H. (1997). Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol. Cell Biol. 17 (3), 1642–1651. doi: 10.1128/MCB.17.3.1642
Yu, C., Li, N., Yin, Y., Wang, F., Gao, S., Jiao, C., et al. (2021). Genome-wide identification and function characterization of GATA transcription factors during development and in response to abiotic stresses and hormone treatments in pepper. J. Appl. Genet. 62, 265–280. doi: 10.1007/s13353-021-00618-3
Yu, R., Chang, Y., Chen, H., Feng, J., Wang, H., Tian, T., et al. (2022). Genome-wide identification of the GATA gene family in potato (Solanum tuberosum l.) and expression analysis. J. Plant Biochem. Biotechnol. 31 (1), 37–48. doi: 10.1007/s13562-021-00652-6
Yuan, Q., Zhang, C., Zhao, T., Meini Yao, M., Xu, X. (2018). A genome-wide analysis of GATA transcription factor family in tomato and analysis of expression patterns. Int. J. Agric. Biol. 20 (6), 1274–1282.
Zhang, K., Jia, L., Yang, D., Hu, Y., Njogu, M. K., Wang, P., et al. (2021). Genome-wide identification, phylogenetic and expression pattern analysis of GATA family genes in cucumber (Cucumis sativus l.). Plants 10 (8), 1626. doi: 10.3390/plants10081626
Zhao, Y., Medrano, L., Ohashi, K., Fletcher, J. C., Yu, H., Sakai, H., et al. (2004). HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in arabidopsis. Plant Cell 16 (10), 2586–2600. doi: 10.1105/tpc.104.024869
Zhu, W., Guo, Y., Chen, Y., Wu, D., Jiang, L. (2020). Genome-wide identification, phylogenetic and expression pattern analysis of GATA family genes in brassica napus. BMC Plant Biol. 20 (1), 543. doi: 10.1186/s12870-020-02752-2
Keywords: potato, heat stress, StGATA2, morphological phenotypes, photosynthesis, transpiration
Citation: Zhu X, Duan H, Jin H, Chen S, Chen Z, Shao S, Tang J and Zhang Y (2023) Heat responsive gene StGATA2 functions in plant growth, photosynthesis and antioxidant defense under heat stress conditions. Front. Plant Sci. 14:1227526. doi: 10.3389/fpls.2023.1227526
Received: 23 May 2023; Accepted: 22 June 2023;
Published: 11 July 2023.
Edited by:
Zemin Wang, Gansu Agricultural University, ChinaReviewed by:
Changbin Gao, Wuhan Academy of Agricultural Sciences, ChinaLei Kang, Hunan Agricultural University, China
Copyright © 2023 Zhu, Duan, Jin, Chen, Chen, Shao, Tang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xi Zhu, zhuxi@catas.cn; Yu Zhang, zhangyu@catas.cn
 Xi Zhu1,2,3*
Xi Zhu1,2,3* Yu Zhang
Yu Zhang