- 1Horticulture Branch, Heilongjiang Academy of Agricultural Sciences, Harbin, Heilongjiang, China
- 2Key Laboratory of Biology and Genetic Improvement of Horticultural Crops (Northeast Region), Ministry of Agriculture and Rural Affairs/National-Local Joint Engineering Research Center for Development and Utilization of Small Fruits in Cold Regions/College of Horticulture & Landscape Architecture, Northeast Agricultural University, Harbin, China
Cold and drought stress considerably suppress the development of plants. In this study, a new MYB (v-myb avian myeloblastosis viral)TF gene, MbMYBC1, was isolated from the M. baccata and located in nucleus. MbMYBC1 has a positive response to low temperature and drought stress. After being introduced into Arabidopsis thaliana, the physiological indicators of transgenic Arabidopsis had corresponding changes under these two stresses, the activities of catalase (CAT), peroxidase (POD) and superoxide dismutase (SOD) increased, electrolyte leakage rate (EL) and the content of proline increased, but the content of chlorophyll decreased. In addition, its overexpression can also activate the downstream expression of AtDREB1A, AtCOR15a, AtERD10B and AtCOR47 related to cold stress and AtSnRK2.4, AtRD29A, AtSOD1and AtP5CS1 related to drought stress. Based on these results, we speculate that MbMYBC1 can respond to cold and hydropenia signals, and can be used in transgenic technology to improve plant tolerance to low temperature and drought stress.
Introduction
Various uncertain factors in the natural environment seriously restrict the development of apple industry and cause serious damage to it, especially low temperature, drought and osmotic stress (Chinnusamy et al., 2006; Liu et al., 2023). When sensing external stress signals, plants will undergo a variety of changes, including physiological and biochemical levels, to reduce their own damage (Han et al., 2018a). Therefore, according to these changes, the damage degree of plants can be roughly known. Stress destroys the integrity of organelles and enhances the permeability of membranes, the more serious the damage of cell membranes, the greater the electrolyte leakage rate (EL) (Blum, 1983). It will also accelerate the degradation of chlorophyll and reduce the photosynthetic rate. During the normal growth of plants, the content of ROS in cells is maintained in a normal state, while stress will destroy the balance of its production and removal (Nandhini et al., 2022). Too much ROS is easy to cause oxidative stress if it is not cleared in time. Superoxide dismutase (SOD), peroxidase (POD) and catalase (CAT) can effectively remove ROS, and their activities can be used to reflect the antioxidant level of plants (Dogru and Çakirlar, 2020; Liang et al., 2022a). Plants can resist the damage of organelles, proteins and cell membranes caused by external stimuli by regulating the accumulation of osmoregulation substances (Gerona et al., 2019; Yao et al., 2022a). In addition, the phenotype of plants will also change significantly.
In order to survive under various environmental conditions, plants have formed a series of stress response mechanisms. It has been proved that transcription factor (TF) can effectively help plants resist adversity (Jiang and Deyholos, 2009; Han et al., 2020a). According to the number of domain repeats, MYB TFs are divided into four subfamilies: 1R-MYB, 2R-MYB, 3R-MYB and 4R-MYB (Li W. et al., 2022). Among them, 2R-MYB TF is the most common in plants and is characterized in many plants (Li et al., 2015). For example, there are 126, 108, 157 and 222 2R-MYB genes in Arabidopsis (Li et al., 2016), grape (Matus et al., 2008), corn (Du et al., 2012) and apple (Cao et al., 2013) respectively. MYB is not only widely involved in various metabolic activities, material synthesis and other biological processes of plants, but also can protect plants from abiotic stresses such as high salt, low temperature, heat and drought (Lin et al., 2021). Studies have shown that soybean GmMYB118 can make plants more adaptable to drought and salinity by regulating osmotic pressure and oxidant and inducing the expression of stress-related genes (Du et al., 2018). In tomato, the overexpression of SlMYB102 can enhance the resistance of plants to cold, and may also participate in CBF and proline synthesis pathway to further improve the adaptability of tomato to stress (Wang et al., 2020).
The process of plant response to stress often involves the participation of multiple genes through multiple channels. The process of plant resistance is generally divided into two steps, first sensing the stress signal and then transmitting it; second, activating the expression of various related genes. CBF/DREB is rapidly induced by low temperature in the cold response pathway and combines with the C-repeat/dehydration response region in the cold response (COR) gene promoter to improve cold resistance (Yamaguchi-Shinozaki and Shinozaki, 1994; Li X. et al., 2022). Drought stress response protein can improve the viability of plants under water shortage conditions through ABA-dependent and ABA-independent pathways (Wassie et al., 2023). Studies have proved that there are ABA response elements in promoters of many genes, such as RAB18, RD22, RD29 (Kazuko and Kazuo, 1993; Latchman, 1997). The expression of proline biosynthesis gene P5CS1 will increase under drought stress, its transcription level can reflect the proline accumulation under drought stress. Moreover, P5CS1 regulation does not depend on ABA signal transduction pathway (Furlan et al., 2020).
Malus baccata (L.) Borkh (M. baccata) is a commonly used grafting rootstock in apple cultivation (Ren et al., 2017), and the cold tolerance and drought tolerance of rootstock have a direct impact on the environmental adaptability of apple, so the study on the stress tolerance of rootstock is of great significance in the process of apple breeding. However, there are few studies on MYB in M. baccata. In this study, we cloned and identified the expression of MbMYBC1 and stress related genes in A. thaliana, and preliminarily clarified the cold and drought resistance mechanism of mountain stator. It has promoted the development of the breeding of M. baccata stress resistant varieties as rootstocks, laid a foundation for the research on the molecular mechanism of cold and drought resistance of apples, and is conducive to the selection of Malus plants breeding genes.
Materials and methods
Cultivation and treatment of M. baccata
M. baccata tissue culture plantlets was put on Murashige and Skoog (MS) medium containing 0.55 mg/L cytokinin (6-BA) and 0.6 mg/L indole butyric acid (IBA) for rapid propagation. After one month, selected robust tissue culture plantlets and transfered them to rooting medium (1.2 mg/L IBA) (Yao et al., 2022b). After rooting, seedlings were put into Hoagland hydroponic culture solution for growth. The room temperature and humidity of tissue culture were maintained at about 25°C and 80%. Changed the hydroponic culture solution regularly. When 7-8 fully developed mature leaves and new strong roots grow out of the hydroponic seedlings, seedlings with roughly the same growth conditions were selected and grouped, 10 seedlings in each group, a total of 5 groups, one of which was the control group without any treatment. The other 4 groups were treated as follows respectively, and the hydroponic seedlings were placed in the 4°C tissue culture room for low temperature stress; The Hoagland hydroponic culture solution with 200 mM NaCl was used for salt stress; Hoagland hydroponic medium containing 20% PEG6000 was used for water stress; The hydroponic seedlings were put into the tissue culture room at 37°C for high temperature stress. The young leaves, mature leaves, roots and stems of all seedlings were sampled after treatment for 0, 1, 3, 6, 9 and 12 h, and stored at -80°C after liquid nitrogen quick freezing (Han et al., 2018b).
Cloning and qRT-PCR expression analysis of MbMYBC1
The total RNA was extracted from the roots, stems and leaves (young leaves and mature leaves) of M. baccata seedlings with EasyPure plant RNA kit (TransGen Biotech, Beijing, China). Then used Trans Script® First-Strand cDNA Synthesis Super Mix (transgen, Beijing) to synthesize the first strand of cDNA. According to the CDs region of MbMYBC1, two pairs of specific primers were designed with Primer 5.0 software. After synthesizing the primers, MbMYBC1-F and MbMYBC1-R were obtained (Supplementary Table S1). The full length of the target gene was obtained by PCR using the first strand cDNA of apple as the template. After detection and purification by agarose gel electrophoresis, the target gene was linked to the cloning vector using pEASY-T1 cloning kit (TransGen Biotech, Beijing, China) and sequenced (BGI, Beijing) (Han et al., 2020a).
To analyze the expression of MbMYBC1, it was detected by qRT-PCR. MbActin was used as a control, which can be stably expressed without being affected by conditions (Modesto et al., 2013), and the gene is amplified from M. baccata tissues. The primers Actin-F and Actin-R were designed according to the sequences published in the GenBank database (Supplementary Table S1). From the partial sequences obtained in this study, MbMYBC1 primer for qRT-PCR was designed, MbMYBC1-qF/qR (Supplementary Table S1). The qPCR reaction system was shown in Supplementary Table S2, and its reaction process was as follows: pre denaturation at 95°C for 5 min, denaturation for 5 s, annealing at 60°C for 1 min, and extension at 72°C for 1 min. Carried out 35 cycles and continued to extend at 72°C for 5 min (Han et al., 2013; Han et al., 2015; Assal and Lin, 2021). The 2−ΔΔCT method was used to analyze the relative transcriptional level data of the target gene (Li Y. et al., 2022).
Bioinformatics analysis of the MbMYBC1
On ExPASy website (https://web.expasy.org/protparam/), the primary structure and various physical and chemical properties of the target protein were predicted (Han et al., 2020b). The domain and tertiary structure of MbMYBC1 protein were predicted on SMART and SWISS-MODEL websites respectively. The sequences of MbMYBC1 were blasted in NCBI database, MYB sequences of several other species with high sequence similarity was select. Compared these sequences with DNAMAN 5.2, and then constructed phylogenetic tree through MEGA7 neighbor connection method. These amino acid sequences were PbMYBC1 (Pyrus bretschneideri, XP_048430460.1), MaMYBC1 (Mercurialis annua, XP_050229005.1), CiMYBC1 (Carya illinoinensis, XP_042940585.1), JcMYBC1, (Jatropha curcas, XP_012088642.1), JrMYBC1 (Juglans regia, XP_035544691.1), CsMYBC1 (Cannabis sativa, XP_021654469.1), MsMYBC1 (Malus sylvestris, XP_050115747.1), EgMYBC1(Eucalyptus grandis, XP_010036386.2), PaMYBC1 (Prunus avium, XP_021808479.1), AtMYBC1 (Arabidopsis thaliana, AT2G40970).
Subcellular localization analysis of the MbMYBC1 protein
The enzyme digestion sites of BamHI and SalI were selected on pSAT6-GFP-N1 vector, and MbMYBC1-ORF was cloned between these two sites to obtain MbMYBC1-GFP fusion protein. The modified red shift green fluorescent protein (GFP) was contained between these two sites. The MbMYBC1-GFP plasmid was injected into the tobacco epidermal cells from the lower epidermis of the leaves using the Agrobacterium tumefaciens injection method, and the empty 35S:: GFP plasmid as the control was also transferred into the leaves (Su et al., 2022). The expression of MbMYBC1-GFP was observed under confocal microscope to determine its location.
Acquisition of transgenic A. thaliana
The 5’ and 3’ ends of MbMYBC1 were linked to the restriction sites of BamHI and SalI by PCR to construct the overexpression vector of Arabidopsis transformation. BamHI and SalI digested PCAM3011 and PCR products and replaced them with GUS gene to connect them. PCAMBIA2300-MbMYBC1 was transferred into GV3101, and then the MbMYBC1 gene was introduced into Colombian ecotype A. thaliana Co1-0 through inflorescence mediation. After screening by 1/2 MS medium containing 50 mg/L kanamycin, the successfully transformed T3 generation A. thaliana was used for subsequent research.
Stress treatment and physiological index determination of transgenic A. thaliana
All A. thaliana (WT, UL, L1, L4, L5) seeds were sowed on the culture medium, and were transferred to a flowerpot containing the same amount of vermiculite and nutrient soil 15 d later, with a tray at the bottom of the flowerpot. When A. thaliana had 8-12 new leaves, they were divided into 3 groups: one group grew normally, one group was treated at -4°C for 12 h, and the other group were stopped watering within 7 d. The growth of plants was resumed for 3 d under normal conditions after stress treatment. The surface changes of each line were observed and the changes of its survival rate and related physiological and biochemical indexes were measured. Three biological replicates and three technical replicates were performed for each sample.
The EL of the sample was determined according to the method of Campos et al. (2003), determination of chlorophyll content by grinding centrifugation with 95% ethanol (Wang et al., 2022). The content of proline in samples was determined by standard curve method according to the description of Ou et al. (2015). The activity of SOD, CAT and POD was determined with the kit of Nanjing Jiancheng Bioengineering Research Institute (Nanjing, China).
Analysis of MbMYBC1 downstream gene expression
The mRNA of A. thaliana grown under 3 conditions was extracted and reverse transcribed to obtain the first strand of cDNA which was used as a template. Using Actin as internal reference gene, several important downstream stress related genes of MYB were detected by qPCR. These genes were AtDREB1A (Lu et al., 2017), AtCOR15a (Steponkus et al., 1998), AtERD10B (Du and Li, 2019), AtCOR47 (Song et al., 2021), AtSnRK2.4 (Nakashima and Yamaguchi-Shinozaki, 2013), AtRD29A (Wu et al., 2022), AtSOD1 (Li J. et al., 2022) and AtP5CS1 (Chen et al., 2015). The reaction procedure and system of qPCR were the same as above methods.
Statistical analysis
SPSS 21.0 software (IBM, Chicago, Illinois, USA) was used for one-way analysis of variables. All test data were the average values obtained after 3 repetitions, and their standard errors (± SE) were calculated. The statistical difference was referred to as significant * p ≤ 0.05 and **p ≤ 0.01.
Results
Bioinformatics analysis of MbMYBC1 gene
The results obtained by ExPASy analysis (Supplementary Figure S1) showed that the open reading frame (ORF) of MbMYBC1 was 948 bp, 315 amino acids (aa) were encoded by MbMYBC1, and the largest proportion of these 18 aa was Pro (13.6%), Ala (10.5%), Ser (8.3%), the theoretical molecular weight (MW) was 34.048 kDa. The aliphatic index of MbMYBC1 protein was 64.22, the theoretical isoelectric point (pI) was 6.34, and the instability index was 62.46. Therefore, it was hydrophilic and unstable.
As shown in Figure 1A, after sequence alignment with 10 other MYBC1 proteins with high homology, it was found that MbMYBC1 protein contained a unique conservative domain of MYB-TF, indicating that it was a MYB protein. The evolutionary tree showed that MbMYBC1 and MsMYBC1 were clustered in the first cluster of the tree, with the closest genetic relationship and the highest homology (Figure 1B). Figure 2A showed the analysis results of the secondary structure of MbMYBC1 protein. It can be seen that it contained 33.9% α-Helix, 6.35% β-coil, 7.3% extended strand and 52.38% random coil (Figure 2B). Its tertiary structure conformed to the predicted secondary structure, which contained HTH region (Figure 2C). These results indicated that MbMYBC1 was a member of 3R-MYB subgroup.
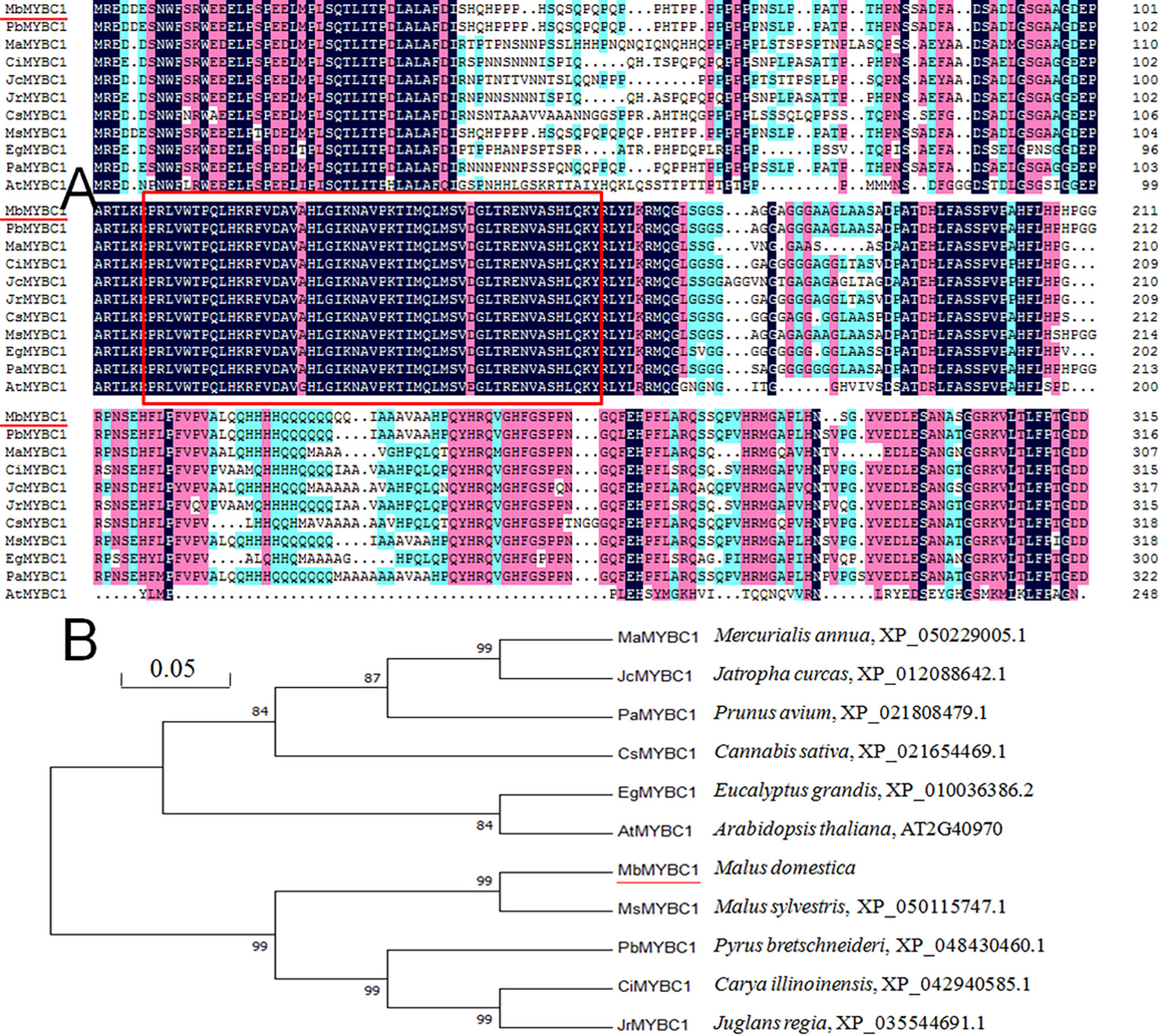
Figure 1 Similarity analysis of amino acid sequences between MbMYBC1 and 10 other MYBs (A) and comparison of genetic relationships (B). The red underline is the target protein, and the part in the box is the MYB structural domain.
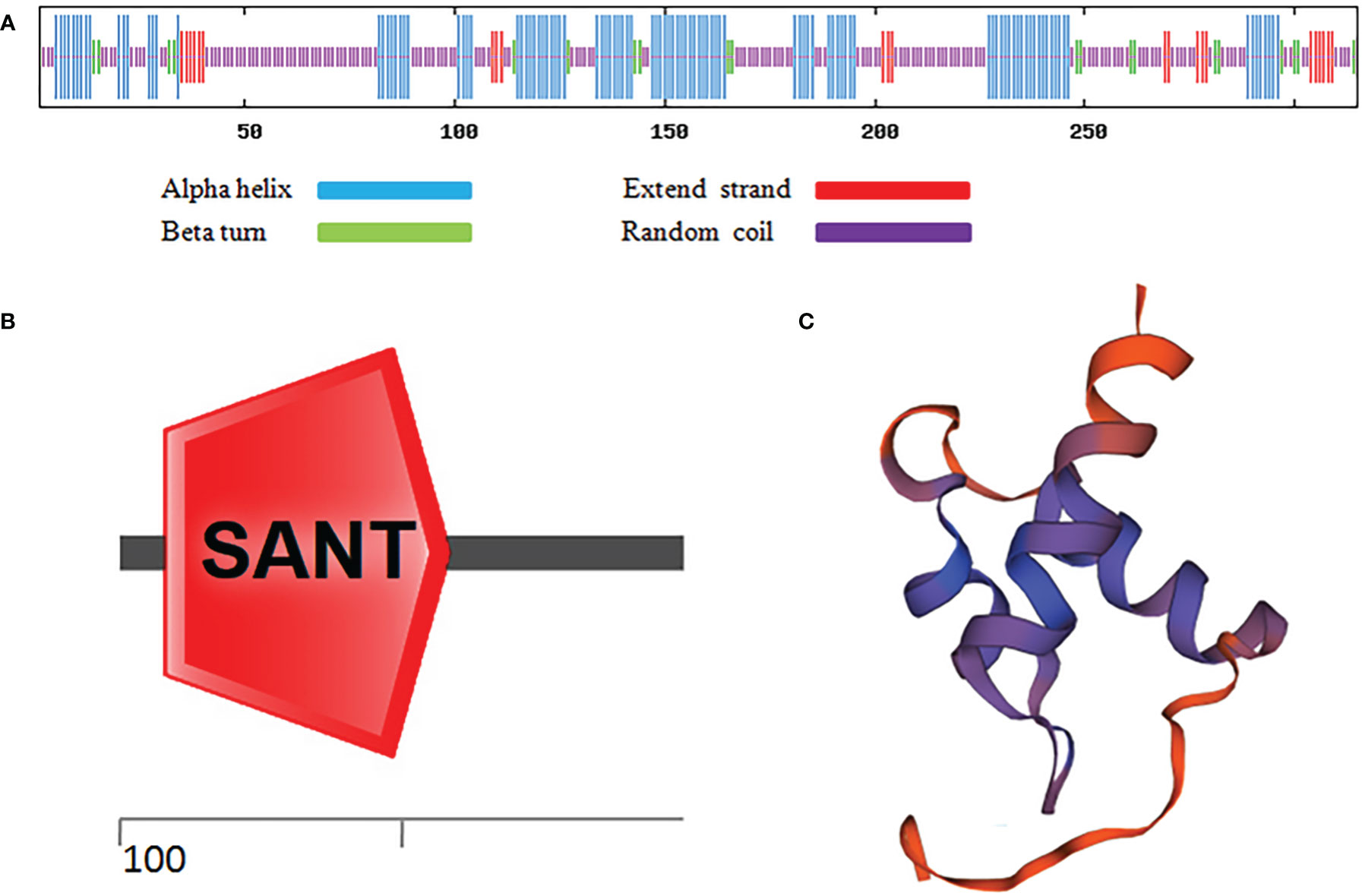
Figure 2 Prediction of Secondary and Tertiary Structure of MbMYBC1 Protein. (A) Predicted protein secondary structure; (B) predicted protein domains; (C) predicted tertiary structure.
Subcellular localization of MbMYBC1 proteins
Figure 3 showed the distribution of 35s::MbMYBC1::GFP under the confocal microscope. The green fluorescence of 35s::GFP protein was distributed in the whole cell (Figure 3B), while the green fluorescence of 35s::MbMYBC1::GFP fusion protein was only distributed in the nucleus (Figure 3D). The nuclear position was finally determined after staining with 4 ‘, 6-diamino-2-phenylindole (DAPI) (Figure 3E). That was to say, it can be preliminarily determined that MbMYBC1 was a nuclear protein.
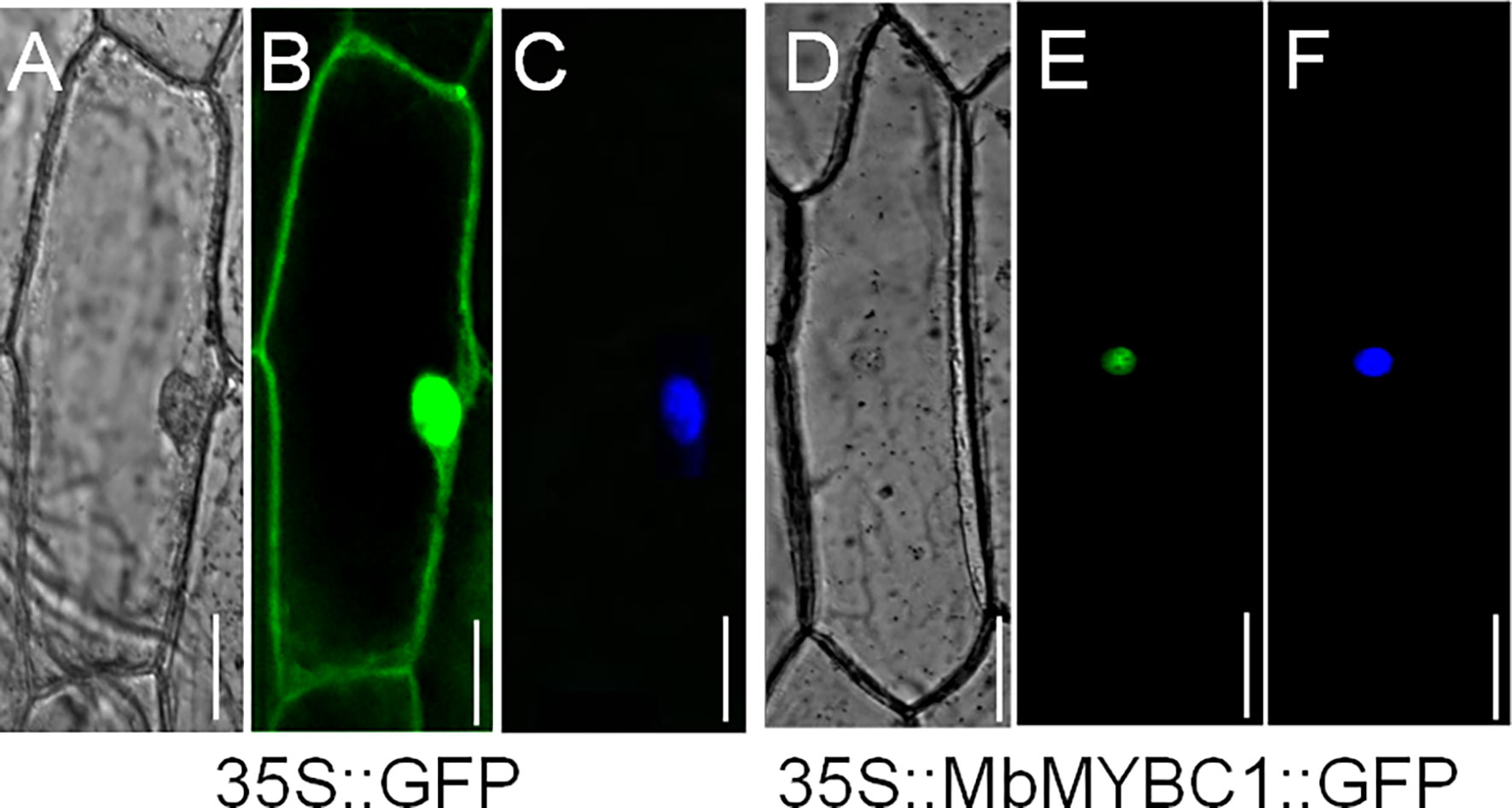
Figure 3 Subcellular localization of MbMYBC1 protein. (A, D) Bright-field images, (B, E) GFP fluorescence, (C, F) the effects after DAPI dyeing. Bar = 50 μm.
Expression level of MbMYBC1 gene
Figure 4 showed the expression of MbMYBC1 in various tissues and the change of expression level in different time periods under various stresses. The results in Figure 4A showed that although MbMYBC1 can be expressed in these organs (root, stem and leaves), there were significant differences in the amount of expression. The expression of MbMYBC1 decreased in turn in young leaves, roots, stems and mature leaves. The expression of MbMYBC1 in new leaves was 6.78 fold higher than that in mature leaves.
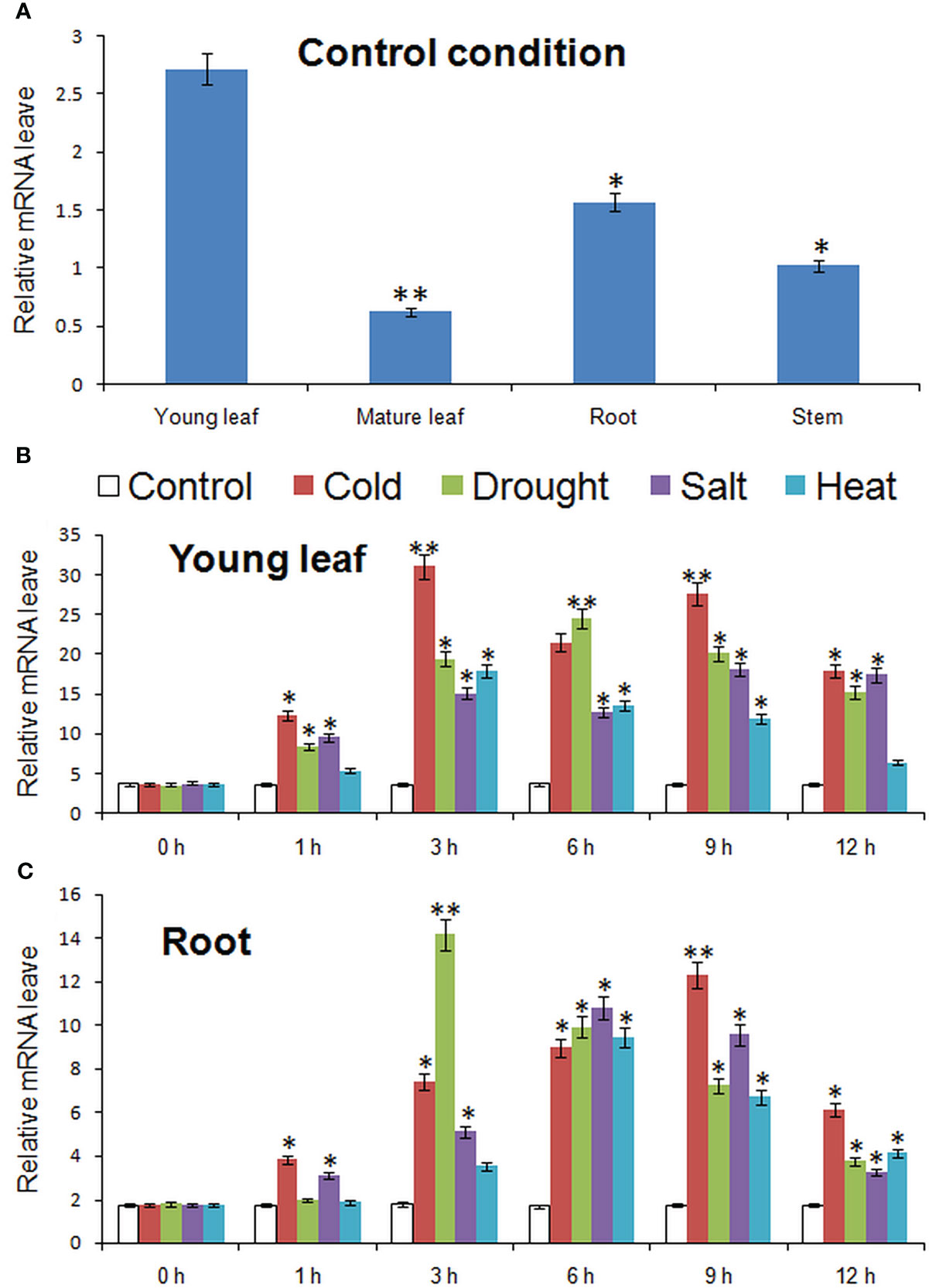
Figure 4 Results of qPCR analysis of MbMYBC1. (A) The expression of MbMYBC1 in different parts of in M. baccata; The expression of MbMYBC1 in new leaves (B) and roots (C) under different stresses. Compared with the Control, the asterisks above the column indicate significant difference and extremely significant difference (*, p ≤0.05; **, p ≤ 0.01).
The expression of MbMYBC1 in young leaves and roots increased first and then decreased with the stress time. Figure 4B showed this result, in young leaves, the stress that made the expression of MbMYBC1 gene reached the peak first was the low and high temperature conditions (3h), and the expression levels under these two conditions were 8.41, 5.89 fold higher than those under normal conditions respectively. Under salt and drought stress, the expression level at 9 and 6 hours was the highest, respectively, which was 4.79 and 6.82 fold of those of the control. But in the root, the peak of expression was reached first under drought (3h), 7.88 times of that of untreated. Under high salt and high temperature conditions, the expression level was the highest at 6 h, 6.22 and 5.34 fold higher than that of untreated ones respectively, and reached the peak at 9 h under low temperature, 7.03 fold higher than that of the control. In addition, from qRT-PCR results, MbMYBC1 was more sensitive to cold and drought stimuli.
Increased tolerance of transgenic A. thaliana to low temperature stress
After cold and drought stress treatment on Arabidopsis, the qRT-PCR results of MbMYBC1 in each line were shown in Figure 5A, in which WT and UL lines were the controls. In the identified 6 T2 generation transformation lines (L1, L2, L3, L4, L5, L6), MbMYBC1 gene had a higher expression level in L1, L4, L5. Therefore, these 3 lines were used to cultivate T3 generation A. thaliana.
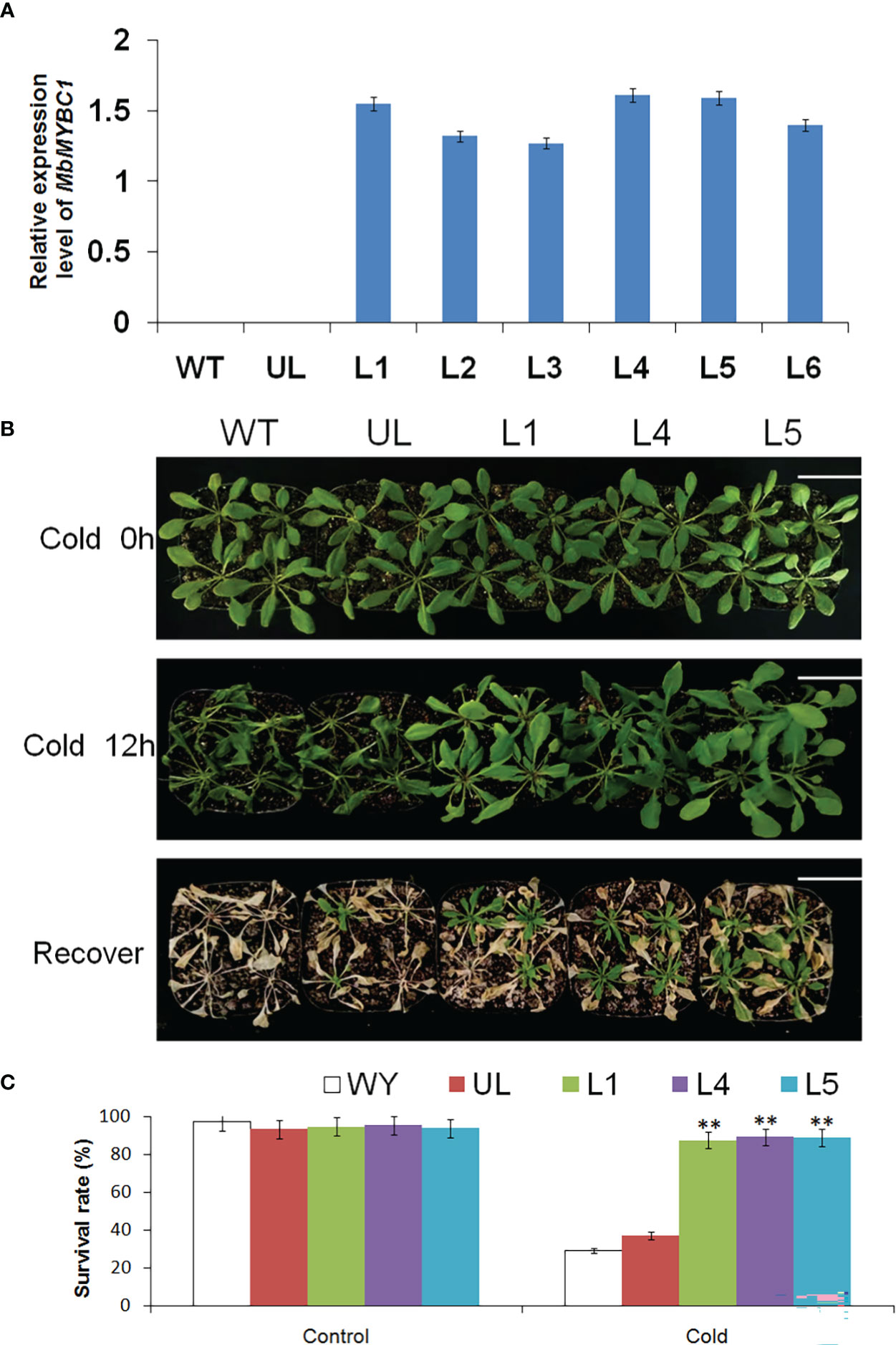
Figure 5 Phenotype and survival rate of A. thaliana under low temperature stress. (A) The relative expression of MbMYBC1 in WT, UL and transgenic A. thaliana. (B) The phenotypes of WT, UL, L1/4/5 lines after treatment at -4°C for 0 h, 12 h and recovery growth. (C) The survival rate of A. thaliana under control and low temperature stress. Bar = 5 cm. (**, p ≤ 0.01).
Figures 5B, C showed the phenotypic changes and survival of A. thaliana after low temperature stress. Without treatment, all Arabidopsis grew well, showing a healthy state. The temperature of its growth environment was set at -4°C. After 12 h of growth, the leaves of WT and UL lines became soft and shrunk, and the plants were short. The transgenic lines suffered less damage, and the changes were not very obvious. After the normal growth temperature was restored, WT and UL almost all dead, and the survival rate was only 29.3, 37.2. However, some transgenic lines still survived, with the survival rates of 87.5, 89.1 and 88.8 respectively. These results indicated that MbMYBC1 played a significant role in improving the cold resistance of A. thaliana.
Without treatment, the related physiological and biochemical indexes of all plants were roughly in the same level. After low temperature stress, there were obvious changes, which can be seen from Figure 6. The EL, content of proline and the activities of SOD, CAT and POD were increased in all Arabidopsis, but the increase was greater in L1/4/5, indicating that the degree of cytoplasmic membrane peroxidation in transgenic lines was relatively low. Chlorophyll content of all lines decreased, but the decrease was greater in WT and UL lines, indicating that they suffered more severe stress and chloroplast structure was severely damaged. It can be inferred that MbMYBC1 can effectively improve the cold tolerance of plants.
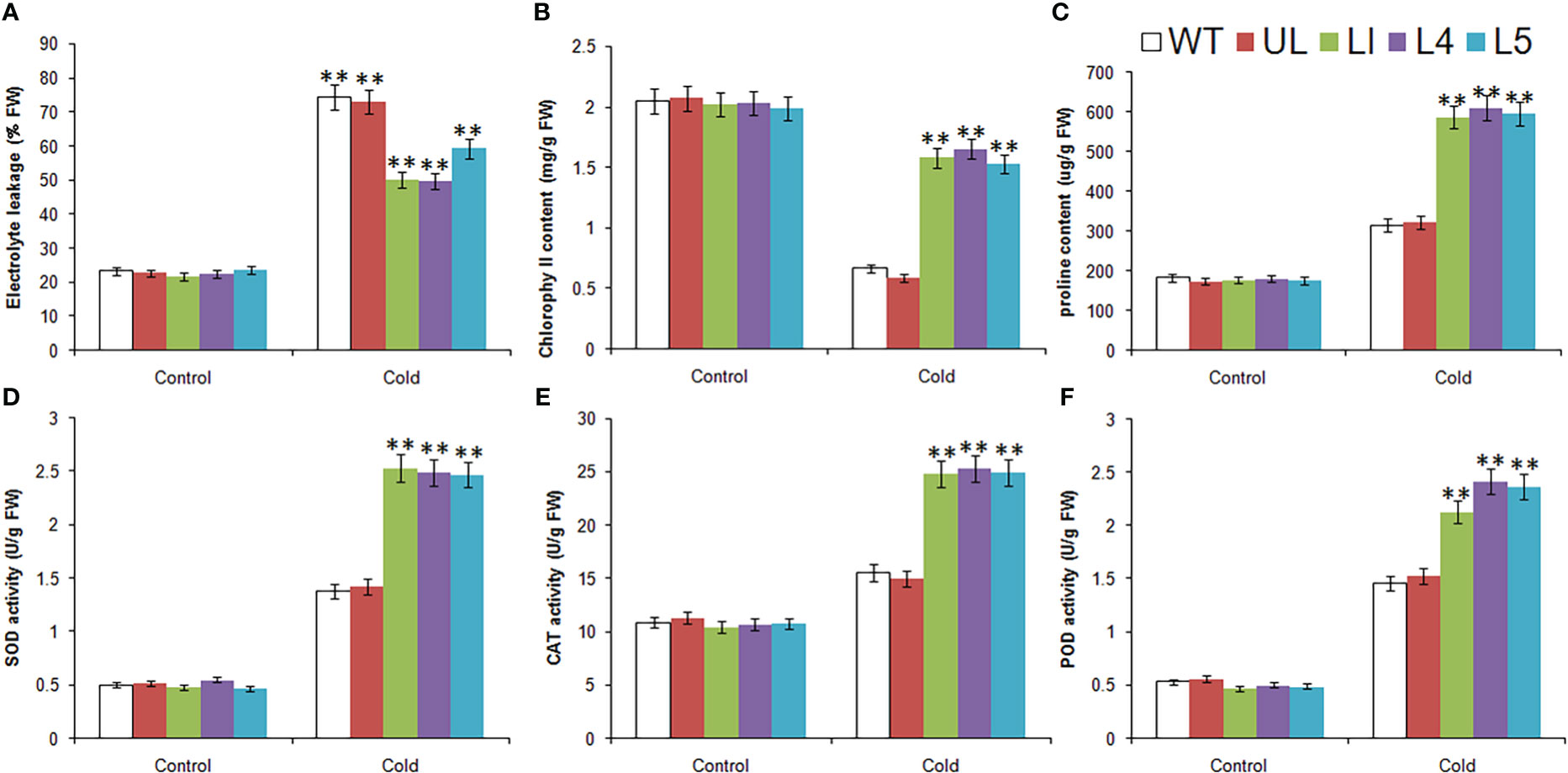
Figure 6 Physiological and biochemical indicatorsin MbMYBC1-OE A. thaliana. (A) EL, (B) Chlorophyll content, (C) proline content, (D) SOD activities, (E) CAT activities, and (F) POD activities. (**, P ≤0.01).The control was the index in the WT. All data were the average of 3 measurements.
Expression analysis of cold resistant downstream genes in MbMYBC1-OE A. thaliana
The expression of four cold stress related target genes downstream of MbMYBC1 was detected by qRT-PCR. The results showed that in Figure 7, these 4 genes were AtDREB1A, AtCOR15a, AtERD10B and AtCOR47. Without -4°C treatment, the expression levels of these genes in all Arabidopsis were close and very low. After cold stress, their expression increased significantly, especially in MbMYBC1-OE A. thaliana, which was much higher than that in UL and WT lines. Therefore, MbMYBC1 may improve the adaptability of plants to low temperature through positive regulation of downstream related target genes.
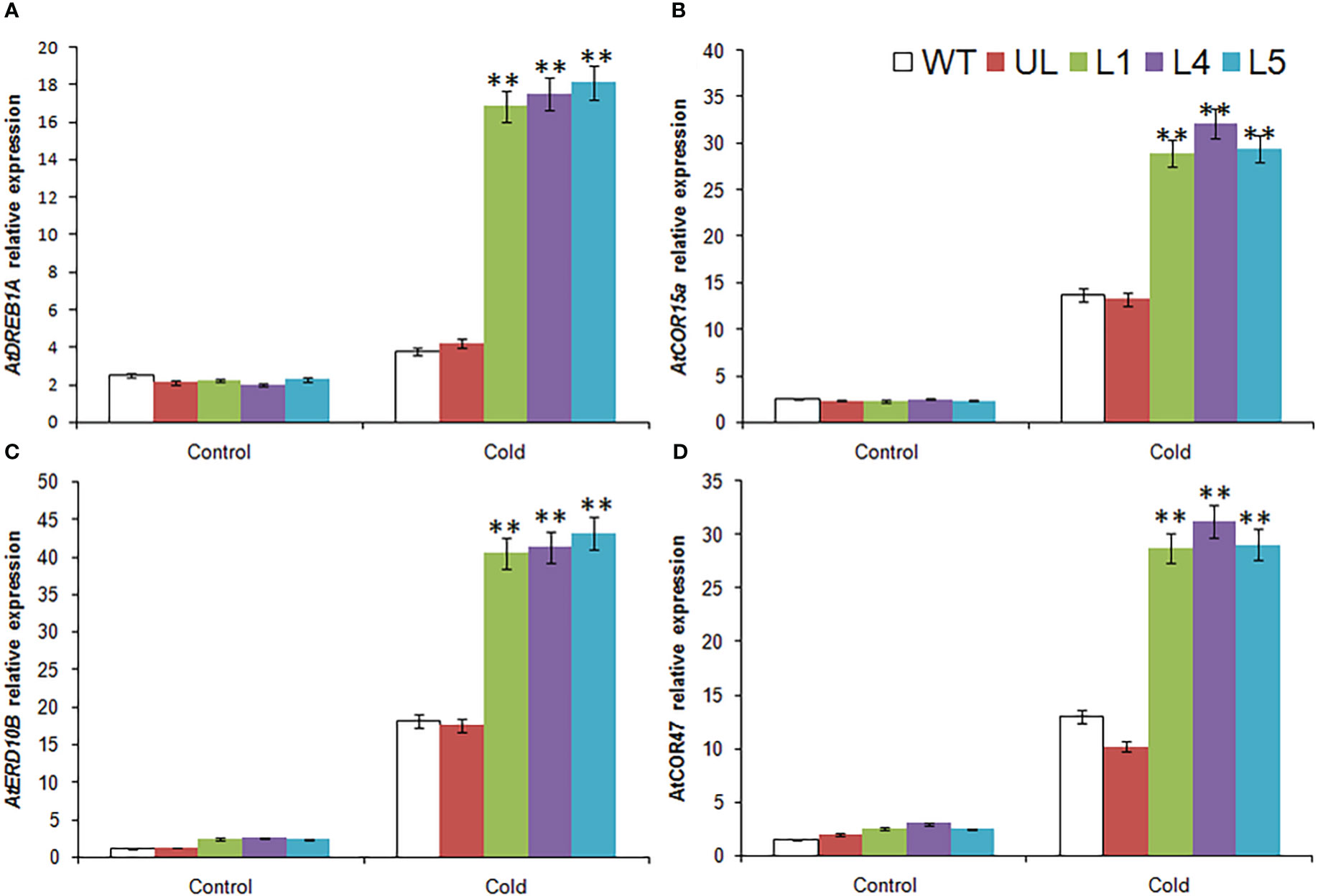
Figure 7 Expression of 4 cold stress related genes in A. thaliana. The expression leave of (A) AtDREB1A, (B) AtCOR15a, (C) AtERD10B and (D) AtCOR47. (**, P ≤0.01). The control was the index in the WT. All data were the average of 3 measurements.
Increased tolerance of transgenic A. thaliana to drought stress
Figure 8 showed the phenotype changes and survival rate of Arabidopsis after drought stress. When growing in a good environment, the appearance of all plants was basically the same, and the leaves were plump. After 7 days of stopping watering, the phenotypes of WT and UL lines had changed significantly, the leaves became smaller and curly, while the changes of transgenic plants were not very obvious. After 3 days of normal watering, only 34.5% and 30.2% of WT and UL survived, while the survival rates of L1, L4 and L5 were 90.7%, 86.4% and 87.4% respectively.
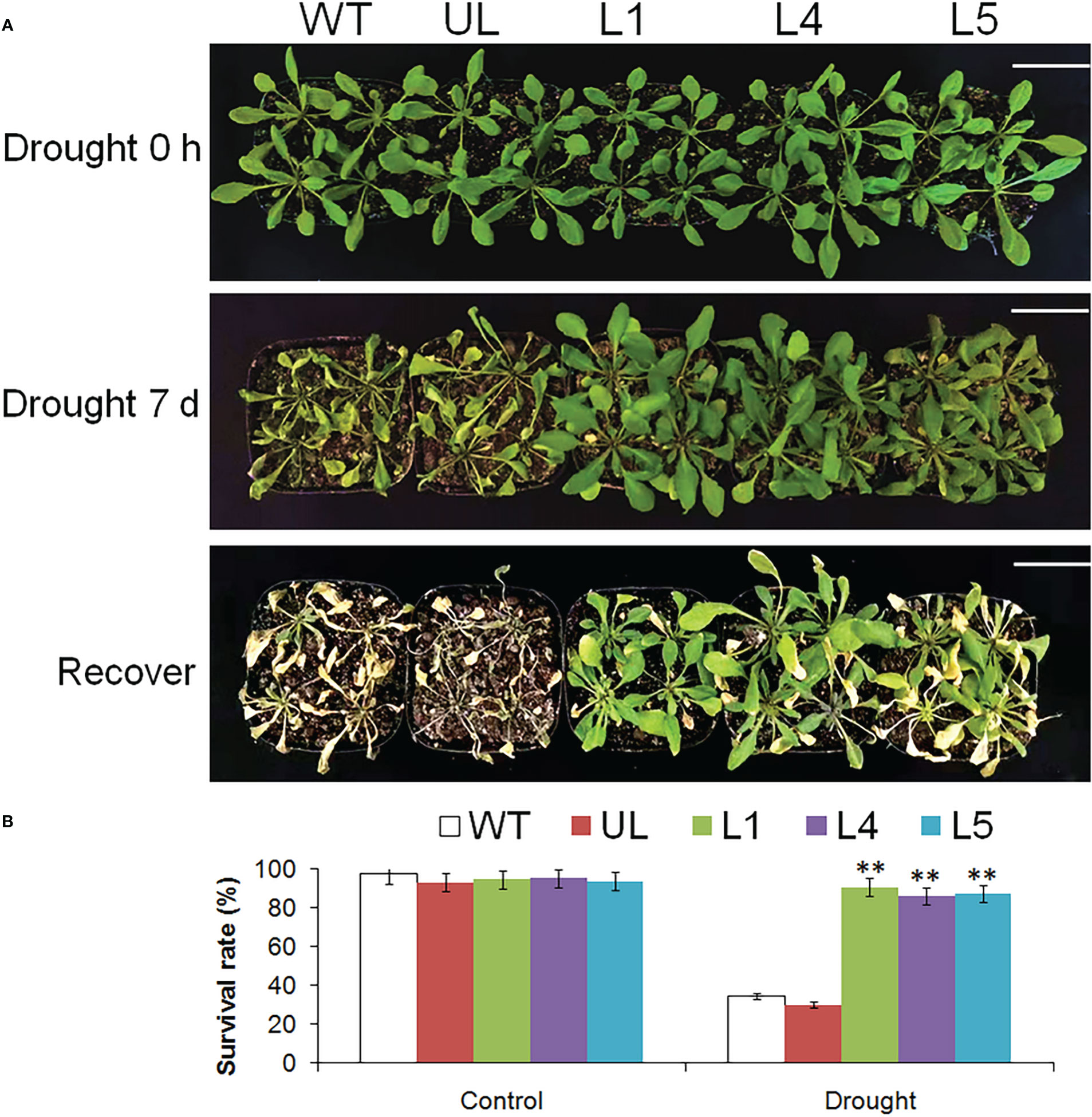
Figure 8 Phenotype and survival rate of A. thaliana. (A) The phenotypes of WT, UL, L1/4/5 lines were observed after 0 and 7 d of stopping watering and recovering growth. (B) The survival rate of A. thaliana under control and drought stress. Bar= 5 cm. (**, p ≤ 0.01).
Under the control condition, the determination results of relevant physiological and biochemical indexes of WT, UL and L1/4/5 lines were basically the same (Figure 9). After drought stress, except for the reduction of chlorophyll content, EL, proline content, SOD, CAT and POD activities were higher than those in the control group, and these indicators were significantly higher in transgenic lines than in WT and UL lines. As for chlorophyll content, it was also higher in transgenic plants, it can be preliminarily inferred that overexpression of MbMYBC1 is helpful to improve the survival rate of plants in arid environments.
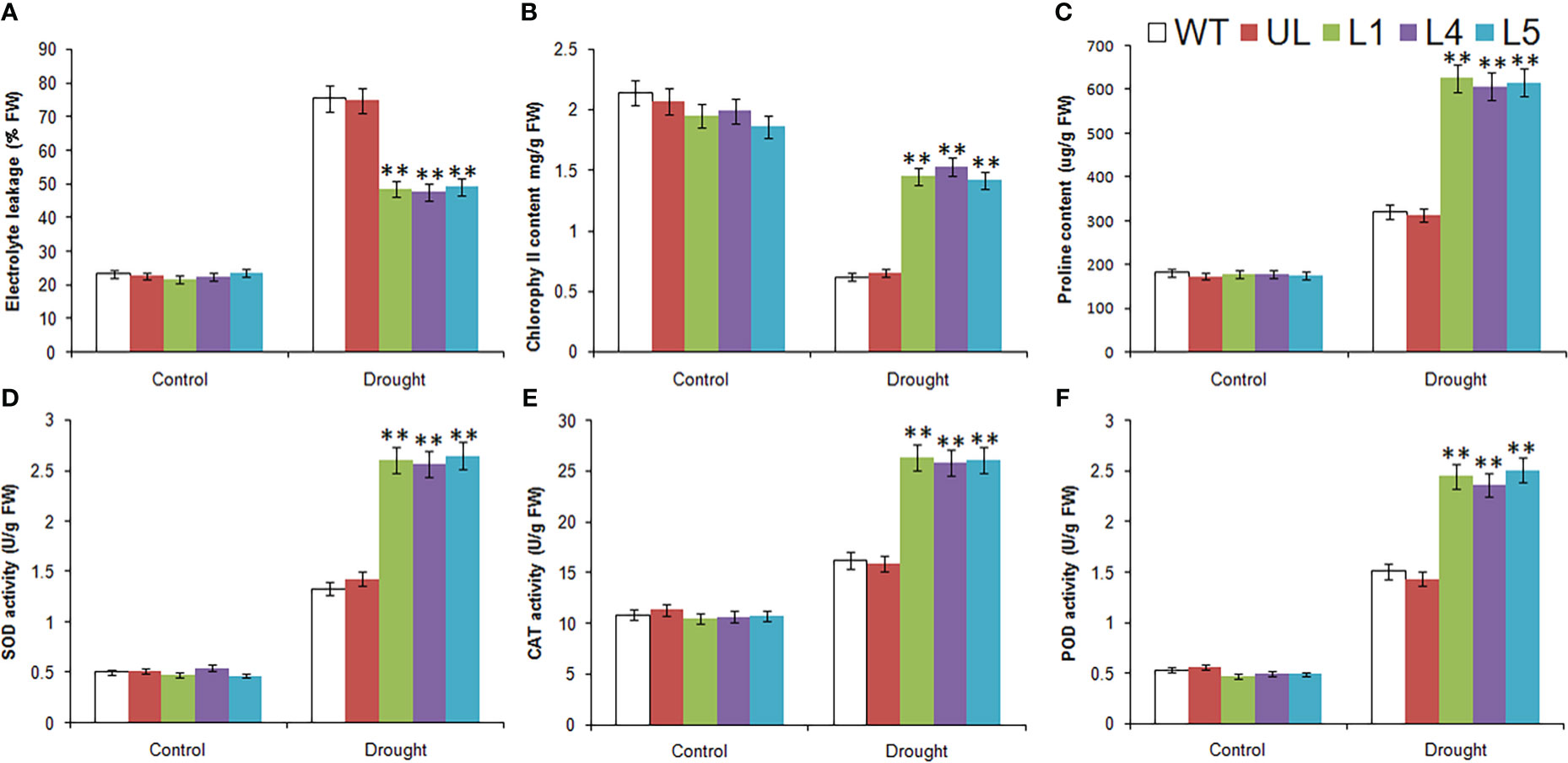
Figure 9 Physiological and biochemical indicatorsin MbMYBC1-OE A. thaliana under drought stress. (A) EL, (B) Chlorophyll content, (C) proline content, (D) SOD activities, (E) CAT activities, and (F) POD activities. (**, P ≤0.01). The control was the index in the WT. All data were the average of 3 measurements.
Expression analysis of drought resistant downstream genes in MbMYBC1-OE A. thaliana
In order to further explore the molecular mechanism of plant drought resistance, the expression levels of several related downstream target genes were detected by qPCR in this study. The results were shown in Figure 10. These 4 genes were AtSnRK2.4, AtRD29A, AtSOD1 and AtP5CS1, respectively. Before watering was stopped, the expression levels of these 4 genes in all plants were at the same low level. After water shortage treatment, their expression levels in all lines were improved to varying degrees, especially in transgenic lines (L1, L4, L5), although they were also increased in WT and UL lines, but the expression level was much lower than that in L1, L4 and L5.
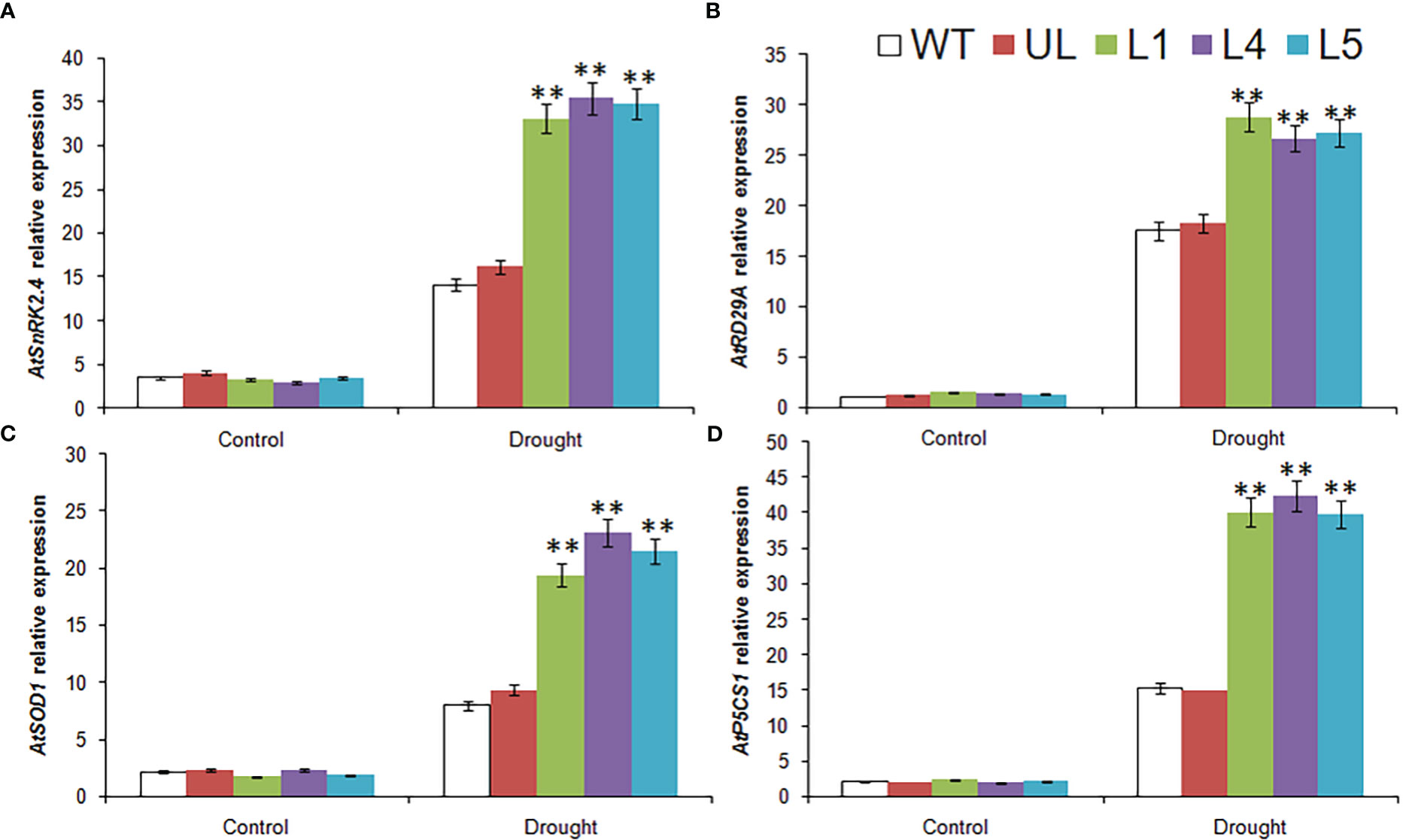
Figure 10 Expression of 4drought stress related genes in A. thaliana. The expression leave of (A) AtSnRK2.4, (B) AtRD29A, (C) AtSOD1 and (D) AtP5CS1. (**, P ≤0.01). The control was the index in the WT. All data were the average of 3 measurements.
Discussion
Because of the adverse changes in the environment, the fixed plants will inevitably suffer from various abiotic stresses, which will restrict the yield and geographical distribution of horticultural crops, as well as economic development (Zhu, 2016; Han et al., 2018c; Tripathi et al., 2022). In order to cope with these environmental factors threatening survival, plants have evolved a variety of adaptive mechanisms, including morphological, physiological and molecular aspects (Yan et al., 2014; Xiong et al., 2020; Liang et al., 2022a). TFs can play a regulatory role at the transcriptional level, enabling plants to adapt to external environmental stress by changing their own structure or metabolic process (Serrano et al., 2014; Qin et al., 2015; Zhang et al., 2018). It can also increase or inhibit the expression of downstream genes by combining with cis acting elements of target genes, so as to improve plant stress resistance (Liu et al., 2019; Liang et al., 2022b).
As one of the largest TF families, MYB has been proved to be involved in the response of plants to cold and drought stress. Dossa et al. (2019) found that the overexpression of SiMYB75 can promote the accumulation of ABA in Arabidopsis, and promote the up regulation of drought stress related genes in its downstream through ABA dependent pathway, so as to improve the tolerance of Arabidopsis to drought stress. Studies have shown that GmMYB118 can up regulate the expression of genes related to drought stress through ABA signal pathway to enhance the viability of transgenic Arabidopsis under drought conditions (Du et al., 2018). Overexpression of GmMYBJ1 can change some physiological traits of plants to adapt to low temperature environment, and can activate the expression of downstream AtRD29B, AtCOR47, AtCOR78, AtP5CS and AtCOR15a, which further enhanced the ability of plants to respond to chill environment (Su et al., 2014).
In our experiment, we cloned and separated MbMYBC1 from M. baccata and characterized it. Sequence analysis and cluster analysis of MbMYBC1 and other MYB proteins showed that they may have similar functions and properties, and the highest homology between MbMYBC1 and MsMYBC1 (Figure 1). The predicted domain and tertiary structure of MbMYBC1 protein also conformed to the function of the proposed transcription factor (Figure 2). The subcellular localization results confirmed that it was a nuclear protein, which was the same as other known MYB proteins (Figure 3).
After qPCR detection of MbMYBC1, it was found that it could be expressed in young leaves, mature leaves, root and stem, but the expression amount was different (Figure 4). Its expression level in young leaves and roots was higher, indicating that the expression of MbMYBC1 was tissue specific. After detecting the expression of MbMYBC1 in young leaves and roots under different conditions, it was found that the expression of this gene could be subjected to cold, drought, salt and high temperature stresses, and the expression level changed with time, showing a trend of first rising and then declining. According to this result, we can know that its expression was also stage specific. In addition, we also found that the expression of MbMYBC1 was more susceptible to the induction of low temperature and drought. In view of this phenomenon, we speculated that MbMYBC1 may play a key role in regulating the response of plants to low temperature and drought stress.
In order to explore the role of MbMYBC1 in plant cold and drought resistance, we detected the changes of phenotypic, physiological and biochemical indicators of transgenic Arabidopsis after cold and drought stress. Plant phenotype is closely related to damage degree (Singh et al., 2021). Cell membrane is the protective barrier of cells, and the MDA content can reflect the permeability of cell membrane under stress (Jungklang et al., 2017). The harsh environment will make plant cells produce excessive ROS, which will cause serious damage to the structural and functional integrity of chloroplasts, affect the synthesis of chlorophyll, and change the activity of antioxidant enzymes (Kavian et al., 2022; Han et al., 2023). An obvious fact was that WT and UL plants grew weaker after stress treatment. However, transgenic lines still had a high survival rate (Figures 5, Figure 8). Moreover, EL, content of chlorophyll and proline, activities of SOD, POD and CAT have changed correspondingly in response to stimuli to adapt to cold and drought (Figures 6, Figure 9). This showed that overexpression of MbMYBC1 can reduce plant damage under low temperature and drought stress.
Overexpression of MbMYBC1 also activated the expression of downstream related target genes (Figures 7, 10). Therefore, based on this result and the proven conclusion, we predicted the possible pathway of this gene in response to cold and drought stress, as show in Figure 11. When subjected to low temperature stress, MbMYBC1 first perceives cold signals and activates the expression of downstream cold response genes DREB1A, CCA1, ERD10B and COR47 through CBF pathway, thus endowing plants with cold tolerance. After receiving drought stimulation, MbMYBC1 transmits signals to downstream SnRK2.4, RD29A, SOD1 containing ABA response elements through ABA dependent pathway to promote their expression, and can also directly activate the expression of P5CS1, giving plants drought tolerance.
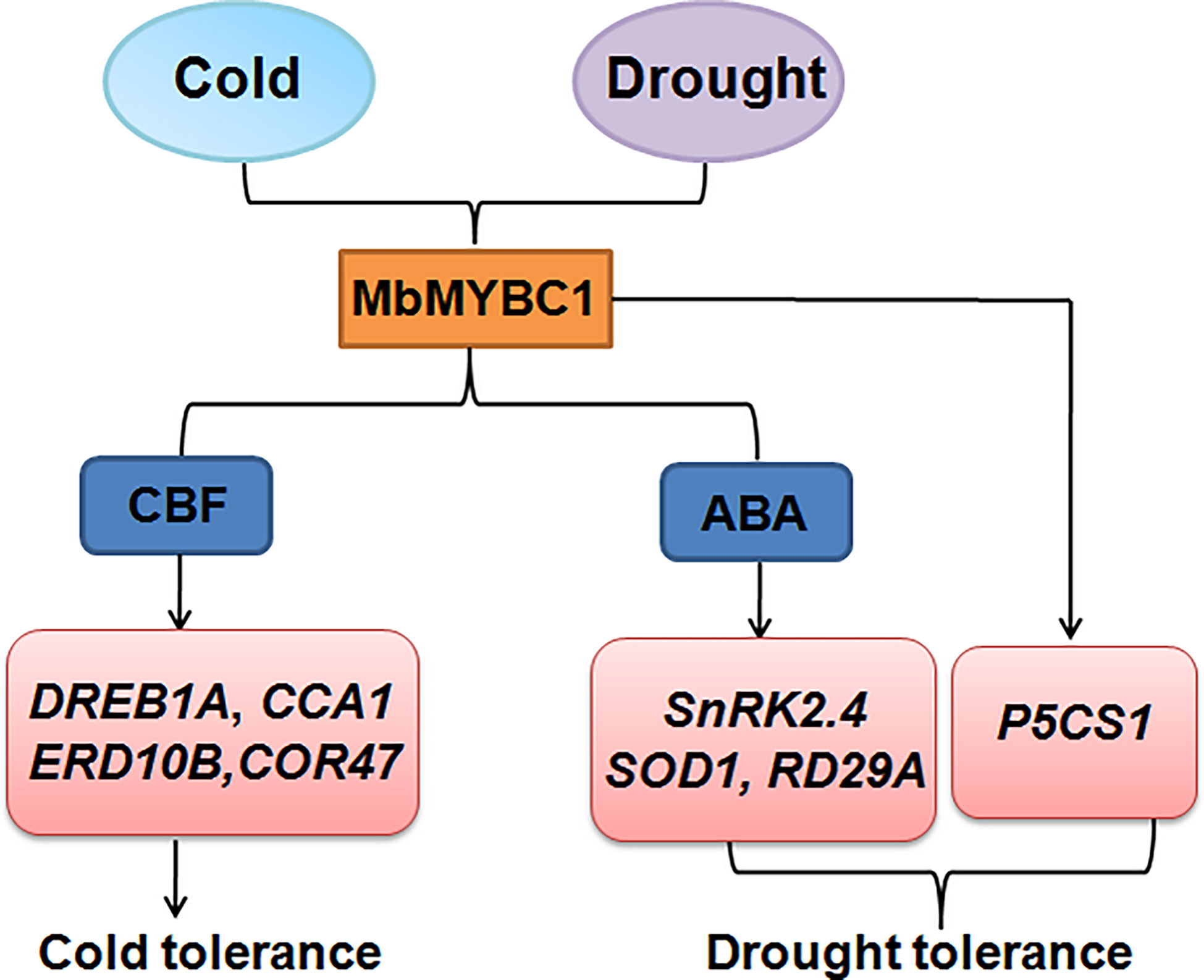
Figure 11 A potential model of MbMYBC1 regulating plant responses to low temperature and drought stress.
Conclusion
We isolated and characterized a new MYB gene, MbMYBC1, which encodes a protein localized in the nucleus. Overexpression of MbMYBC1 can change the relevant physiological and biochemical indicators and also regulate the expression of stress-related genes so as to improve the survival ability of plants under cold and drought conditions. This provides a theoretical basis and candidate genes for improving apple quality.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
WL contributed to the conception of the study, TW, YW, and XL performed the experiment, contributed significantly to analysis and manuscript preparation, JH and DH and performed the data analyses and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by the National Natural Science Foundation of China (32172521), the Natural Science Fund Joint Guidance Project of Heilongjiang Province (LH2022C097), the Collaborative Innovation System of Agricultural Bioeconomy in Heilongjiang Province and Heilongjiang Province Research Institute Research Expenses Project (CZKYF2023-1-B021).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1141446/full#supplementary-material
Supplementary Figure 1 | Gene sequence and amino acid sequence of MbMYBC1. The underscore marked the conservative sequence of MYB.
References
Assal, N., Lin, M. (2021). PCR procedures to amplify GC-rich DNA sequences of mycobacterium bovis. J. Microbiol. Meth. 181, 106121. doi: 10.1016/j.mimet.2020.106121
Blum, A. (1983). Genetic and physiological relationships in plant breeding for drought resistance. Agric. Water Manag. 7 (1–3), 195–205. doi: 10.1016/0378-3774(83)90083-5
Campos, P. S., Quartin, V., Ramalho, J. C., Nunes, M. A. (2003). Electrolyte leakage and lipid degradation account for cold sensitivity in leaves of coffea sp plants. J. Plant Physiol. 160, 283–292. doi: 10.1078/0176-1617-00833
Cao, Z. H., Zhang, S. Z., Wang, R. K., Zhang, R. F., Hao, Y. J. (2013). Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PloS One 8, e69955. doi: 10.1371/journal.pone.0069955
Chen, Y., Li, L. L., Zong, J. Q., Chen, J. B., Guo, H., Guo, A. G., et al. (2015). Heterologous expression of the halophyte zoysia matrella h+-pyrophosphatase gene improved salt tolerance in Arabidopsis thaliana. Plant Physiol. Bioch. 91, 49–55. doi: 10.1016/j.plaphy.2015.04.004
Chinnusamy, V., Zhu, J., Zhu, J. (2006). Salt stress signaling and mechanisms of plant salt tolerance. Genet. Eng. (N. Y.) 27, 141–177. doi: 10.1007/0-387-25856-6_9
Dogru, A., Çakirlar, H. (2020). Effects of leaf age on chlorophyll fluorescence and antioxidant enzymes activity in winter rapeseed leaves under cold acclimation conditions. Braz. J. Bot. 43, 11–20. doi: 10.1007/s40415-020-00577-9
Dossa, K., Mmadi, M. A., Zhou, R., Liu, A. L., Yang, Y. X., Diouf, D., et al. (2019). Ectopic expression of the sesame MYB transcription factor SiMYB305 promotes root growth and modulates ABA-mediated tolerance to drought and salt stresses in Arabidopsis. AoB Plants 12, plz081. doi: 10.1093/aobpla/plz081
Du, H., Feng, B. R., Yang, S. S., Huang, Y. B., Tang, Y. X. (2012). The R2R3-MYB transcription factor gene family in maize. PloS One 7, e37463. doi: 10.1371/journal.pone.0037463
Du, Z., Li, J. (2019). Expression, purification and molecular characterization of a novel transcription factor KcCBF3 from kandelia candel. Protein Expres. Purif. 153, 26–34. doi: 10.1016/j.pep.2018.08.006
Du, Y., Zhao, M., Wang, C., Gao, Y., Wang, Y., Liu, Y., et al. (2018). Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 18, 320. doi: 10.1186/s12870-018-1551-7
Furlan, A. L., Bianucci, E., Giordano, W., Castro, S., Becker, D. F. (2020). Proline metabolic dynamics and implications in drought tolerance of peanut plants. Plant Physiol. Bioch. 151, 566–578. doi: 10.1016/j.plaphy.2020.04.010
Gerona, M. E. B., Deocampo, M. P., Egdane, J. A., Ismail, A. M., Dionisio-Sese, M. L. (2019). Physiological responses of contrasting rice genotypes to salt stress at reproductive stage. Rice Sci. 26, 207–219. doi: 10.1016/j.rsci.2019.05.001
Han, D., Ding, H., Chai, L., Liu, W., Zhang, Z., Hou, Y., et al. (2018a). Isolation and characterization of MbWRKY1, a WRKY transcription factor gene from Malus baccata (L.) borkh involved in drought tol chinnusamy erance. Can. J. Plant Sci. 98, 1023–1034. doi: 10.1139/cjps-2017-0355
Han, D., Du, M., Zhou, Z., Wang, S., Li, T., Han, J., et al. (2020a). An NAC transcription factor gene from Malus baccata, MbNAC29, increases cold and high salinity tolerance in Arabidopsis. In Vitro Cell. Dev-Pl. 56, 588–599. doi: 10.1007/s11627-020-10105-9
Han, D., Han, J., Yang, G., Wang, S., Xu, T., Li, W. (2020b). An ERF transcription factor gene from Malus baccata (L.) borkh, MbERF11, affects cold and salt stress tolerance in Arabidopsis. Forests 11, 514. doi: 10.3390/f11050514
Han, J., Li, X., Li, W., Yang, Q., Li, Z., Cheng, Z., et al. (2023). Isolation and preliminary functional analysis of FvICE1, involved in cold and drought tolerance in fragaria vesca through overexpression and CRISPR/Cas9 technologies. Plant Physiol. Bioch 196, 270–280. doi: 10.1016/j.plaphy.2023.01.048
Han, D., Shi, Y., Yu, Z., Liu, W., Lv, B., Wang, B., et al. (2015). Isolation and functional analysis of MdCS1: A gene encoding a citrate synthase in Malus domestica (L.) borkh. Plant Growth Regul. 75, 209–218. doi: 10.1007/s10725-014-9945-5
Han, D., Yang, G., Xu, K., Shao, Q., Yu, Z., Wang, B., et al. (2013). Overexpression of a Malus xiaojinensis Nas1 gene influences flower development and tolerance to iron stress in transgenic tobacco. Plant Mol. Biol. Rep. 31, 802–809. doi: 10.1007/s11105-012-0551-2
Han, D., Zhang, Z., Ding, H., Wang, Y., Liu, W., Li, H., et al. (2018c). Molecular cloning and functional analysis of MbWRKY3 involved in improved drought tolerance in transformed tobacco. J. Plant Interact. 13, 329–337. doi: 10.1080/17429145.2018.1478994
Han, D., Zhang, Z., Ni, B., Ding, H., Liu, W., Li, W., et al. (2018b). Isolation and functional analysis of MxNAS3 involved in enhanced iron stress tolerance and abnormal flower in transgenic Arabidopsis. J. Plant Interact. 13, 433–441. doi: 10.1080/17429145.2018.1499145
Jiang, Y., Deyholos, M. K. (2009). Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69, 91–105. doi: 10.1007/s11103-008-9408-3
Jungklang, J., Saengnil, K., Uthaibutra, J. (2017). Effects of water-deficit stress and paclobutrazol on growth, relative water content, electrolyte leakage, proline content and some antioxidant changes in curcuma alismatifolia gagnep. cv. Chiang mai pink. Saudi J. Biol. Sci. 24, 1505–1512. doi: 10.1016/j.sjbs.2015.09.017
Kavian, S., Safarzadeh, S., Yasrebi, J. (2022). Zinc improves growth and antioxidant enzyme activity in aloe vera plant under salt stress. S. Afr. J. Bot. 147, 1221–1229. doi: 10.1016/j.sajb.2022.04.011
Kazuko, Y. S., Kazuo, S. (1993). Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Genet. Genomics 236, 331–340. doi: 10.1007/bf00277130
Latchman, D. S. (1997). Transcription factors: An overview. Int. J. Biochem. Cell B. 29(12), 1305–1312. doi: 10.1016/S1357-2725(97)00085-X
Li, X., Cheng, X., Li, J., Qiao, X., Li, L., Yu, L., et al. (2016). Genome-wide identification,evolution and functional divergence of MYB transcription factors in Chinese white pear (Pyrus bretschneideri). Plant Cell Physiol. 57, 824–847. doi: 10.1093/pcp/pcw029
Li, X., Liang, X., Li, W., Yao, A., Liu, W., Wang, Y., et al. (2022). Isolation and functional analysis of MbCBF2, a Malus baccata (L.) borkh CBF transcription factor gene, with functions in tolerance to cold and salt stress in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 23, 9827. doi: 10.3390/ijms23179827
Li, C., Ng, C. K. Y., Fan, L. M. (2015). MYB transcription factors,active players in abiotic stress signaling. Environ. Exp. Bot. 114, 80–91. doi: 10.1016/j.envexpbot.2014.06.014
Li, Y., Zhong, J., Huang, P., Shao, B., Li, W., Liu, W., et al. (2022). Overexpression of MxFRO6, a FRO gene from Malus xiaojinensis, increases iron and salt tolerance in Arabidopsis thaliana. In Vitro Cell. Dev-Pl. 58, 189–199. doi: 10.1007/s11627-022-10256-x
Li, W., Zhong, J., Zhang, L., Wang, Y., Song, P., Liu, W., et al. (2022). Overexpression of a Fragaria vesca MYB transcription factor gene (FvMYB82) increases salt and cold tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 23, 10538. doi: 10.3390/ijms231810538
Li, J., Zhou, H., Xiong, C., Peng, Z., Du, W., Li, H., et al. (2022). Genome-wide analysis R2R3-MYB transcription factors in xanthoceras sorbifolium bunge and functional analysis of XsMYB30 in drought and salt stresses tolerance. Ind. Crop Prod. 178, 114597. doi: 10.1016/j.indcrop.2022.114597
Liang, X., Li, Y., Yao, A., Liu, W., Yang, T., Zhao, M., et al. (2022a). Overexpression of MxbHLH18 increased iron and high salinity stress tolerance in arabidopsis thaliana. international. Int. J. Mol. Sci. 23, 8007. doi: 10.3390/ijms23148007
Liang, X., Luo, G., Li, W., Yao, A., Liu, W., Xie, L., et al. (2022b). Overexpression of a Malus baccata CBF transcription factor gene, MbCBF1, increases cold and salinity tolerance in Arabidopsis thaliana. Plant Physiol. Bioch. 192, 230–242. doi: 10.1016/j.plaphy.2022.10.012
Lin, S., Singh, R. K., Moehninsi, Navarre, D. A. (2021). R2R3-MYB transcription factors, StmiR858 and sucrose mediate potato flavonol biosynthesis. Hortic. Res. 8, 25. doi: 10.1038/s41438-021-00463-9
Liu, Y., Yang, H., Wen, F., Bao, L., Zhao, Z., Zhong, Z. ,. M. (2023). Chitooligosaccharide-induced plant stress resistance. Carbohyd. Polym. 302, 120344. doi: 10.1016/j.carbpol.2022.120344
Liu, W., Zhao, B., Chao, Q., Wang, B., Zhang, Q., Zhang, C., et al. (2019). Function analysis of ZmNAC33, a positive regulator in drought stress response in Arabidopsis. Plant Physiol. Bio. 145, 174–183. doi: 10.1016/j.plaphy.2019.10.038
Lu, X., Yang, L., Yu, M., Lai, J., Wang, C., McNeil, D., et al. (2017). A novel zea mays ssp. mexicana l. MYC-type ICE-like transcription factor gene ZmmICE1, enhances freezing tolerance in transgenic Arabidopsis thaliana. Plant Physiol. Bioch. 113, 78–88. doi: 10.1016/j.plaphy.2017.02.002
Matus, J. T., Aquea, F., Arce-Johnson, P. (2008). Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Bio. 8, 83. doi: 10.1186/1471-2229-8-83
Modesto, P., Peletto, S., Pisoni, G., Cremonesi, P., Castiglioni, B., Colussi, S., et al. (2013). Evaluation of internal reference genes for quantitative expression analysis by real-time reverse transcription-PCR in somatic cells from goat milk. J. Dairy Sci. 96, 7932–7944. doi: 10.3168/jds.2012-6383
Nakashima, K., Yamaguchi-Shinozaki, K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32, 959–970. doi: 10.1007/s00299-013-1418-1
Nandhini, K., Saraswathi, R., Premalatha, N. (2022). Identification of drought tolerant entries based on stress tolerant indices and physiological traits in RIL population of cotton (Gossypium hirsutum). Crop Design. 1, 100014. doi: 10.1016/j.cropd.2022.100014
Ou, L., Wei, G., Zhang, Z., Dai, X., Zou, X. (2015). Effects of low temperature and low irradiance on the physiological characteristics and related gene expression of different pepper species. Photosynthetica 53, 85–94. doi: 10.1007/s11099-015-0084-7
Qin, Y., Tian, Y., Liu, X. (2015). A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 464, 428–433. doi: 10.1016/j.bbrc.2015.06.128
Ren, J., Li, X., Mao, J., Zuo, C., Zhao, X., Chen, B. (2017). Physiological and quantitative phosphoproteome analyses of drought stress-induced mechanisms in Malus baccata (L.) borkh. Biochem. Syst. Ecol. 72, 47–55. doi: 10.1016/j.bse.2017.04.006
Serrano, M., Coluccia, F., Torres, M., L'Haridon, F., Métraux, J. P. (2014). The cuticle and plant defense to pathogens. Front. Plant Sci. 5, 274. doi: 10.3389/fpls.2014.00274
Singh, A., Jones, S., Ganapathysubramanian, B., Sarkar, S., Mueller, D., Sandhu, K., et al. (2021). Challenges and opportunities in machine-augmented plant stress phenotyping. Trends Plant Sci. 26, 53–69. doi: 10.1016/j.tplants.2020.07.010
Song, Q., Wang, X., Li, J., Chen, T., Liu, Y., Yang, X. (2021). CBF1 and CBF4 in solanum tuberosum l. differ in their effect on low-temperature tolerance and development. Environ. Exp. Bot. 185, 104416.
Steponkus, P. L., Uemura, M., Joseph, R. A., Gilmour, S. J., Thomashow, M. F. (1998). Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 95, 14570–14575. doi: 10.1073/pnas.95.24.14570
Su, L., Li, J., Liu, D., Ying, Z., Zhang, H., Li, X., et al. (2014). A novel MYB transcription factor, GmMYBJ1, from soybean confers drought and cold tolerance in Arabidopsis thaliana. Gene 538, 46–55. doi: 10.1016/j.gene.2014.01.024
Su, Y., Wang, P., Lu, S., Chen, B. (2022). Molecular cloning, bioinformatics analysis, and transient expression of MdAux/IAA28 in apple (Malus domestic). Gene Rep. 26, 101464. doi: 10.1016/j.genrep.2021.101464
Tripathi, D., Singh, M., Pandey-Rai, S. (2022). Crosstalk of nanoparticles and phytohormones regulate plant growth and metabolism under abiotic and biotic stress. Plant Stress 6, 100107. doi: 10.1016/j.stress.2022.100107
Wang, M., Hao, J., Chen, X., Zhang, X. (2020). SlMYB102 expression enhances low-temperature stress resistance in tomato plants. PeerJ 8, e10059. doi: 10.7717/peerj.10059
Wang, G., Zeng, F., Song, P., Sun, B., Wang, Q., Wang, J. (2022). Effects of reduced chlorophyll content on photosystem functions and photosynthetic electron transport rate in rice leaves. J. Plant Physiol. 272, 153669. doi: 10.1016/j.jplph.2022.153669
Wassie, M., Song, S. R., Cao, L. W., Chen, L. (2023). A medicago truncatula calcineurin b-like protein, MtCBL13 confers drought sensitivity in arabidopsis through ABA-dependent pathway. Environ. Exp. Bot. 206, 105141. doi: 10.1016/j.envexpbot.2022.105141
Wu, F., Muvunyi, B. P., Yan, Q., Kanzana, G., Ma, T., Zhang, Z., et al. (2022). Comprehensive genome-wide analysis of polyamine and ethylene pathway genes in cleisto genes songorica and CsSAMDC2 function in response to abiotic stress. Environ. Exp. Bot. 202, 105029. doi: 10.1016/j.envexpbot.2022.105029
Xiong, C., Zhao, S., Yu, X., Sun, Y., Li, H., Ruan, C., et al. (2020). Yellowhorn drought-induced transcription factor XsWRKY20 acts as a positive regulator in drought stress through ROS homeostasis and ABA signaling pathway. Plant Physiol. Bioch. 155, 187–195. doi: 10.1016/j.plaphy.2020.06.037
Yamaguchi-Shinozaki, K., Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 6, 251–264.
Yan, H., Jia, H., Chen, X., Hao, L., An, H., Guo, X. (2014). The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 55, 2060–2076. doi: 10.1093/pcp/pcu133
Yao, C., Li, X., Li, Y., Yang, G., Liu, W., Shao, B., et al. (2022b). Overexpression of a Malus baccata MYB transcription factor gene MbMYB4 increases cold and drought tolerance in arabidopsis thaliana. Int. J. Mol. Sci. 23, 1794. doi: 10.3390/ijms23031794
Yao, C., Li, W., Liang, X., Ren, C., Liu, W., Yang, G., et al. (2022a). Molecular cloning and characterization of MbMYB108, a Malus baccata MYB transcription factor gene, with functions in tolerance to cold and drought stress in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 23, 4846. doi: 10.3390/ijms23094846
Zhang, A., Wang, W., Tong, Y., Li, M., Grierson, D., Ferguson, I., et al. (2018). Transcriptome analysis identifies a zinc finger protein regulating starch degradation in kiwifruit. Plant Physiol. 178, 850–863. doi: 10.1104/pp.18.00427
Keywords: Malus baccata (L.) Borkh, MbMYBC1, transgenic technology, cold stress, drought stress
Citation: Liu W, Wang T, Wang Y, Liang X, Han J and Han D (2023) MbMYBC1, a M. baccata MYB transcription factor, contribute to cold and drought stress tolerance in transgenic Arabidopsis. Front. Plant Sci. 14:1141446. doi: 10.3389/fpls.2023.1141446
Received: 10 January 2023; Accepted: 06 February 2023;
Published: 16 February 2023.
Edited by:
Qing Yang, Beijing Forestry University, ChinaReviewed by:
Jian Jin, Northeast Institute of Geography and Agroecology (CAS), ChinaKun Xiao, Shanghai Jiao Tong University, China
Copyright © 2023 Liu, Wang, Wang, Liang, Han and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Deguo Han, deguohan@neau.edu.cn; Wanda Liu, liuwanda@haas.cn
 Wanda Liu1*
Wanda Liu1* Deguo Han
Deguo Han