- State Key Laboratory of Rice Biology, China National Rice Research Institute, Hangzhou, China
The hull (palea and lemma) is the specific organ of grass florets. Although many genes related to the hull development have been cloned, the genetic mechanisms behind the development are still unclear, and the evolutionary relationship has different explanations and heated arguments between the palea and lemma. In this study, we found a specific mr1 mutant with a reduced palea, showing an enlarged mrp and degraded bop. Phenotype observations and molecular evidences showed that the bop was converted to the mrp-like organ. Our findings first reveal that the bop and mrp are homologous structures, and the palea and lemma are the same whorl floral organs. MR1 may prevent the transformation of the bop into mrp by regulating the expressions of hull identity genes. Meantime, the mr1 mutant showed altered grain size and grain quality, with defective physical and chemical contents. MR1 was controlled by a single recessive gene and was finally located on chromosome 1, with a physical distance of 70 kb. More work will be needed for confirming the target gene of MR1, which would contribute to our understanding of grain formation and the origin between the lemma, bop, and mrp.
Introduction
The rice hull, such as palea and lemma, is the unique floral organ of the outer whorls in grass (Xu et al., 2021). They are located in the outer layer of floret structure, which can provide photosynthetic products for internal organs in the early stage, and protect seeds from diseases and insect pests after ripening. The hull size is also one of the determinants of seed size. Therefore, the hull development is very important for the formation of rice yield and quality.
The flowers of typical eudicots are composed of sepals, petals, stamens, and pistils. According to the genetic and molecular analyses of Arabidopsis thaliana and Antirrhinum majus, Coen and Meyerowitz proposed an ABC model of floral organ characteristics in the early 1990s (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994). The classical ABC model suggests that there are three kinds of genes A, B, and C to control the development of four whorls floral organs, the same class of genes control the two adjacent whorls. That is to say, the sepals, petals, stamens, and pistils of four floral organs are determined by A, AB, BC, and C genes, respectively (Yanofsky et al., 1990; Mandel et al., 1992; Goto and Meyerowitz, 1994). Gradually, due to the identification of class D genes that regulate ovule development and the class E genes that regulate the development of internal three whorls of floral organs, ABC model is further extended to ABCDE model (Colombo et al., 1995; Pelaz et al., 2000; Theissen and Saedler, 2001). With our deep-going understanding of the flower development of monocots, we found that in monocots such as rice, the shape and function of the stamens and pistils of the inner whorls are similar to those of eudicots, and the ABCDE model is also applicable to them; however, the shape and function of the hull and lodicules of the outer whorls are quite different from those of eudicots.
In Arabidopsis, the development of the outer whorls of floral organs is mainly regulated by class A and E genes (Coen and Meyerowitz, 1991; Angenent et al., 1995; Pelaz et al., 2000). Class A genes include APETALA1 (AP1) and APETALA2 (AP2), they specifically regulate the development of sepals and petals in the outer two whorls of floral organs. Mutations in the class A genes will lead to the homeotic transformation from sepals and petals to pistils and stamens (Angenent et al., 1995). There are four class E genes in Arabidopsis, SEPALLATA1/2/3/4 (SEP1/2/3/4). At first, it is found that SEP1/2/3 redundantly regulates the characteristics of three inner whorls, and later it is found that SEP1/2/3 also redundantly regulates the development of sepals together with SEP4 (Pelaz et al., 2000).
In rice, there are class A and E genes homologous to Arabidopsis. OsMADS14/15/18/20 are the orthologous genes of AP1 in Arabidopsis. OsMADS14 is mainly expressed in the inflorescence and developing seeds (Pelucchi et al., 2002), the overexpression of OsMADS14 can lead to a significant advance in rice flowering. The degenerative palea (dep) mutant is caused by a single nucleotide mutation of OsMADS15 gene, which shows that the lemma is elongated and the body of palea (bop) is degenerated, leaving only two marginal regions of palea (mrp), which indicates that OsMADS15 is necessary to maintain the bop and the development of the lemma (Wang et al., 2010). The overexpression of OsMADS18 leads to the early flowering and the early initiation of axillary meristem, which indicates that OsMADS18 can initiate differentiation of branches and leaves (Fornara et al., 2004). OsMADS20 is only expressed in the stems and developing seeds, and it is significantly different from the expression pattern of class A genes in Arabidopsis. Class E genes include two main types: SEP1/2/4 (LOFSEP) and SEP3 (Malcomber and Kellogg, 2005). SEP1/2/4 genes include OsMADS1/LHS1, OsMADS5, and OsMADS34, and SEP3 genes include OsMADS7/OsMADS45 and OsMADS8/OsMADS24. OsMADS1 plays a key role in the late developmental stages of floral organs, and it first expressed in the meristem of spikelet (Chung et al., 1994; Prasad et al., 2001). Among several mutants of OsMADS1 gene, the palea and lemma elongated in varying degrees, indicating that OsMADS1 plays an important role in the development of palea and lemma (Jeon et al., 2000). The complete loss of OsMADS1 function resulted in the complete homeotic transformation of the three inner organs (lodicules, stamens, and pistils) into the palea and lemma-like structures (Agrawal et al., 2005). The sterile lemma of osmads34 mutant was elongated, while the palea and lemma were normal, but in the osmads34 and osmads1 double mutant, the palea and lemma were more elongated and leafed than the osmads1 single mutant, which indicated that OsMADS34 and OsMADS1 redundantly regulate the development of palea and lemma (Gao et al., 2010). The overexpression of OsMADS5 promoted the early flowering phenotype, but did not affect the morphology of flower (Jeon et al., 2000). OsMADS7 and OsMADS8 can interact with other proteins such as OsMADS13, OsMADS16, and OsMADS18 (Favaro et al., 2003; Kater et al., 2006). And OsMADS7 and OsMADS8 have similar interaction patterns with OsMADS1. But it is still not clear whether the genes OsMADS5/7/8 play a role in the regulation of palea and lemma development.
In this study, we identified a mutant with abnormal palea development, named marginal region 1 (mr1). The mr1 mutant showed a defective fusion palea with an enlarged mrp and a degraded bop, first revealing that the bop and mrp are homologous organs. The mr1 mutant also showed altered grain size and quality, with defectively physical and chemical contents. Further, we performed genetic analysis and fine mapping of the mr1 mutant traits, which tried to detect the MR1 gene and its function. The present and future work will contribute to our understanding of grain formation and organ origin.
Materials and Methods
Plant Materials
The mr1 mutant was identified in the ethyl methanesulfonate (EMS) mutant population of Shuangkezao (SKZ) (cv. indica), and the mutant character was inherited stably after multiple generations of self-bred and selection. The mr1 mutant was crossed with Nipponbare to construct a mapping population. The resulting F1 grains were sown and self-crossed to construct the F2 population, which used for gene mapping and genetic analysis. All these materials were planted under natural conditions in the paddy fields of China National Rice Research Institute.
Morphological and Histological Analysis
In the flowering stage, we selected the fresh spikelets of wild-type and mr1 mutant and dissected them with tweezers, then analyzed the phenotypic characteristics of them by NIKON SMZ1500 stereoscope.
Paraffin sections were used for histological analysis, according to the method of Ren et al., 2019b. We selected the spikelets of wild-type and mr1 mutant at the heading stage and fixed them in mixed solution (50% absolute ethanol, 0.9 mol/L acetic acid, and 3.7% formaldehyde) at 4°C for more than 16 h, and dehydrated them with different concentrations of ethanol. These materials were then infiltrated with xylene and embedded in paraffin. A rotary microtome was used to cut the paraffin into 8 μm thick pieces and the slides were put at 42°C for several days. We chose some perfect slides to stain with safranine and fast green, and then dehydrated them with different concentrations of ethanol. These stained slides were infiltrated with xylene and covered with neutral resins. At last, we observed these slides under an optical microscope.
Scanning Electron Microscopy
According to the method described by Mizukami and Ma, 1992, the scanning electron microscopy (SEM) analysis was performed. We selected the young spikelets of wild-type and mr1 at the stage of floral organ differentiation and fixed them in 3% glutaraldehyde solution at 4°C for about 24 h. Then the materials were dehydrated with different concentrations of ethanol, and incubated in the mixture of ethanol and isoamyl acetate for 1 h. The materials were dried and wrapped with gold powder. These coated samples were finally observed under a HITACHI S-3500 SEM with a −30°C cooled stage.
Fine Mapping of the Mutant Gene
Among the progenies from the cross between the mr1 mutant and Nipponbare, we selected a total of 558 F2 plants which were showed the mutant phenotype for mapping of the MR1 gene (Supplementary Table 1). The simple sequence repeat (SSR) markers we used were obtained from the GRAMENE website.1 And for fine mapping, we developed more new in/del primers by comparing the sequences of the indica 93-11 and japonica Nipponbare. Primer sequences used are listed in Supplementary Table 2.
RNA Extraction and Gene Expression Analysis
The roots, culms, leaves, internodes, inflorescences at various developmental stages, and all floral organs (pistils, stamens, lodicules, paleae, and lemmas) of the wild-type and mr1 mutant were collected, the total RNA was extracted from these tissues or organs using the RNeasy Plant Mini Kit (Axygen). The 2 μg total RNA was reverse transcribed to cDNA by using the Reverse Transcriptase Kit (Invitrogen) with genomic DNA eraser (Takara) in a 20 μl reaction volume. For the quantitative reverse transcription-PCR (qRT-PCR) analysis, the cDNA samples were used as templates after being diluted 10-fold. The qRT-PCR was carried out on a StepOne-Plus System (Applied Biosystems) using the SYBR Green PCR Mix Kit (Applied Biosystems) (Ren et al., 2019b). At least three biological replicates were carried out, and the relative expression levels were quantified using a relative quantitation method. Primer sequences used for qRT-PCR are listed in Supplementary Table 2.
In situ Hybridization
Healthy young panicles from the wild-type and mr1 mutant were put in 70% mixed solution (50% absolute ethanol, 0.9 mol/L acetic acid, and 3.7% formaldehyde), then dehydrated in gradient concentrations of ethanol and xylene, and finally embedded in paraffin. Hybridization and immunological detection of these samples were performed as previous description (Ren et al., 2018). The probes were prepared and labeled by a DIG RNA Labeling Kit from Roche according to the manufacturer’s recommendations. Primer sequences used for in situ hybridization are listed in Supplementary Table 2.
Analysis of Grain Quality
The mature grains were air-dried at room temperature and then were crushed to rice powder for physicochemical properties analysis. The total starch content was examined by using a starch assay kit (Megazyme) (Cai et al., 2018). The amylose content measurement was performed according to the description by Liu et al. (2009), and the total protein content measurement was performed according to the description by Kang et al. (2005). To analyze the gelatinization temperature of the samples, we used a differential scanning calorimeter (DSC1). First, we put 5 mg rice powder into an aluminum cup and fixed it with 10 μl distilled water. Then we used a machine to seal the cup. At last, we put the cup into the DSC1, and it run under the 10°C min–1 heating rate and in the temperature range of 35–100°C. According to the method described by Cai et al. (2018), we examined the swelling and gelatinization characteristics of endosperm starch in urea solutions. All measurements contained three biological replications. Analysis of variance (ANOVA) was used to test whether there are significant differences between these samples.
Results
Phenotype of the mr1 Mutant
In the wild-type, the floret includes a lemma, a palea, two lodicules, six stamens, and a pistil from outer to inner (Figures 1A–C). At the seedling and vegetative stage, the phenotype of the mr1 mutant was normal, and the flowering period was also not different from that of wild-type. However, the mr1 mutant appeared abnormal after heading stage. Compared to the wild-type, the mr1 mutant showed reduced paleae, and it was prone to splitting (Figures 1A,B,D,E), but the lemma was not altered (Figures 1A,B,D,E). Meantime, the stamens of mr1 mutant were reduced and no differences were found between the wild-type and mr1 mutant in the other floral organs (Figures 1C,F). According to our measurements and statistics, the palea, bop and mrp of mr1 mutant were narrower than that of wild-type, and the lemma had no significant changes (Figures 1G–I).
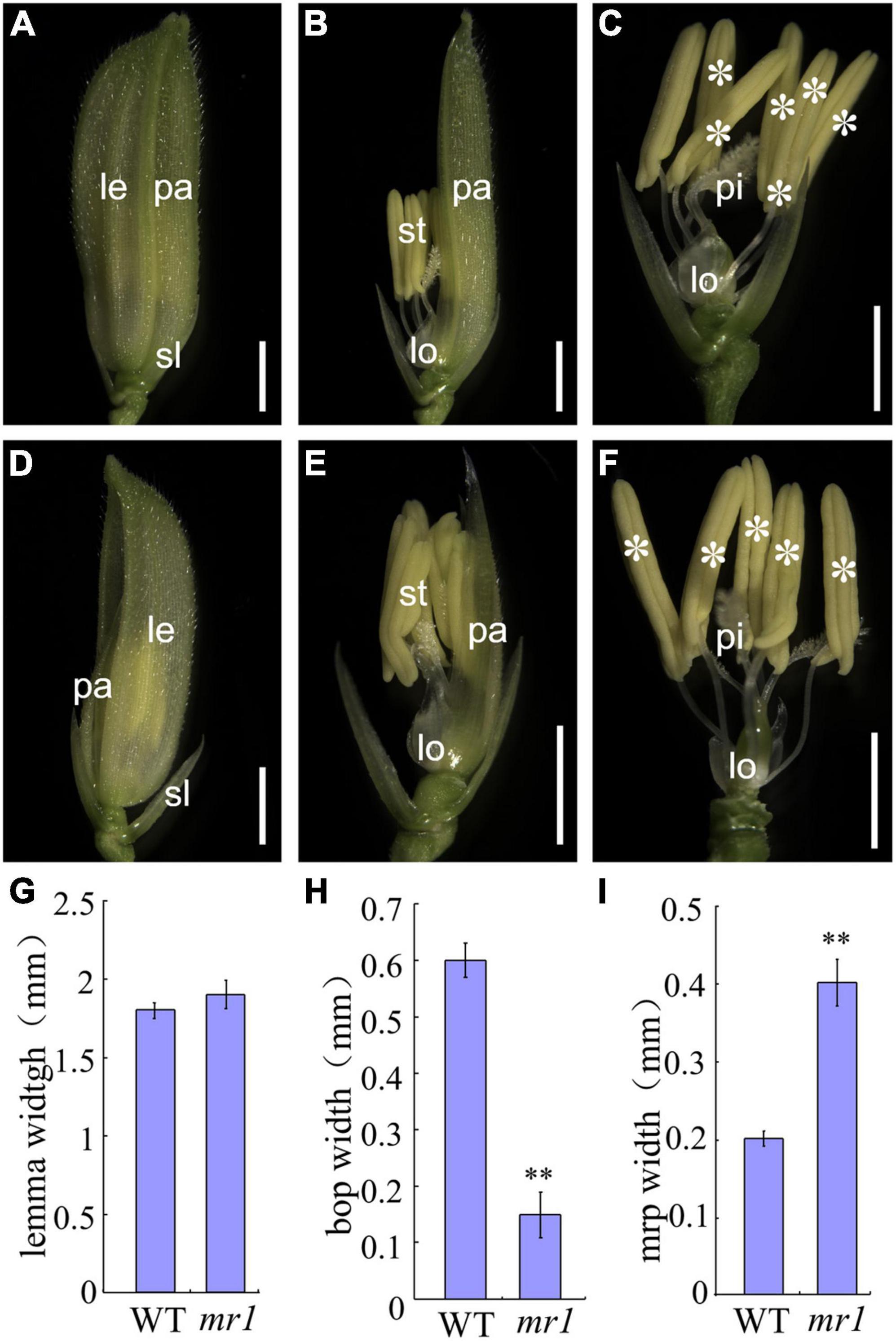
Figure 1. Phenotypes of wild-type and mr1 florets. (A) Wild-type floret. (B) Wild-type floret without lemma. (C) Wild-type floret without lemma and palea. (D) mr1 floret. (E) mr1 floret without lemma. (F) mr1 floret without lemma and palea. (G) Lemma width. (H) Bop width. (I) mrp width. le, lemma; pa, palea; sl, sterile lemma; lo, lodicule; st, stamen; pi, pistil. Asterisk indicates the stamen. Bars, 2 mm in (A–F). **Significant difference at p < 0.01 compared with the wild-type by Student’s t-test. Error bars indicate SD.
For histological analysis, we further carried out scanning electron microscopy (SEM) and paraffin sections for the florets of wild-type and mr1 mutant (Figure 2). We found that in the normal floret, there were five vascular bundles in the lemma, and there were three in the palea (Figures 2A–E,K). And the palea comprised of two mrps and a body of palea (bop), which was different from the lemma (Figures 2B–E,K). As the outer whorls of floral organs, palea and lemma had their distinctive cellular morphology (Figures 2A–E). They both comprised of silicified cells (sc), non-silicified cells (nsc), fibrous sclerenchyma (fs), and spongy parenchymatous cells (spc) (Figures 2D,E,K). In addition, the mrp differed from the bop, showing a distinctive smooth epidermis but lacking silicified thickening epicuticular cells (Figures 2B–E,K). Because of this unique mrp, the lemma and palea appeared a well-interlocked structure (Figures 2B–E,K).
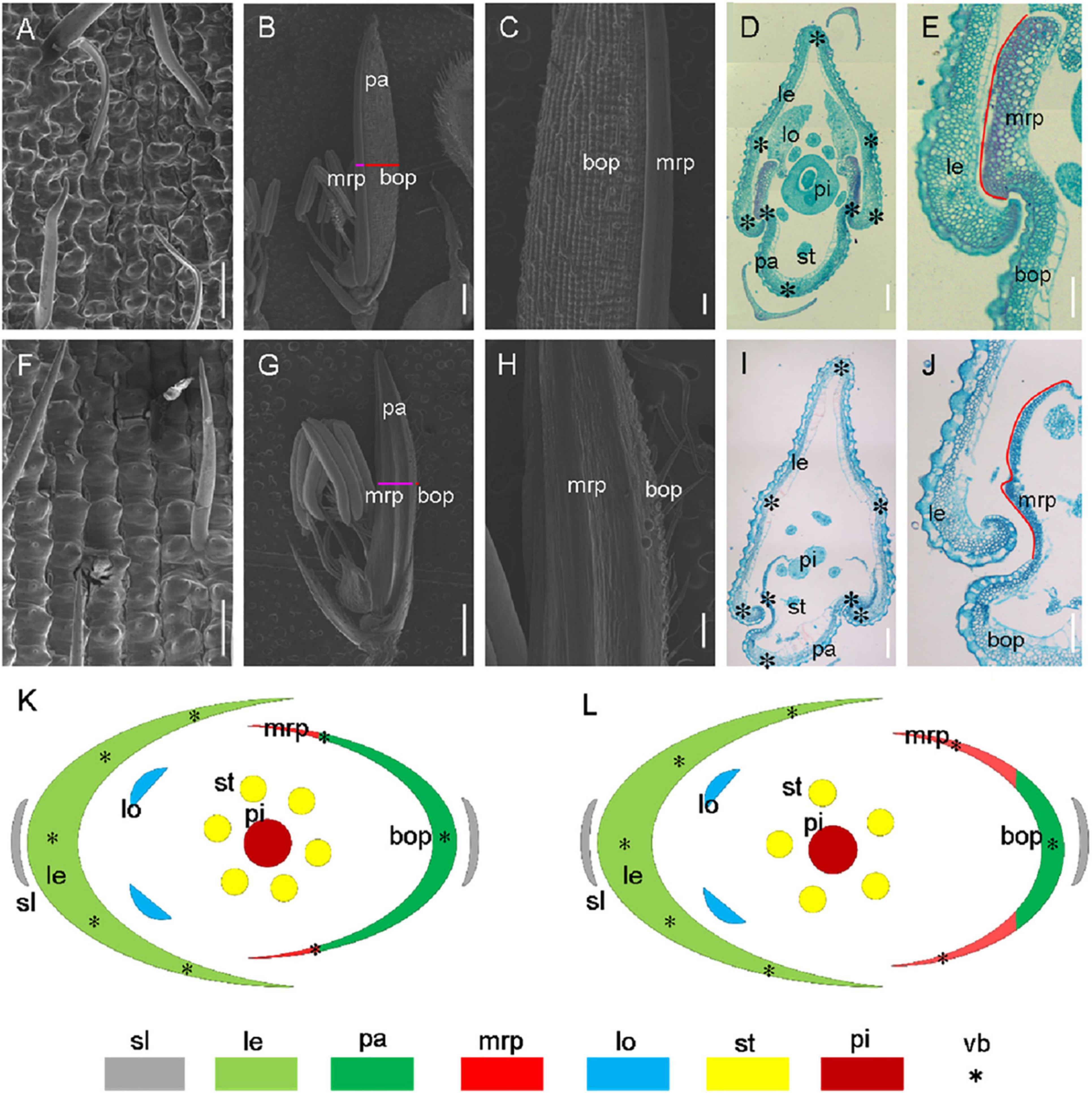
Figure 2. Histological analysis of hull in the wild-type and the mr1 mutant. (A) Epiderm of the wild-type lemma. (B) Epiderm of the wild-type palea. (C) Enlarged palea in the wild-type. (D) Transverse section of wild-type floret. (E) Interlocked structure of wild-type hull. (F) Epiderm of the mr1 lemma. (G) Epiderm of the mr1 palea. (H) Enlarged palea in the mr1 mutant. (I) Transverse section of mr1 floret. (J) Interlocked structure of mr1 hull. (K) Diagram of wild-type spikelet. (L) Diagram of mr1 spikelet. le, lemma; pa, palea; lo, lodicule; st, stamen; pi, pistil; bop, body of palea; mrp, marginal region of palea; sl, sterile lemma; vb, vascular bundle. Black asterisks indicate the vascular bundles. Red lines indicate the mrp. Bars, 100 μm in (A,E,F,J); 1 mm in (B,G); 200 μm in (C,D,H,I).
In the mr1 mutant, the mrp was enlarged, and the bop was reduced compared with the wild-type (Figures 2F–J,L). However, the number of vascular bundles was not altered in the mr1 palea, and the mr1 lemma appeared normal vascular bundles (Figure 2I). And the reduced palea resulted in the poor interlocked between the mrp and lemma, so the hull was prone to splitting (Figures 2I,J,L). These results indicated that in the mr1 mutant, the lemma kept normal phenotype, while the palea was significantly reduced due to the decrease of bop and increase of mrp. In addition, no obvious histocytological defects were observed for three inner whorls of floral organs except stamen number in the wild-type and mr1 mutant (Figures 2B–D,I,J,L).
Analysis of Floral Organ Development at the Early Stage
The development of early differentiation stages of wild-type and mr1 mutant florets was observed by SEM, the division of developmental stages we used refers to Ikeda et al. (2004). At the Sp4 stage, the primordia of lemma and palea of the wild-type floret began to develop, the primordia of lemma developed earlier, with a bulge on the top; and the lemma and palea were semicircular, and the marginal tissues were interconnected to form a circular structure, which held the floral meristem inside (Figure 3A). At this stage, no obvious differences were found between the wild-type and mr1 mutant (Figure 3E). During the Sp5 to Sp6 stages, the stamen primordia of the wild-type florets began to differentiate, and the visible spherical stamen primordia were formed synchronously except the stamen adjacent to the lemma (Figure 3B). The primordia of lemma and palea further differentiated, and their marginal tissues began to interlock (Figure 3B). We also observed that the mr1 palea was smaller than the wild-type in these stages (Figure 3F). At the Sp7 stage, the primordia of lemma and palea of the wild-type developed normally and presented a semi-closed state, wrapping the primordia of inner floral organs (Figure 3C). At Sp8, the primordia of lemma and palea of the wild-type was completely closed, and the inner whorls organs were completely wrapped (Figure 3D). Whereas, during the Sp7 and Sp8 stages, the mr1 florets showed significantly smaller palea than that of the wild-type (Figures 3G,H). These results suggested that the mr1 palea decreased from the early stage of floral organ development.
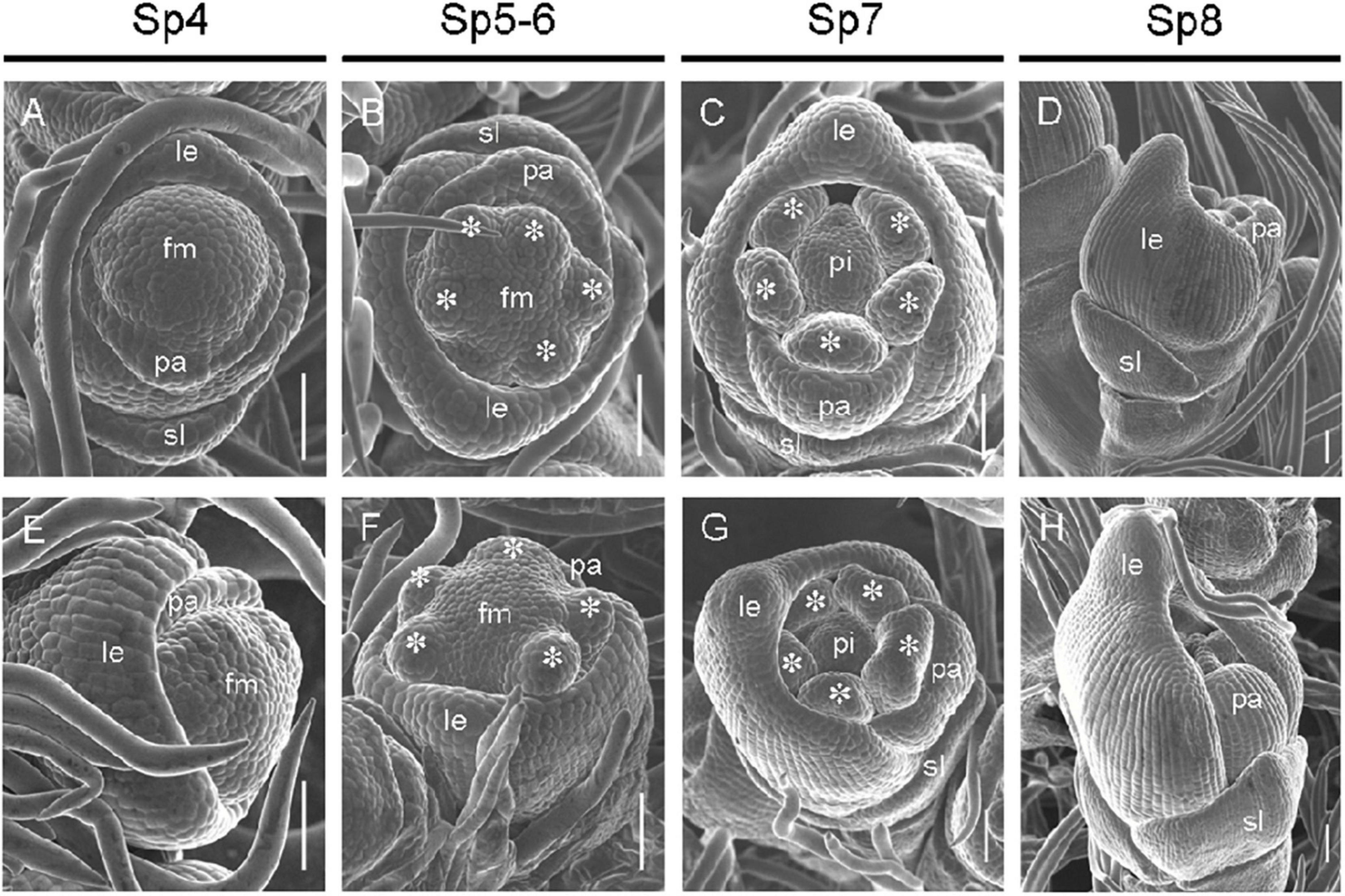
Figure 3. Florets at early developmental stages in the wild-type and the mr1 mutant. (A–D) Wild-type floret. (A) Sp4. (B) Sp5-6. (C) Sp7. (D) Sp8. (E–H) mr1 floret. (E) Sp4. (F) Sp5-6. (G) Sp7. (H) Sp8. fm, floral meristem; sl, sterile lemma; le, lemma; pa, palea; pi, pistil. Asterisk indicates the stamen. Bars, 50 μm.
Expression of Marker Genes in the Floret
To further examine whether the identity of palea has changed, we used qRT-PCR to investigate the expression levels of several marker genes, such as the hull (palea and lemma) marker genes OsMADS1, OsMADS14, and OsMADS15, the lemma marker gene DROOPING LEAF (DL), and the mrp marker gene OsMADS6 in the palea and lemma of the wild-type and mr1 mutant. The results exhibited that the OsMADS14 expression showed no significant difference between the wild-type and mr1 mutant (Figure 4A). The expression patterns of OsMADS1 and OsMADS15 in the mr1 lemmas were consistent with that of wild-type lemmas, respectively, while the expression levels of OsMADS1 and OsMADS15 in the mr1 paleae were obviously reduced than that of wild-type paleae, respectively (Figure 4A). DL showed no expression difference in the wild-type and mr1 mutant, while an increase expression of OsMADS6 was found in the mr1 mutant (Figure 4A). And, we detailedly detected the expression of OsMADS6 in the bop and mrp. Compared with the wild-type, the OsMADS6 transcript was increased in the bop of mr1 mutant, whereas its expression was not obviously altered in the mrp of mr1 mutant (Figure 4B). The lower expressions of OsMADS1 and OsMADS15 in the mr1 paleae may be caused by the small bop, while the higher expression of OsMADS6 in the mr1 paleae was due to the enlarged mrp.
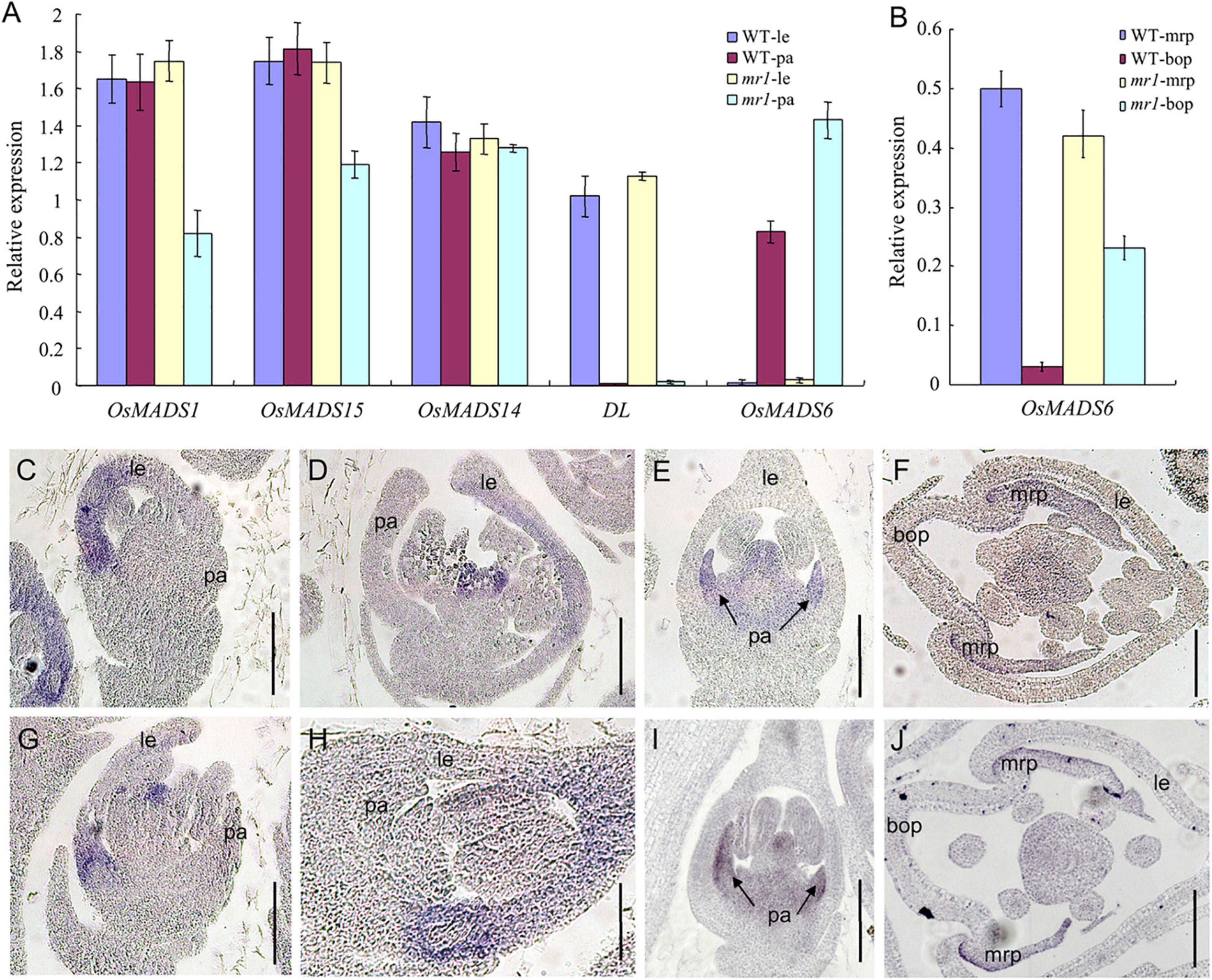
Figure 4. Expression of floral organ genes in the wild-type and the mr1 mutant. (A) Relative expression f genes in the wild-type and the mr1 floral organs. (B) Relative expression f genes in the bop and mrp of wild-type and the mr1 mutant. (C,D) DL expression in the wild-type floret. (E,F) OsMADS6 expression in the wild-type floret. (G,H) DL expression in the mr1 floret. (I,J) OsMADS6 expression in the mr1 floret. le, lemma; pa, palea; mrp, marginal region of palea; bop, body of palea. Bars, 50 μm.
Next, we further investigate the temporal and spatial expression patterns of the DL and OsMADS6 gene in the wild-type and mr1 mutant florets by in situ hybridization.
In the wild-type, DL expression was found in the lemma and pistil at stage SP7, then DL mRNAs were found in the lemma at stage Sp8 (Figures 4C,D,G,H). In contrast, we observed no obvious differences of DL expression between the wild-type and mr1 mutant at similar stages (Figures 4C,D,G,H). At stages Sp7 and Sp8, in flowers of the wild-type, OsMADS6 showed similar expression patterns with the mr1 mutant; its signals were present in the mrp and pistil (Figures 4E,F,I,J). In general, these results indicated that the identity of mrp was not disturbed in the mr1 mutant, and the lemma had the similar identity in the wild-type and mr1 mutant.
Genetic Analysis
The mr1 mutant was crossed with Nipponbare, all F1 plants appeared the wild-type phenotype, showing that this mutation was a recessive trait. The phenotype of mr1 was separated in F2 population. The total number of F2 population was 2,367, including 1,809 normal plants and 558 mutant plants (Supplementary Table 1). The separation ratio was 3.242:1. By the chi-square test, the separation ratio was consistent with the theoretical ratio of 3:1 (Supplementary Table 1). Thus, we reached a conclusion that the trait of mr1 mutant was controlled by a single recessive gene.
Mapping of MR1
In order to identify the MR1 gene, we performed a map-based cloning method. A total of 558 mutant plants in F2 population of mr1/Nipponbare cross were used for gene mapping. A total of 240 pairs of SSR markers with an average distribution of 12 chromosomes were selected for polymorphism analysis, 112 of which showed polymorphisms between two parents, SKZ and Nipponbare. Using these 112 pairs of SSR markers for linkage analysis, we found that the marker B1-15 on chromosome 1 was obviously linked to the mr1 phenotype. Then 558 mutant plants in the F2 population were investigated with this marker, and 32 recombinants were found. Other 20 SSR markers, which were located near the B1-15, were examined with the same approach. Then, the MR1 gene was initially located between the two polymorphic markers, B1-15 and B1-16, and they had 32 and 25 recombinants, respectively (Figure 5A). To further determine the location of the MR1 gene, 16 new in/del primers were developed based on the sequences downloaded from the rice genomic databases (Nipponbare for japonica and 93-11 for indica), and eight of them exhibited the polymorphisms. All 558 mutants were screened with these eight pairs of primers, and the recombinants were 17, 11, 8, 4, 1, 1, 3, and 12, respectively (Figure 5B). Finally, the MR1 gene was located in about 70 kb region between the markers ID8 and ID6 (Figure 5B). In this region, eight open frames were shown, and no known genes were reported (Figure 5C and Supplementary Table 3). ORF1 encoded a protein phosphatase 2C. ORF2 encoded a ZOS1-11-C2H2 zinc finger protein. ORF3, ORF4, and ORF8 encoded expression proteins. ORF5 and ORF7 encoded translation initiation factors. ORF6 encoded an OsFBX16-F-box domain protein. We next sequenced the nucleotide sequences of all genes. Unfortunately, we examined the DNA sequence, and failed to identify the nucleotide changes between wild-type and mr1 mutant.
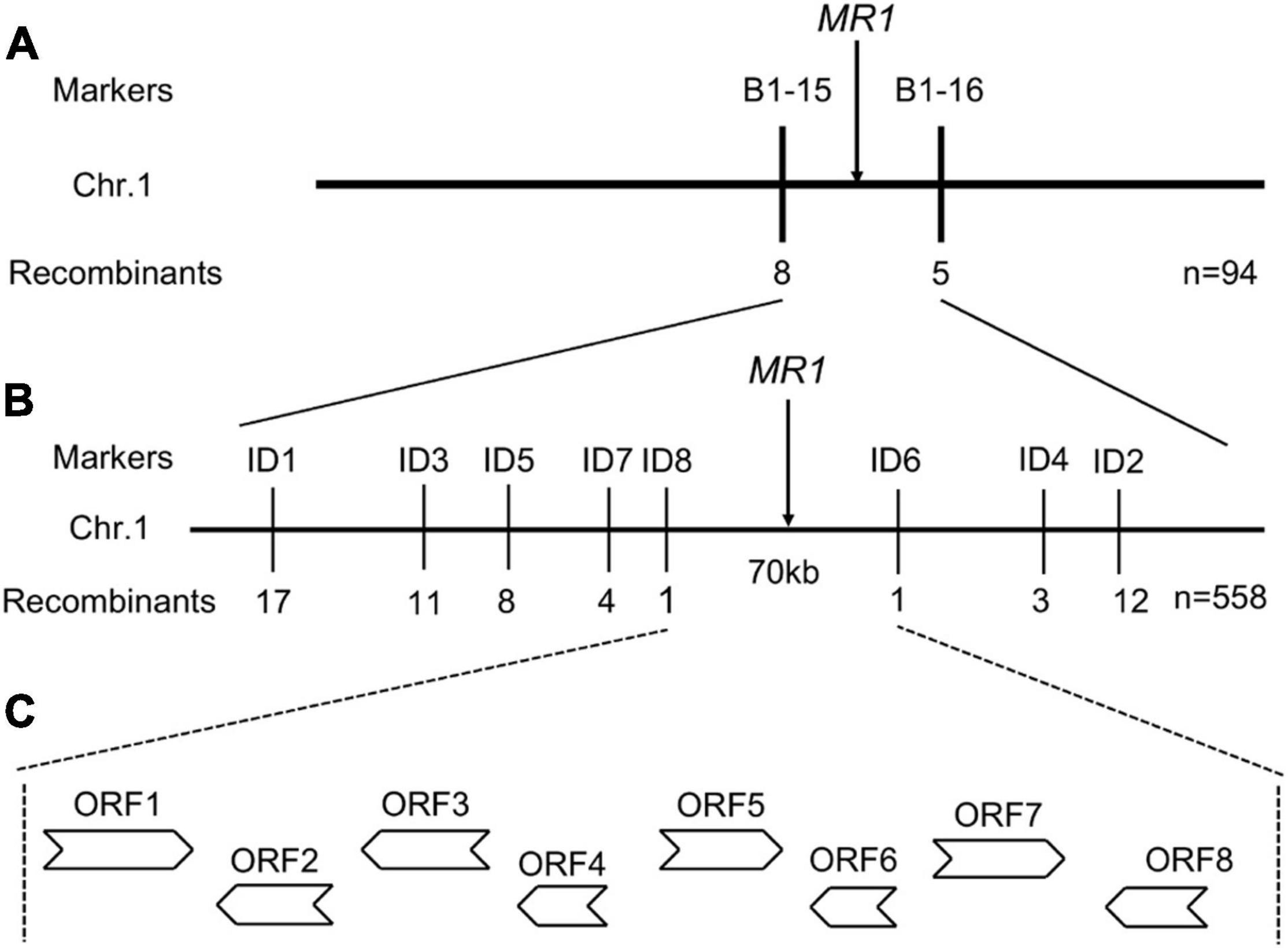
Figure 5. Fine mapping of the MR1 gene. (A) Genetic linkage map for the primary location of the MR1 gene using 94 F2 recessive plants. (B) Fine physical map of the MR1 gene using 558 F2 recessive plants. (C) ORFs in the target region. ORFs indicate open reading frames.
The Expression Regulation of Genes Involved in Palea Development
In consideration of the mr1 mutant displayed defects in palea development, we detected whether MR1 regulates the expression levels of known genes related to palea development in rice. We examined MADS-box genes comprising class A gene (DEP/OsMADS15) and class E gene (MFO1/OsMADS6), and other palea-related genes (MFS1, CFO1, CCP1, DP1, REP1, OPB, AH2, and FON4). We used qRT-PCR to analyze the expression levels of these genes in the wild-type and the mr1 mutant young panicles less than 2 cm (Figure 6). Most tested genes displayed significant changes in the mr1 mutant except REP1 and CCP1 (Figure 6). These results indicated that the reduced palea in the mr1 mutant may be associated with the changes of expression levels of these palea development-related genes, and MR1 determined the palea fate by possibly promoting MFO1 and CFO1, or suppressing DEP, MFS1, DP1, OPB, AH2, and FON4.
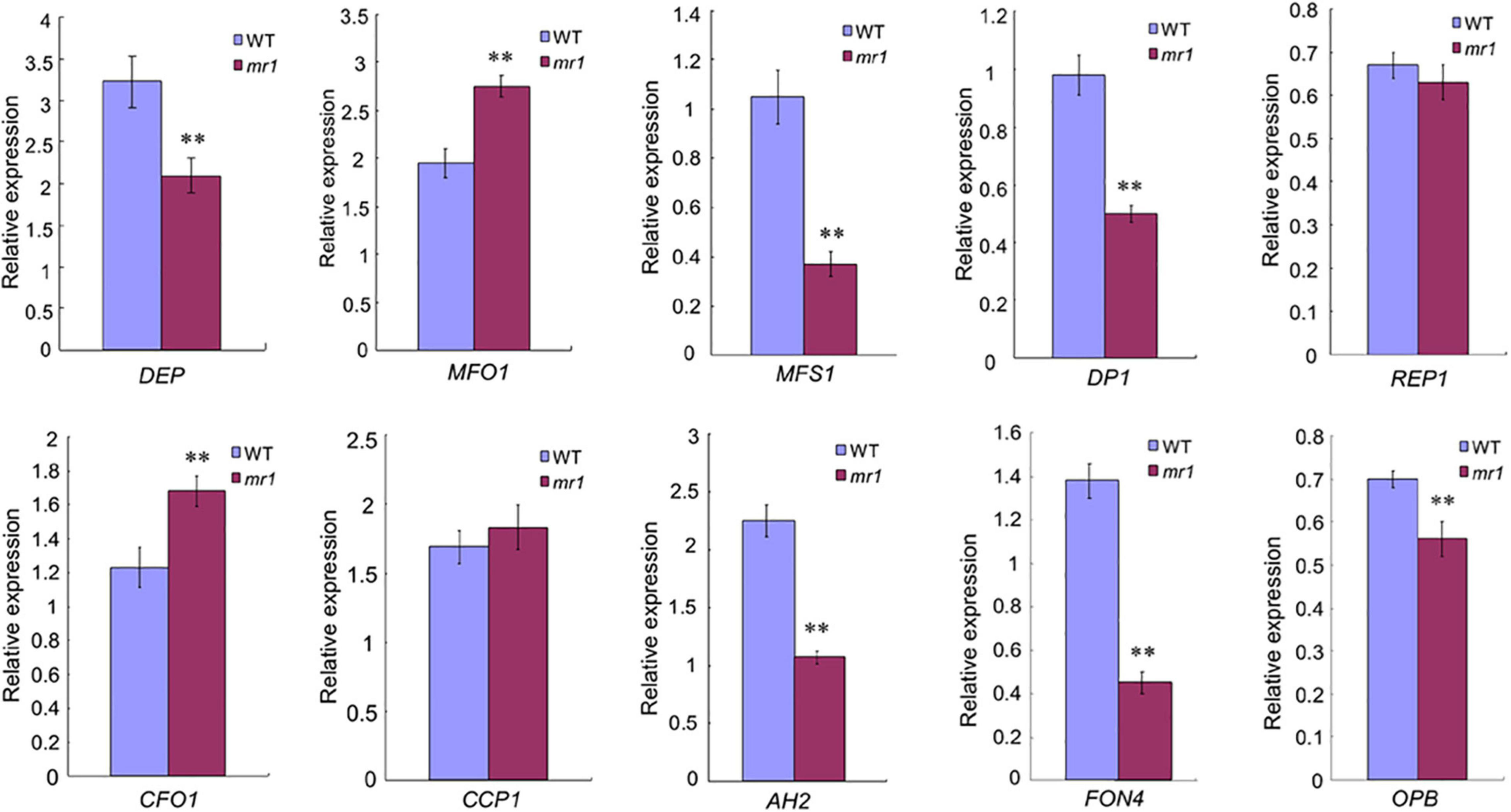
Figure 6. Expression analysis of palea identity-associated genes by qRT-PCR. **Significant difference at p < 0.01 compared with the wild-type by Student’s t-test. Error bars indicate SD.
mr1 Changed Grain Size and Quality
The length and 1,000-grain weight of grains in the mr1 mutant were significantly decreased, but the grain width was not changed than that of the wild-type, respectively (Figures 7A–G). However, the length, width, and 1,000-grain weight of brown rice in the mr1 mutant were significantly reduced than that of wild-type, respectively (Figures 7E–J). We examined several genes that determined grain size by modulating cell proliferation and expansion (Yu et al., 2020; Hao et al., 2021). Compared with the wild-type, the transcript levels of BIG1, BIG2, GW2, GS3, GL3, GIF1, and BSG1, were significantly altered in the dls1 mutant (Supplementary Figure 1), implying that the mutated MR1 disturbed the expressions of grain size-related genes.
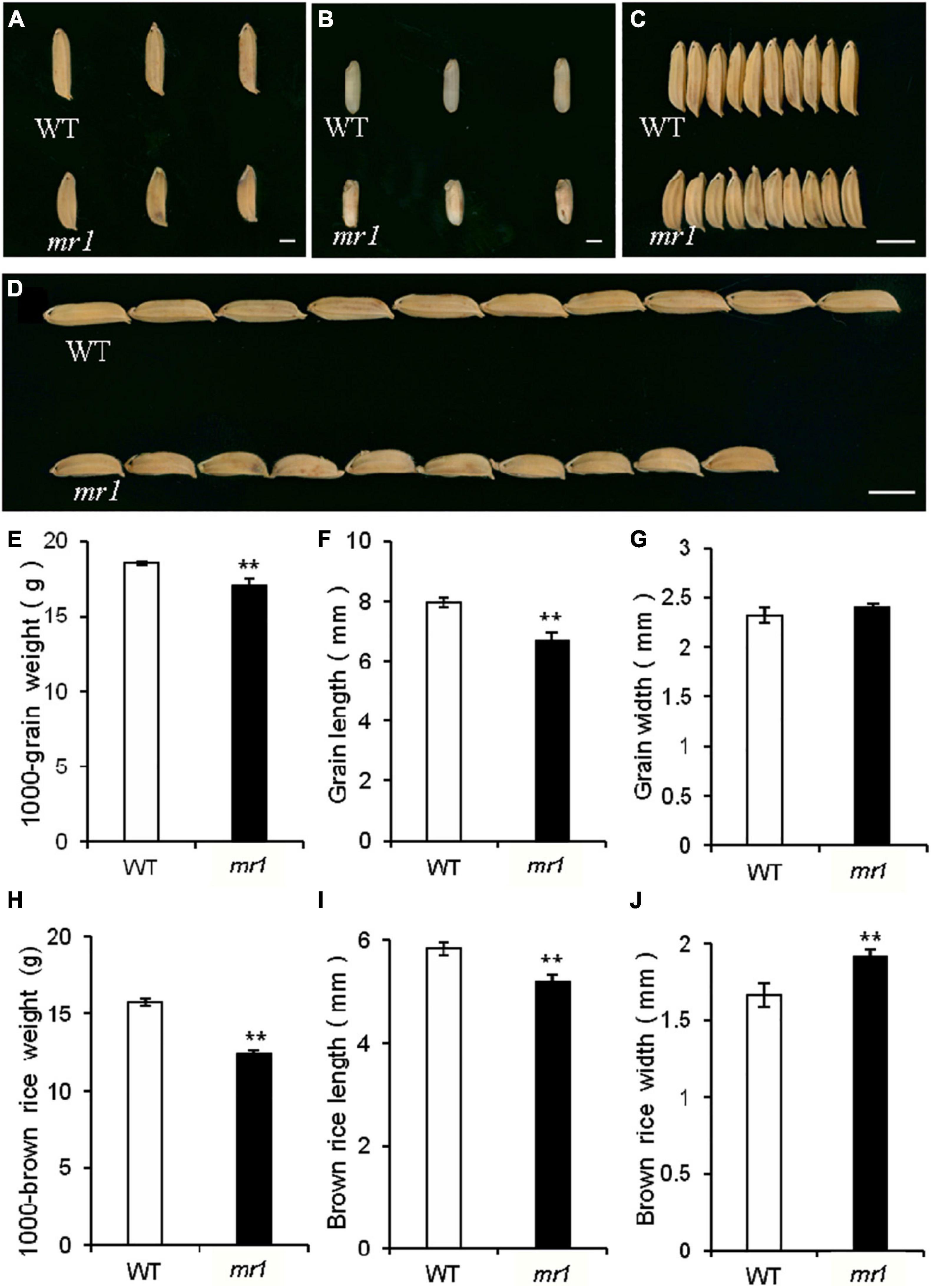
Figure 7. Grains of the wild-type and the mr1 mutant. (A) Grains of the wild-type and mr1 mutant. (B) Brown rice of the wild-type and mr1 mutant. (C–D) Grains of the wild-type and mr1 mutant. (E–J) Length, width, and weight of grains and brown rice in the wild-type and mr1 mutant. Scale bar, 2 mm in (A,B); 5 mm in (C,D). **Significant difference at p < 0.01 compared with the wild-type by Student’s t-test. Error bars indicate SD.
We also investigated several physicochemical properties of starch (Figures 8A–K). In the mr1 mutant, the amylose content was increased, and the soluble sugar content was decreased that of the wild-type, respectively (Figures 8H,I). And, the total starch and total protein contents of mr1 grains were not influenced than that of the wild-type, respectively (Figures 8G,J). The starch solubility in urea solutions was also measured to examine the gelatinization properties. The powered rice samples were mixed with various concentrations (0–9 mol L–1) of urea solutions. The results exhibited that there are obvious differences in (3–9) mol L–1 urea between the wild-type and mr1 starch, and the mr1 starch was more difficult to gelatinize than the wild-type starch (Figure 8K). We could conclude that the physicochemical properties of starch were obviously different between the wild-type and mr1 endosperm, indicating that MR1 could affect grain quality.
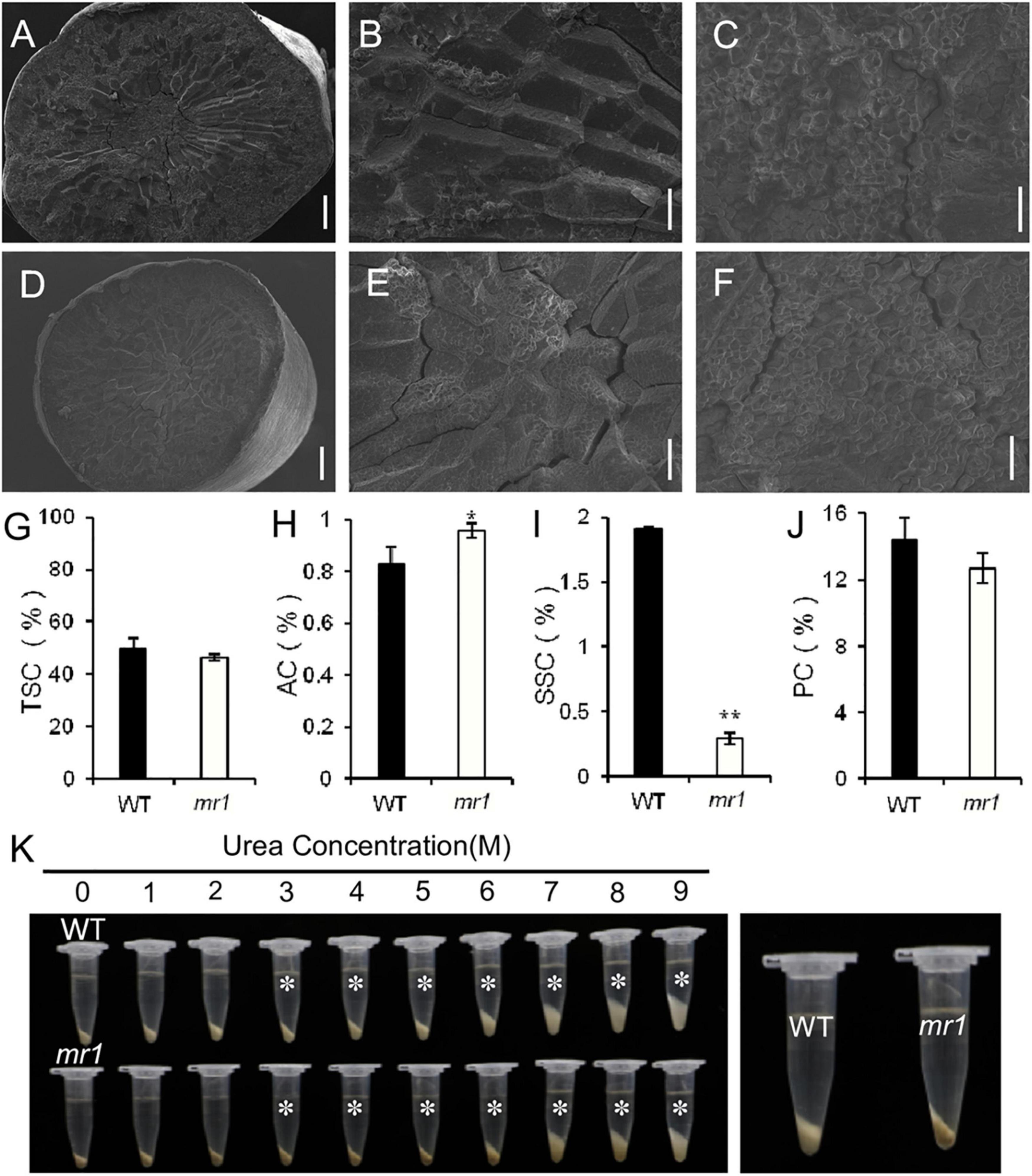
Figure 8. Characteristics of grain and starch in the wild-type and mr1 mutant. (A–F) SEM analysis of transverse sections of brown rice in the wild-type and mr1 mutant. (A–C) Brown rice of the wild-type. (D–F) Brown rice of the mr1 mutant. (G–J) Total starch, amylose, soluble sugar, and protein content in the wild-type and mr1 mutant. (K) Gelatinization characteristics of starch in urea solutions (1–9 M). Asterisks indicate the starch of mr1 endosperm is more difficult to gelatinize in 3–9 M urea solution than that of WT. The most significant difference was observed for 3 M urea (right). Bars, 500 μm in (A,D); 100 μm in (B,E); 20 μm in (C,F). *Significant difference at p < 0.05 compared with the wild-type by Student’s t-test. **Significant difference at p < 0.01 compared with the wild-type by Student’s t-test. Error bars indicate SD.
Discussion
MR1 Regulates Palea Development
As an indispensable part of developmental biology, flower development has been widely studied, and the ABCDE model, which successfully interprets the formation of four whorls in flower, has also been widely accepted. Nevertheless, grasses have highly specialized flowers and inflorescence structures, which makes it hard to explain the process of flower development with the knowledge of other model plants. In this study, the palea of mr1 mutant was reduced while all the other floral organs (pistil, lodicule, and lemma) were still normal except stamen number. Polymorphic markers were used to locate the gene related to the mutation on the long arm of chromosome 1, and there is no report about the related genes of mutation character is located near this position. Therefore, we suggested that MR1 may be a new gene related to the development of palea in rice.
The origin and identity of the palea and lemma remain a topic of heated discussion. There are some genes that affect the development of both palea and lemma. For example, DEGENERATED HULL1 (DH1) encodes a protein with a LOB domain. In the dh1 mutant, about most florets could not develop normal lemma and palea, but degenerated into membranous or fibrous organs, and with the loss of lodicules, stamens, and pistils (Li et al., 2008). TRIANGULAR HULL1 (TH1) gene encodes a DUF640 domain transcription factor, the development of palea and lemma was affected both in transverse and longitudinal direction, and they became smaller and thicker in the th1-1 mutant (Li et al., 2012). Oryza sativa JAGGED (OsJAG) encodes a C2H2 zinc finger domain transcription factor, the loss of function leads to the malformation of all floral organs, such as lemma and palea degenerated and bent inward, lodicules elongated, pistils deformed, and all stamens became pistil-like organs (Duan et al., 2010). In our study, the mr1 mutant specially affected palea development, but the lemma development is not influenced, so we speculate that although these two organs have some similarities in morphology, they have different identities and may be regulated by different genetic pathways. Firstly, the palea was usually considered to be a prophyll, and the flower occurs in the axil of it (Ren et al., 2013), and the lemma was widely considered to be a true bract surrounding the spikelet (Luo et al., 2005; Zheng et al., 2015). Secondly, they were different in structure and size. The palea had three vascular bundles, while the lemma had five, so the palea was smaller than the lemma. And the palea was composed of bop and mrp, while the lemma had only one component. In addition, there were several genes, such as OsMADS15, MFO1, DP1, and CFO1, showing that there are lots of genes which only affected the development of palea, but not that of lemma. All these facts are consistent with our idea.
Furthermore, in a barley mutant called leafy lemma, its lemma was completely transformed into a vegetative leaf with ligule, blade, and sheath, while the palea and other floral organs did not change at all, suggesting that the lemma in grass may not be part of the floret perianth (Pozzi et al., 2000). And in the same barley mutant, the palea and lemma exhibited different phenotypes, indicating that palea and lemma are not homologous in barley. In rice, there is a floral organ identity gene SUPERWOMAN1 (SPW1), it belongs to class B genes and is homologous to APETALA3 in Arabidopsis. The lodicules of spw1 mutant transformed into palea-like organs instead of lemma (Nagasawa et al., 2003), which indicating that the palea of rice may be equivalent to the sepal of eudicot in evolution. Here, in the mr1 florets, the palea was reduced, while the lemma maintained its identity, which seemed to confirm the view that the palea and lemma have different origins. However, some other researchers believed that the palea, and even the lemma, should be equivalent to the sepals of eudicot (Ambrose et al., 2000; Goto et al., 2001). Now, there is not corresponding mutants to support such a hypothesis, so it is still not confirmed.
There are various bop or mrp defective mutants in previous studies. MOSAIC FLORAL ORGANS1 (MFO1) and CHIMERIC FLORAL ORGANS1 (CFO1) both belong to the MADS-box genes, their mutants showed enlarged and silicified mrp, which makes the mutant palea look like bop or lemma-like organ (Ohmori et al., 2009; Sang et al., 2012). RETARDED PALEA1 (REP1) belongs to TCP gene family, in the rep1 mutant, the bop was decreased and the development of it was delayed, but in the REP1 overexpression plants, the mrp overgrew (Yuan et al., 2009). DEPRESSED PALEA1 (DP1) encodes an AT-hook DNA-binding protein, MULTI-FLORET SPIKELET1 (MFS1) encodes an AP2/ERF protein, both the bop of dp1 and mfs1 mutant was completely lost, and only two marginal regions were preserved (Jin et al., 2011; Ren et al., 2013). In recent studies, ABNORMAL HULL2 (AH2) and FLORAL ORGAN NUMBER4 (FON4) were proved to be able to identify the bop (Ren et al., 2019a,b). In the ah2 and fon4 mutants, the bop size was changed, while the mrp was not altered. In our study, detailed observations revealed that the mr1 mutant bore an enlarged mrp and a reduced bop, which suggests that the bop was partly converted to mrp, and MR1was required for bop and mrp identity. Molecular evidences also showed that the small bop of mr1 mutant indeed had the mrp identity. These findings suggested that the bop and mrp were homologous organs and a fused organ. Together, more mutants will be needed to further confirming the relationship of the lemma, bop, and mrp.
In this study, the MR1 gene was fine mapped on chromosome 1 with a physical distance of 70 kb. In this region, there were eight genes, and none of them had been cloned to be related to palea development. But according to the results of sequencing and alignment of these ORF genes between the SKZ and mr1 mutant, we failed to identify the MR1 gene. So we speculated that the mr1 mutant phenotypes might be caused by the intergenic enhancer or repressor and the alteration of promoter sequence or gene expression in the target region. Obviously, this hypothesis needed more experimental evidences to be verified. For instance, we can sequence and compare the promoter regions or 5′ and 3′ UTR sequences, and detect the expression levels of candidate genes. We believe that the further cloning and functional analysis of the MR1 gene would be helpful to enrich the molecular mechanism of rice palea development, and provide abundant evidences to reveal the relationship between its hull (palea and lemma) and the floral organs of eudicots, as well as the evolutionary relationship between the bop, mrp, and lemma.
MR1 Affects Grain Size and Quality
We also observed the mr1 mutant produced smaller defective grains compared with the wild-type. Several known genes that control grain size by modulating cell proliferation and expansion have been reported. BIG1, BIG2, GW2, GS3, GL3, GIF1, and BSG1 transcripts were changed in the mr1 mutant than in the wild-type. In addition, we found that the mr1 grains had low SSC, and high AC and the mr1 starch was more difficult to gelatinize than that of the wild-type. These findings revealed that the mutation of MR1 reduced the grain size and quality, which may be caused by defective paleae. Taken together, these results suggested that MR1 had a possible effort to improve the grain yield and quality.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
DR designed the research. DR, WX, WL, XY, and DZ performed the research. DR and WX wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (32071993 and 91735304).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.864099/full#supplementary-material
Footnotes
References
Agrawal, G. K., Abe, K., Yamazaki, M., Miyao, A., and Hirochika, H. (2005). Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol. Biol. 59, 125–135. doi: 10.1007/s11103-005-2161-y
Ambrose, B. A., Lerner, D. R., Ciceri, P., Padilla, C. M., Yanofsky, M. F., and Schmidt, R. J. (2000). Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5, 569–579. doi: 10.1016/s1097-2765(00)80450-5
Angenent, G. C., Franken, J., Busscher, M., van Dijken, A., van Went, J. L., Dons, H. J., et al. (1995). A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell 7, 1569–1582. doi: 10.1105/tpc.7.10.1569
Cai, Y. C., Li, S. F., Jiao, G. A., Sheng, Z. H., Wu, Y. W., Shao, G. N., et al. (2018). OsPK2 encodes a plastidic pyruvate kinase involved in rice endosperm starch synthesis, compound granule formation and grain filling. Plant Biotechnol. J. 16, 1878–1891. doi: 10.1111/pbi.12923
Chung, Y. Y., Kim, S. R., Finkel, D., Yanofsky, M. F., and An, G. (1994). Early flowering and reduced apical dominance result from ectopic expression of a rice MADS box gene. Plant Mol. Biol. 26, 657–665. doi: 10.1007/BF00013751
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Colombo, L., Franken, J., Koetje, E., van Went, J., Dons, H. J., Angenent, G. C., et al. (1995). The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 7, 1859–1868. doi: 10.1105/tpc.7.11.1859
Duan, Y. L., Diao, Z. J., Liu, H. Q., Cai, M. S., Wang, F., Lan, T., et al. (2010). Molecular cloning and functional characterization of OsJAG gene based on a complete-deletion mutant in rice (Oryza sativa L.). Plant Mol. Biol. 74, 605–615. doi: 10.1007/s11103-010-9703-7
Favaro, R., Pinyopich, A., Battaglia, R., Kooiker, M., Borghi, L., Ditta, G., et al. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15, 2603–2611. doi: 10.1105/tpc.015123
Fornara, F., Parenicová, L., Falasca, G., Pelucchi, N., Masiero, S., Ciannamea, S., et al. (2004). Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes. Plant Physiol. 135, 2207–2219. doi: 10.1104/pp.104.045039
Gao, X. C., Liang, W. Q., Yin, C. S., Ji, S. M., Wang, H. M., Su, X., et al. (2010). The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 153, 728–740. doi: 10.1104/pp.110.156711
Goto, K., and Meyerowitz, E. M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560. doi: 10.1101/gad.8.13.1548
Goto, K., Kyozuka, J., and Bowman, J. L. (2001). Turning floral organs intoleaves, leaves into floral organs. Curr. Opin. Genet. Dev. 11, 449–456. doi: 10.1016/s0959-437x(00)00216-1
Hao, J. Q., Wang, D. K., Wu, Y. B., Huang, K., Duan, P. G., Li, N., et al. (2021). The GW2-WG1-OsbZIP47 pathway controls grain size and weight in rice. Mol. Plant 14, 1266–1280. doi: 10.1016/j.molp.2021.04.011
Ikeda, K., Sunohara, H., and Nagato, Y. (2004). Developmental course of inflorescence and spikelet in rice. Breed. Sci. 54, 147–156. doi: 10.1270/jsbbs.54.147
Jeon, J. S., Jang, S., Lee, S., Nam, J., Kim, C., Lee, S. H., et al. (2000). leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12, 871–884. doi: 10.1105/tpc.12.6.871
Jin, Y., Luo, Q., Tong, H. N., Wang, A. J., Cheng, Z. J., Tang, J. F., et al. (2011). An AT-hook gene is required for palea formation and floral organ number control in rice. Dev. Biol. 359, 277–288. doi: 10.1016/j.ydbio.2011.08.023
Kang, H. G., Park, S., Matsuoka, M., and An, G. (2005). White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J. 42, 901–911. doi: 10.1111/j.1365-313X.2005.02423.x
Kater, M. M., Dreni, L., and Colombo, L. (2006). Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J. Exp. Bot. 57, 3433–3444. doi: 10.1093/jxb/erl097
Li, A., Zhang, Y., Wu, X., Tang, W., Wu, R., Dai, Z., et al. (2008). DH1, a LOB domain-like protein required for glume formation in rice. Plant Mol. Biol. 66, 491–502. doi: 10.1007/s11103-007-9283-3
Li, X. J., Sun, L. J., Tan, L. B., Liu, F. X., Zhu, Z. F., Fu, Y. C., et al. (2012). TH1, a DUF640 domain-like gene controls lemma and palea development in rice. Plant Mol. Biol. 78, 351–359. doi: 10.1007/s11103-011-9868-8
Liu, L. L., Ma, X. D., Liu, S. J., Zhu, C. L., Jiang, L., Wang, Y. H., et al. (2009). Identification and characterization of a novel Waxy allele from a Yunnan rice landrace. Plant Mol. Biol. 71, 609–626. doi: 10.1007/s11103-009-9544-4
Luo, Q., Zhou, K. D., Zhao, X. F., Zeng, Q. C., Xia, H. A., Zhai, W. X., et al. (2005). Identification and fine mapping of a mutant gene for palealess spikelet in rice. Planta 221, 222–230. doi: 10.1007/s00425-004-1438-8
Malcomber, S. T., and Kellogg, E. A. (2005). SEPALLATA gene diversification: brave new whorls. Trends Plant Sci. 10, 427–435. doi: 10.1016/j.tplants.2005.07.008
Mandel, M. A., Gustafson-Brown, C., Savidge, B., and Yanofsky, M. F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. doi: 10.1038/360273a0
Mizukami, Y., and Ma, H. (1992). Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71, 119–131. doi: 10.1016/0092-8674(92)90271-d
Nagasawa, N., Miyoshi, M., Sano, Y., Satoh, H., Hirano, H., Sakai, H., et al. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130, 705–718. doi: 10.1242/dev.00294
Ohmori, S., Kimizu, M., Sugita, M., Miyao, A., Hirochika, H., Uchida, E., et al. (2009). MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21, 3008–3025. doi: 10.1105/tpc.109.068742
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., and Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Pelucchi, N., Fornara, F., Favalli, C., Masiero, S., Lago, C., and Pè Enrico, M. (2002). Comparative analysis of rice MADS-box genes expressed during flower development. Sexual Plant Reproduction 15, 113–122. doi: 10.1007/s00497-002-0151-7
Pozzi, C., Faccioli, P., Terzi, V., Stanca, A. M., Cerioli, S., Castiglioni, P., et al. (2000). Genetics of mutations affecting the development of a barley floral bract. Genetics 154, 1335–1346. doi: 10.1093/genetics/154.3.1335
Prasad, K., Sriram, P., Kumar, C. S., Kushalappa, K., and Vijayraghavan, U. (2001). Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev. Genes Evol. 211, 281–290. doi: 10.1007/s004270100153
Ren, D. Y., Li, Y. F., Zhao, F. M., Sang, X. C., Shi, J. Q., Wang, N., et al. (2013). MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice. Plant Physiol. 162, 872–884. doi: 10.1104/pp.113.216044
Ren, D. Y., Xu, Q. K., Qiu, Z. N., Cui, Y. J., Zhou, T. T., Zeng, D. L., et al. (2019a). FON4 prevents the multi-floret spikelet in rice. Plant Biotechnol. J. 17, 1007–1009. doi: 10.1111/pbi.13083
Ren, D. Y., Cui, Y. J., Hu, H. T., Xu, Q. K., Rao, Y. C., Yu, X. Q., et al. (2019b). AH2 encodes a MYB domain protein that determines hull fate and affects grain yield and quality in rice. Plant J. 100, 813–824. doi: 10.1111/tpj.14481
Ren, D. Y., Yu, H. P., Rao, Y. C., Xu, Q. K., Zhou, T. T., Hu, J., et al. (2018). ‘Two-floret spikelet’ as a novel resource has the potential to increase rice yield. Plant Biotechnol. J. 16, 351–353. doi: 10.1111/pbi.12849
Sang, X. C., Li, Y. F., Luo, Z. K., Ren, D. Y., Fang, L. K., Wang, N., et al. (2012). CHIMERIC FLORAL ORGANS1, encoding a monocot-specific MADS box protein, regulates floral organ identity in rice. Plant Physiol. 160, 788–807. doi: 10.1104/pp.112.200980
Theissen, G., and Saedler, H. (2001). Plant biology. floral quartets. Nature 409, 469–471. doi: 10.1038/35054172
Wang, K. J., Tang, D., Hong, L. L., Xu, W. Y., Huang, J., Li, M., et al. (2010). DEP and AFO regulate reproductive habit in rice. PloS Genet 6:e1000818. doi: 10.1371/journal.pgen.1000818
Weigel, D., and Meyerowitz, E. M. (1994). The ABCs of floral homeotic genes. Cell 78, 203–209. doi: 10.1016/0092-8674(94)90291-7
Xu, Q. K., Yu, X. Q., Cui, Y. J., Xia, S. S., Zeng, D. L., Qian, Q., et al. (2021). LRG1 maintains sterile lemma identity by regulating OsMADS6 expression in rice. Sci. China Life Sci. 7, 1190–1192. doi: 10.1007/s11427-020-1816-x
Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldmann, K. A., and Meyerowitz, E. M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. doi: 10.1038/346035a0
Yu, X., Xia, S., Xu, Q., Cui, Y., Gong, M., Zeng, D., et al. (2020). ABNORMAL FLOWER AND GRAIN 1 encodes OsMADS6 and determines palea identity and affects rice grain yield and quality. Sci. China Life Sci. 63, 228–238. doi: 10.1007/s11427-019-1593-0
Yuan, Z., Gao, S., Xue, D. W., Luo, D., Li, L. T., Ding, S. Y., et al. (2009). RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol. 149, 235–244. doi: 10.1104/pp.108.128231
Keywords: rice (Oryza sativa L.), mr1 mutant, palea, organ origin, grain size and quality
Citation: Xie W, Liu W, Yu X, Zeng D and Ren D (2022) Fine Mapping of Rice Specific MR1, a Gene Determines Palea Identity. Front. Plant Sci. 13:864099. doi: 10.3389/fpls.2022.864099
Received: 04 February 2022; Accepted: 17 February 2022;
Published: 24 May 2022.
Edited by:
Ran Xu, Hainan University, ChinaReviewed by:
Jian Sun, Shenyang Agricultural University, ChinaBanpu Ruan, Hangzhou Normal University, China
Copyright © 2022 Xie, Liu, Yu, Zeng and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Deyong Ren, rendeyong616@163.com
†These authors have contributed equally to this work
 Wei Xie†
Wei Xie† Dali Zeng
Dali Zeng Deyong Ren
Deyong Ren