- 1Center of Plant Molecular Biology (ZMBP), University of Tübingen, Tübingen, Germany
- 2Plant and Crop Sciences Division, School of Biosciences, University of Nottingham, Sutton Bonington Campus, Loughborough, United Kingdom
- 3School of Life Sciences, Huaiyin Normal University, Huai'an, China
- 4Department of Molecular Genetics and Biotechnology, Yuriy Fedkovych Chernivtsi National University, Chernivtsi, Ukraine
- 5Laboratory of Molecular Epigenetics, Institute of Biophysics, Academy of Sciences of the Czech Republic, Brno, Czechia
The history of rDNA research started almost 90 years ago when the geneticist, Barbara McClintock observed that in interphase nuclei of maize the nucleolus was formed in association with a specific region normally located near the end of a chromosome, which she called the nucleolar organizer region (NOR). Cytologists in the twentieth century recognized the nucleolus as a common structure in all eukaryotic cells, using both light and electron microscopy and biochemical and genetic studies identified ribosomes as the subcellular sites of protein synthesis. In the mid- to late 1960s, the synthesis of nuclear-encoded rRNA was the only system in multicellular organisms where transcripts of known function could be isolated, and their synthesis and processing could be studied. Cytogenetic observations of NOR regions with altered structure in plant interspecific hybrids and detailed knowledge of structure and function of rDNA were prerequisites for studies of nucleolar dominance, epistatic interactions of rDNA loci, and epigenetic silencing. In this article, we focus on the early rDNA research in plants, performed mainly at the dawn of molecular biology in the 60 to 80-ties of the last century which presented a prequel to the modern genomic era. We discuss – from a personal view – the topics such as synthesis of rRNA precursor (35S pre-rRNA in plants), processing, and the organization of 35S and 5S rDNA. Cloning and sequencing led to the observation that the transcribed and processed regions of the rRNA genes vary enormously, even between populations and species, in comparison with the more conserved regions coding for the mature rRNAs. Epigenetic phenomena and the impact of hybridization and allopolyploidy on rDNA expression and homogenization are discussed. This historical view of scientific progress and achievements sets the scene for the other articles highlighting the immense progress in rDNA research published in this special issue of Frontiers in Plant Science on “Molecular organization, evolution, and function of ribosomal DNA.”
Introduction
Cytologists in the twentieth century recognized the nucleolus as a common structure in all eukaryotic cells, using both light and electron microscopy. During the winter of 1931, Barbara McClintock observed that in interphase nuclei of maize the nucleolus was formed in association with a specific chromosomal region, normally located at the end of chromosome 6, which she called the nucleolar organizer region (NOR; McClintock, 1934). It took, however, almost 40 years before the composition of the NOR was deciphered (i.e., rRNA genes) with the aid of chromosome in situ hybridization techniques in the seventies (Gall, 1981). The molecular structure and function of the ribosomal RNA genes, which are located in or around the nucleolus, were analyzed after the structure of DNA, the triplet code and the central dogma “DNA makes RNA makes protein” were established. In early days of rDNA research, general molecular biological principles were being established rapidly. For example, experiments with cultured cells showed that radioactive amino acids were first polymerized in the cytosol, in association with ribosomes, which identified them as the subcellular sites of protein synthesis. The frog Xenopus became a crucial model system for studying the ribosomal RNA genes (rDNA) when Brown and Gurdon (1964) showed that the arrested development and eventual death of Xenopus anucleolate mutant embryos was due to their inability to make new rRNA. Birnstiel et al. (1966) demonstrated that these mutants lacked the rDNA, which was subsequently isolated from the wild type and shown to be composed of multiple copies of alternating 18S and 28S rDNA cistrons (Birnstiel et al., 1968). Distinct plant ribosomal DNA satellites had been noted around the same time by Matsuda and Siegel (1967) and characterized subsequently by Goldberg et al. (1972) and Bendich and Anderson (1974). In the mid to late 1960s the synthesis of nuclear-encoded rRNA was the only system in multicellular organisms where transcripts of known function could be isolated, and their synthesis and processing could be studied.
In this retrospective article, we review research carried out in plants from the 60 to 80-ties of the last century focusing on nuclear encoded ribosomal RNA genes as a prequel to recent genomic activities (reviewed in Volkov et al., 2004, 2007; Layat et al., 2012; Sáez-Vásquez and Delseny, 2019; Appels et al., 2021 – this issue). The continuing rise in publications on rDNA in plants witnessed over the past 60 years (Figure 1) is testimony to its continuing importance.
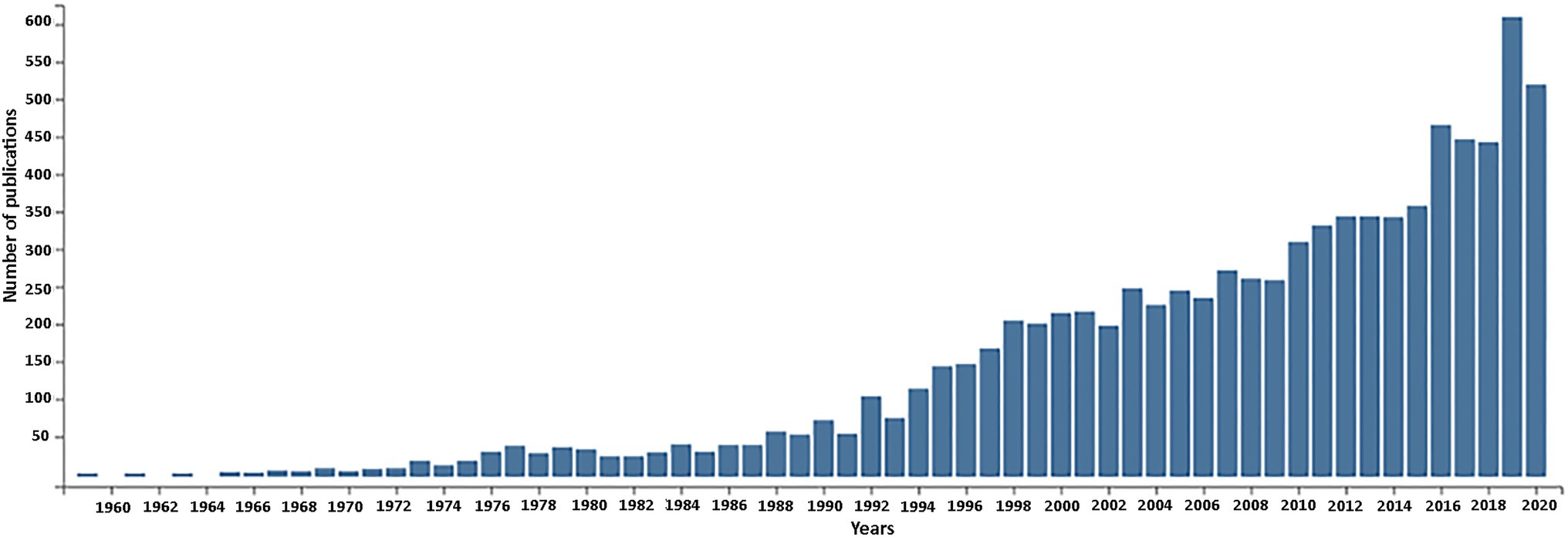
Figure 1. Publications related to plant nuclear rDNA research over the timespan of 1960–2020. The number of retrieved publications is shown in 2-year increments. The total number of publications returned by the Web of Science database was 8,948. Key words used for searched fields: rRNA or rDNA and plant with following filters: no animal, no fungal, no chloroplast (plastom), no mitochondrion.
General Organization of Nuclear-Encoded 35S rDNA, rRNA Precursor, and 5S rDNA: The Beginning of Plant Molecular Research
Many scientists in the 1960s felt that the existence of the tough plant cell wall surrounding a small amount of cytoplasm and a large vacuole containing many secondary products made it very difficult to carry out molecular research on plants. This was certainly felt by some people in the Max Planck Society in Germany and also in the United Kingdom and United States, but plant molecular research began to flourish as methods to overcome these difficulties were developed. Research had been restarted rapidly at the University of Tübingen after the end of World War II. At the Botanical Institute of the University, most people were engaged in studies of the “Biological Clock” and circadian rhythms with Prof. Erwin Bünning (a leading plant physiologist and one of the founders of plant research in Tübingen, (Chandrashekaran, 2006), when a new assistant and later lecturer, Gerhard Richter, arrived. He had spent a research stay performing molecular research in the lab of the biochemist James F. Bonner in Pasadena, the United States. At that time in Tübingen, people from the Max Planck Institute were involved in codon studies and how messenger RNA transported the “Bauanleitung” (“contruction manual”) of proteins to the ribosomes, the sites for protein synthesis. So, the environment was prepared for molecular biology, and Gerhard Richter had no problem in convincing Prof. Bünning to establish new laboratories for this kind of research and, most importantly, for working with radioactive substances. Vera Hemleben (VH) thought it would be interesting to work with higher plants and to study nucleic acid synthesis in dark-grown seedlings of beans which could be cultivated under semi-sterile conditions. Other PhD students got involved, and 32P-phosphate radioactively labeled nucleic acids were isolated and separated on MAK [methylated albumin on Kieselgur (“silica”)] columns; we isolated the ribosomal RNA fractions and determined the GC-content of the 18S and 25S rRNA (Hemleben-Vielhaben, 1966). Other researchers, e.g., Joe Key now in Athens, Georgia, did similar work (Leaver and Key, 1970). VH decided later to work with Lemna perpusilla, the small aquatic monocot plant, which could be cultivated under completely sterile conditions. This was necessary for radioactive pulse-labeling and pulse-chase experiments to follow the fate of the newly synthesized RNA and to characterize the nuclear encoded rRNA precursor, which was 2.3 × 106 Da in size (Hemleben, 1972). From the late 1960s the polycistronic transcription unit, the rRNA precursor, was studied in several different eukaryotic systems. Studies with animals established that the rRNA genes were transcribed as a polycistronic precursor, of variable size in different organisms (45S in humans and 35S in plants), which subsequently underwent endonucleolytic cleavage and methylation before being incorporated, together with ribosomal proteins, into nascent ribosome subunits and transported from the nucleolus to the cytosol (Figure 2). Cytosolic ribosomes, formed by 18S (in the small 40S ribosome subunit), 5.8S and 25S plus 5S rRNA (in the large 60S subunit) and ribosomal proteins, were isolated and were, of course, essential constituents in the popular wheat germ in vitro protein synthesis system.
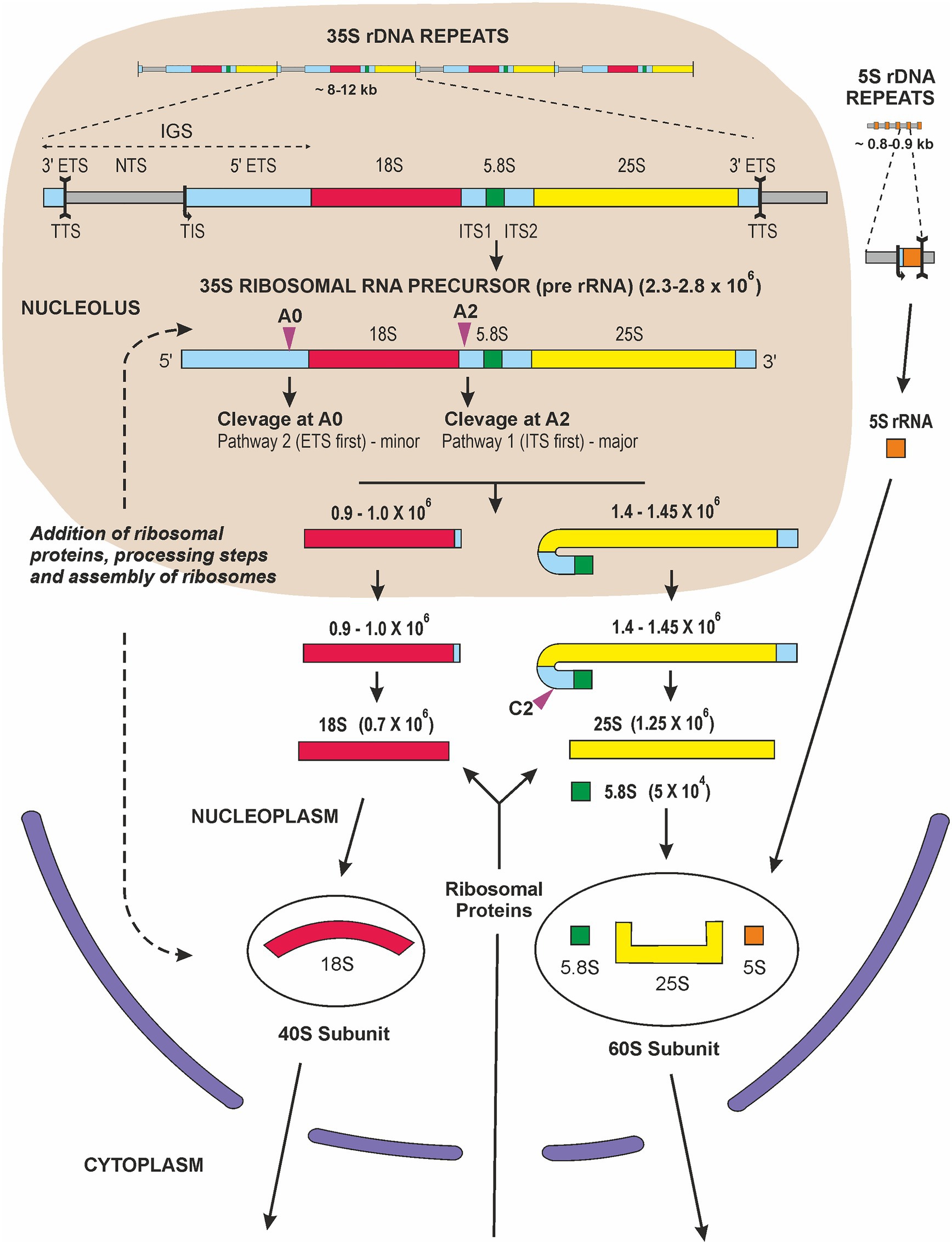
Figure 2. Graphic representation of ribosome biogenesis in eukaryotic cells (adapted from (Grierson, 1984; Sáez-Vásquez and Delseny, 2019). Transcription of rDNA requires RNA Pol I activity and a subset of general transcription factors. The primary transcript, precursor-rRNA (35S in plants and yeast or 45S in mammals), encodes three rRNAs and is first co-transcriptionally processed into the mature 18S, 5.8S, and 25S/28S rRNA. This processing steps involve multiple endonucleolytic and exonucleolytic cleavages (violet arrowheads) occurring in the nucleolus [A0 site or P site according to nomenclature of (Sáez-Vásquez and Delseny, 2019)], A2 site and the nucleoplasm (C2). The 18S rRNAs assemble with ribosomal proteins of the small 40S ribosomal subunit, RPSs, while 5.8S, 25S/28S and 5S rRNA assemble with ribosomal proteins, RPLs, forming the large 60S ribosomal subunit. The 5S rRNA is transcribed in the nucleoplasm by RNA Pol III and imported into the nucleolus. Assembly and transport of ribosomal particles from nucleolus to cytoplasm requires hundreds of specific 40S and 60S RBFs (Ribosome Biogenesis Factors). The 40S and 60S ribosomal subunits finally join to form translationally competent ribosomes in the cytoplasm. Sizes of individual rRNA molecules are in daltons.
In September 1968, Don Grierson joined Ulrich Loening’s laboratory in Edinburgh to study the synthesis of rRNA in primary leaves of mung bean seedlings (Grierson et al., 1970; Grierson and Loening, 1972, 1974). Ulrich had developed methods for extracting undegraded RNA, fractionating it on the basis of size in polyacrylamide gels and determining the molecular weights with great precision (Loening, 1969). Ulrich built electrophoresis tanks from Perspex sheets, with platinum wire electrodes, insisted on redistilling phenol for use in RNA extraction, recrystallizing acrylamide and using deoxygenated monomer solutions to get consistent polymerization and electrophoresis results. Disposable plastic ware and automatic microsyringes were the subject of dreams. Ulrich also devised a novel gel-scanner and apparatus for slicing gels and automated detection of radioactivity in the slices. These methods generated worldwide interest and attracted collaborators and many visitors to the laboratory. The synthesis and processing of a polycistronic precursor (pre-rRNA) was studied in several plants, including mung bean leaves and roots (Grierson et al., 1970; Grierson and Loening, 1974), pea roots and cultured artichoke cells (Fraser and Loening, 1974), carrot (Leaver and Key, 1970), cultured sycamore cells (Cox and Turnock, 1973). In general, the observations were similar: when seedlings, plant tissues, or organs were incubated with 3H-uridine or 32P-phosphate for short periods of time the radioactivity was incorporated into distinct macromolecular transcripts. On polyacrylamide gels these could be seen above a polydisperse array of RNA molecules, presumably mRNAs, nascent molecules and processing products. Molecular weight estimates for the largest molecules ranged from 2.3 to 2.8 × 106 Da. At slightly later times, radioactivity was also detected in molecules of around 1.4 and 0.75 × 106 Da and pulse-chase experiments, kinetics of accumulations and comparison of the nucleotide composition of these molecules all supported the conclusion that the initial transcript was a large polycistronic molecule that included one 18S, 5.8S and 25S transcript, together with “transcribed spacer” RNA. Subsequent ribonucleolytic cleavage gave rise to the immediate rRNA precursors, each slightly larger than the mature rRNAs. Aggregates and breakdown products of rRNAs could also be distinguished (Grierson and Loening, 1974), and electrophoresis under denaturing conditions in formamide gels gave lower molecular weight estimates of the size of the initial transcripts in carrot, parsley, and sycamore (Seitz and Seitz, 1979). The size characterization of individual pre-RNA molecules and the complexities of the rRNA maturation pathway stimulated further research. The processing of plant pre-rRNA, updated to show recent findings, is illustrated in Figure 2 and the enzymes catalyzing individual RNA cleavage steps (A0, A1, A2 and C2 sites) have now been identified (Tomecki et al., 2017). Moreover, plant small nucleolar RNA (snoRNAs) that are thought to take part in pre-rRNA cleavage events were identified (Brown and Shaw, 1998). It became clear that the cleavage events are compartmentalized, some occurring in the nucleolus (A0, and A2) with others take place in the nucleoplasm (C2; Figure 2). Finally, although key steps of plant pre-rRNA processing seem to be similar to that of other eukaryotes (Grierson, 1984) notable differences exist between yeast, plant and animal pre-RNA pathways. For example, the analysis of ribosome biogenesis in plants revealed two alternative processing pathways coexisting in plants (Weis et al., 2015; Sáez-Vásquez and Delseny, 2019). A major pathway 1 is initiated by ITS1 cleavage (A2 site, Figure 2) and subsequent removal of the 5'-ETS, which is comparable to the human processing pathway. Pathway 2 starts with the 5'-ETS removal (cleavage at the A0 site, Figure 2) followed by the ITS1 cleavage which leads to the separated assembly of the pre-40S and pre-60S ribosomal subunits. This pathway is reminiscent of rRNA processing in yeast. In addition, 5'-ETS processing is initiated by exoribonucleolytic trimming of the 5'-end by XRNs in Arabidopsis thaliana (Zakrzewska-Placzek et al., 2010).
Other rRNA transcription units were found transcribed from the DNA in chloroplasts, containing their smaller 16S and 23S rRNAs, components of the distinct 70S ribosomes, and a further class of ribosomes in the mitochondria, although their synthesis was not studied in such detail. Double labeling experiments showed a stable polycistronic precursor of rRNA in leaves, which was distinct from, and larger than that in roots (Grierson and Loening, 1972). This RNA species was subsequently shown to be synthesized by chloroplasts, although the conclusion that it represented a polycistronic chloroplast rRNA precursor was not unanimous (Hartley and Ellis, 1973; Grierson and Loening, 1974). This may have been because chloroplasts were believed to have been derived from blue-green bacteria during the course of evolution and prokaryotic rRNAs had been found to be monocistronic (Adesnik and Levinthal, 1969; Dahlberg and Peacock, 1971; Grierson and Smith, 1973; Seitz and Seitz, 1973). Of course, the similarity between the rRNAs of bacteria and chloroplasts had evolutionary significance, as is now widely recognized (Gray, 2017). Similarities between bacteria and these cellular organelles had been noted in the 19th and early 20th centuries but this idea was not given much credence until Lynn Margulis (then Lynn Sagan) published her account of what was described as “perhaps the first unified theory of eukaryogenesis,” proposing, as is now widely accepted, that “mitochondria and plastids might have originated endosymbiotically from prokaryotic progenitors” (Gray, 2017).
The discovery and application of restriction enzymes by Werner Arber, Daniel Nathans, and Hamilton O. Smith, who were jointly awarded the Nobel Prize in Physiology or Medicine in 1978 and the work of Sir Kenneth Murray (department of Molecular Biology, University of Edinburgh), in combination with development of other new technologies for transformation of bacteria with foreign DNA, paved the way for gene cloning and experimental gene transfer between organisms. In Tübingen, now at the Genetics department, the VH group planned to study gene transfer in higher plants with the genetically well-defined Matthiola incana. Therefore, highly 3H-labeled total DNA was supplied to plant seedlings, and we found integration of this foreign DNA into the nuclei (Hemleben et al., 1975; Leber and Hemleben, 1979). We reported these results at the first Plant Molecular Biology Conference in Liege in 1974. Here, we met Don Grierson, and we organized a 1-year project in 1975/76, supported by an EMBO Long-Term Fellowship to DG. During a meeting in Edinburgh (a summer school run by Ulrich Loening and the late Max Birnstiel in Edinburgh in 1975), we had learned how to separate DNA on Actinomycin D-CsCl gradients and to separate the often highly repeated rRNA genes (rDNA) from the main-band DNA in animal systems (see Birnstiel et al., 1968). Don had also done this with mung beans for his PhD (Grierson, 1972), and this opened up the possibility to purify plant rDNA (Grierson and Hemleben, 1977; Hemleben et al., 1977) and later to characterize it by restriction enzyme analysis, which we carried out firstly during a research stay at Joe Key’s lab in Athens/Georgia in 1978 (Friedrich et al., 1979).
The DNA-content of animals and plants (for plants see: Nagl et al., 1979; Wenzel and Hemleben, 1982a; Michael and Van Buren, 2020) appeared to vary enormously, and the question was: What is the explanation for repetitive genome components and which sequences are redundant? The mature, purified and radioactively labeled rRNAs were used as probes in classical liquid phase hybridization experiments with genomic DNA, showing often enormously high numbers of the tandemly repeated genes for the 18S, 5.8S and 25S (Matsuda and Siegel, 1967; Bendich and Anderson, 1974; Ingle et al., 1975; Wenzel and Hemleben, 1982a). Matsuda and Siegel (1967) further showed that the amount of rDNA cistrons (units) in the nuclear DNA varied among tobacco, pumpkin, pinto beans and Chinese cabbage plants over a 10-fold range. Therefore, it became clear that, besides the highly repetitive satellite DNA, the tandemly arranged and highly redundant rRNA genes contribute to this variability (Marazia et al., 1980; Rogers and Bendich, 1987). Our first chromatin and methylation studies with Matthiola incana and Brassica pekinensis showed that most of the rRNA genes were not transcriptionally active. The rDNA-containing chromatin was not accessible to DNase I digestion, and most of the rRNA genes appeared highly methylated (Leber and Hemleben, 1979; Leweke and Hemleben, 1982; Wenzel and Hemleben, 1982b). At that time, this gene silencing phenomenon was not yet widely called “epigenetics,” although there was a department of Epigenetics established at the Edinburgh University. Transcriptional regulation of nuclear encoded rRNA genes by methylation and demethylation, respectively, was observed also by other researchers (Flavell et al., 1988; Thompson and Flavell, 1988). In Tübingen, we were working with Cucurbitaceae species, which were known for an enormously high number of ribosomal RNA genes (Bendich and Anderson, 1974) and in the 1990s a PhD student Ramon Torres-Ruiz was able to identify the pattern and degree of methylation in the rDNA of Cucurbita pepo (Torres-Ruiz and Hemleben, 1994).
The nuclear ribosomal DNA could, as described, be separated from the main nuclear DNA by Act D-CsCl gradient ultracentrifugation followed by restriction enzyme mapping. This enabled in the early 1980s cloning of plant rDNA using specific gene probes for the 18S, 5.8S, and 25S rRNA and for the internal transcribed (ITS1 and IT2), external transcribed (ETS) and the non-transcribed (NTS) regions of the intergenic spacer (IGS; Figure 2), which allowed subsequent rDNA sequencing. Early on, large DNA fragments, especially those containing internal repeated DNA elements (subrepeats) identified later in the 35S rDNA IGS (35S IGS), were difficult to clone in plasmid vectors, but they could be analyzed by restriction endonucleases and often showed length heterogeneity even in single individuals (Ganal and Hemleben, 1986; Rogers et al., 1986; Yokota et al., 1989). Later, we and others were able to clone the complete large IGS and to characterize this region by DNA sequencing using at that time the radioactive sequencing methods (Sanger et al., 1977). Of course, the polymerase chain reaction (PCR) developed by the biochemist Karl Mullis, who in 1993 shared the Nobel Prize in Chemistry, facilitated enormously this procedure. Length heterogeneity of the rDNA repeats was due mostly to different numbers of subrepeats in the 35S IGS upstream or downstream of the transcription initiation site, TIS (Appels et al., 1986; Ganal et al., 1988; Rathgeber and Capesius, 1990; Borisjuk and Hemleben, 1993; King et al., 1993; Zentgraf and Hemleben, 1993; Figure 2, upper part). Interestingly, it appeared that in some plants (e.g., some Vigna sp.) these repeated elements of the 35S IGS formed independent highly amplified satellite DNA genome components (Unfried et al., 1991; Macas et al., 2003; Lim et al., 2004b; Jo et al., 2009; Kirov et al., 2018). In contrast, 35S DNA amplification of satellites within the 5S rDNA loci is rare. Nevertheless, a 170-bp satellite sequence (termed jumper) apparently invaded 5S rDNA in the evolutionary history of the Phaseolus genus (Ribeiro et al., 2017). Of note, only a single satellite monomer is found in the 5S IGS while there may be thousands of copies outside of 5S rDNA loci, forming pericentromeric and subtelomeric domains. This may suggest that the rDNA intergenic spacers have relatively frequently hosted non-coding satellites but their expansion is limited to one or a few copies, probably due to selection constrains imposed on spacer lengths.
In chromosomes, the 5S rRNA genes occur either as long tandem repeats of regularly spaced units (S-type arrangement) or as solitary insertions within the 35S rDNA intergenic spacer (L-type arrangement). The S-type arrangement is the most frequent organization of 5S rDNA in angiosperms (Ellis et al., 1988; Hemleben and Werts, 1988; Scoles et al., 1988; Röser et al., 2001) while, so far, the L-type arrangement has been detected only in some member of the Asteraceae family (Garcia et al., 2009, 2010; Souza et al., 2019). The L-type arrangement is more typical for plants that diverged early during angiosperm evolution (Capesius, 1997; Wicke et al., 2011) and some groups of gymnosperms (Galian et al., 2012; Garcia and Kovarik, 2013). The 5S rDNA was apparently invaded by an LTR transposon in the early evolutionary history of angiosperms, giving rise to Cassandra retrotransposons (Kalendar et al., 2008), which are now widespread in modern species. Truncated incomplete copies of 35S rDNA seem to be scattered in genomes of both plants (Tulpová et al., 2020) and animals (Robicheau et al., 2017) and likely represent remnants of former NORs.
Transcriptional Regulation of rRNA Genes
The nuclear encoded 18S, 5.8S, and 25S ribosomal RNA genes were already known in yeast and animals to be transcribed by RNA polymerase I, and this was confirmed in plants in Joe Key’s lab (Lin et al., 1976) and in Tübingen (Grossmann et al., 1979, 1980). Functional studies identified the putative transcription initiation (TIS) and transcription termination sites (TTS) for plant rDNA (Delcasso-Tremousaygue et al., 1988; Gerstner et al., 1988; Gruendler et al., 1989; Schiebel and Hemleben, 1989; Zentgraf and Hemleben, 1992, 1993; Figure 2, upper part). In addition, the repeated elements upstream or downstream of the TIS obviously had an enhancer function (reviewed in Hemleben and Zentgraf, 1994). Under the current view, the nucleoprotein complex responsible for transcription initiation of 45S (35S) rRNA is composed of numerous protein components (Nucleolar Remodelling Complex (NoRC), UBF, histone acetyltransferases, helicases, and RNA polymerase I, among others) and several species of noncoding RNA (Bersaglieri and Santoro, 2019; Yan et al., 2019). The RNA polymerase I holoenzyme has been purified to apparent homogeneity by biochemical approaches, maintaining the capacity to initiate rDNA transcription (Sáez-Vásquez and Pikaard, 2000). Nucleolin, a relatively abundant nucleolar structural protein, seems to be involved in selection of rDNA variants for transcription in Arabidopsis (Pontvianne et al., 2010).
The 5S rRNA genes are transcribed by RNA polymerase III and their promoter elements and termination sites were putatively described for plants (Hemleben and Werts, 1988). Transcriptional regulation of the multigenic 5S rDNA in Arabidopsis has been extensively studied in Sylvette Tourmentes’s laboratory (University of Clermont-Ferrand, France), confirming the originally described 5S rRNA internal and external elements involved in regulation of the gene’s transcription (reviewed by Layat et al. (2012). Later, in Roman Volkov’s laboratory, conservation of the putative external promoter elements in the 5S rDNA intergenic spacer (5S IGS) was demonstrated for several families of angiosperms (Volkov et al., 2001, 2017; Tynkevich and Volkov, 2014, 2019; Ishchenko et al., 2018, 2020).
While the major advances in our knowledge of rDNA regulation were achieved in yeast (Moss, 2004) and animals (rat, mouse and Xenopus; (Grummt et al., 1985; Pikaard and Reeder, 1988), the research on plant rDNA also made significant progress over the years (Weis et al., 2015; Sáez-Vásquez and Delseny, 2019). Now it is widely accepted that in addition to transcriptional regulation of individual rDNA repeat units, the entire rDNA arrays (NOR) are targets of regulation as exemplified by studies in Arabidopsis (Mohannath et al., 2016) and wheat (Handa et al., 2018). The findings described recently by (Sims et al., 2021), sequencing entire rDNA arrays and deciphering their higher structure promise further deeper insights into the functional organization and molecular regulation of plants rDNA loci. By applying a combination of long- and short-read sequencing the authors revealed clustering of rDNA domains in Arabidopsis NOR2 and expression of several variants of rRNAs with their tissue-specific integration into active ribosomes (Sims et al., 2021). In leaf tissue of the ecotype Columbia-0 (reference genome), the rDNA of NOR4 on chromosome 4 is usually more active than that on chromosome 2 (NOR2; Chandrasekhara et al., 2016). However, NOR4 is not always dominant and many natural populations of Arabidopsis thaliana show considerable epigenetic variability, i.e., dominant expression of NOR4, NOR2, or codominant expression of both loci (Rabanal et al., 2017).
The duckweed species Spirodela polyrhiza and S. intermedia might emerge as a promising new model to study rDNA regulation (in addition to Arabidopsis) because of their compact rDNA loci, composed of no more than a hundred 35S repeated units (Michael et al., 2017; Hoang et al., 2020), even fewer than the 100–200 rDNA copies in yeast (Salim et al., 2017), and an order of magnitude lower than rRNA gene copies in other plants (Wang et al., 2019). The copy number of 5S rDNA is estimated to be about 170 in Landoltia punctuata (Chen et al., 2021) – this issue. This exceptionally low copy number makes the duckweed rDNA locus relatively simple to access applying the third-generation sequencing platforms for ultra-long sequencing reads (Jung et al., 2019).
Molecular Evolution of rDNA Loci
Although it was initially believed that the rRNA genes are quite conserved, it turned out in the 1980s that the regions of the 5' and 3’ ETS, NTS, and ITS1 and 2 are characterized by a huge intra- and interspecies variability. This made them highly suitable as markers for our phylogenetic and molecular evolution studies at population or interspecies levels (Torres et al., 1989; Grebenstein et al., 1998; Jobst et al., 1998; Volkov et al., 2003, 2010; Denk et al., 2005; Grimm et al., 2005; Komarova et al., 2008; Schlee et al., 2011). Table 1 summarizes individual 35S rDNA subregions used for phylogenetic markers.
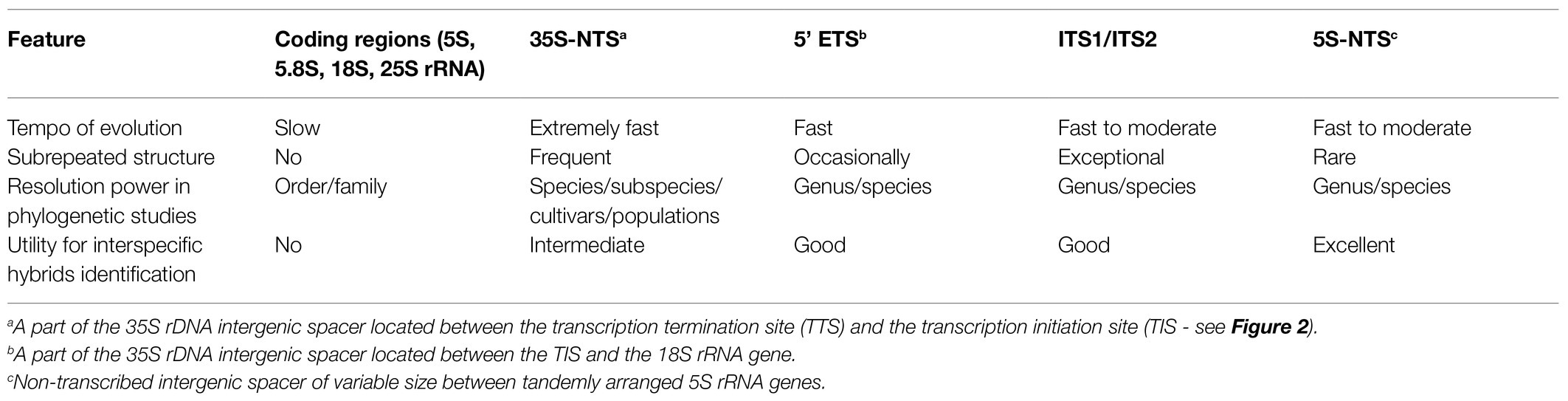
Table 1. Characteristics of individual subregions of plant rDNA units and their relevance for phylogenetic analysis.
Similarly, the 5S IGS appeared very useful for clarifying phylogenetic relationships at low taxonomic levels (Röser et al., 2001; Volkov et al., 2001; Denk and Grimm, 2010; Tynkevich and Volkov, 2019; Ishchenko et al., 2021) and for identification of interspecific hybrids (Garcia et al., 2020). This contrasts with the really strongly conserved sequences of the mature 5S, 5.8 S, 18S, and 25S rRNA coding regions. However, variable segments of 18S and 25S coding regions (“expansion segments”) evolve faster than the conserved stems. These features of rRNA coding regions can be used for phylogenetic studies particularly at higher taxonomic levels (Poczai and Hyvonen, 2010; Soltis and Soltis, 2016). The secondary structure of rRNA transcripts is another layer of phylogenetic information in addition to primary sequence (Noller et al., 1981; Selig et al., 2008). Currently, a public database of ITS2 secondary structure models comprise more than 80 thousand sequences (Selig et al., 2008). Consequently, various rDNA regions have been studied worldwide in the 1990s until the present day as phylogenetic markers in population, species, genus, and higher systematic order studies and helped to solve the phylogenetic relationships between organisms. Public databases storing biological information about rDNA loci (Szymanski et al., 1998; Selig et al., 2008; Cantara et al., 2011; Garcia et al., 2012; Quast et al., 2012) represent a valuable source for structural, functional and phylogenetic studies.
In the 1990s Nikolai (NB) and Ljudmilla Borisjuk and later Roman Volkov (RV) and Irina Panchuk from the Ukraine joined the VH laboratory, NB and RV as Alexander v. Humboldt fellows. They had already started to study plant rDNA, and we had a very successful cooperation over the years working mostly on several genera of Solanaceae (Borisjuk et al., 1994, 1997; Volkov et al., 1999a,b; Komarova et al., 2004). In 1983–1985, RV and NB worked in the group of Andrey S. Antonov at Moscow University (Russia). NB worked on the characterization of genomes of Solanaceae and Brassicaceae somatic hybrids generated by protoplast fusion in the Lab of Yuri Gleba in Kyiv (Ukraine). RV was interested in describing rearrangements of repeated sequences in natural allopolyploids, particularly in the genus Nicotiana, which includes several allopolyploids and aneuploids. These young researchers decided to perform the 35S rDNA restriction mapping for several artificial and natural allopolyploids. They used as probes for Southern hybridization 18S and 25S rRNA from maize and a fragment of 25S coding sequence of lemon isolated by Volodymyr Kolosha (Pushchino, Russia) under the supervision of Tengiz Beridze (Georgia). Mapping experiments revealed that generally the interspecies and inter-tribal hybrids obtained by protoplast fusion or sexual crossing inherited a combination of parental rDNA (Gleba et al., 1988; Borisjuk and Miroshnichenko, 1989; Miroshnichenko et al., 1989). Additionally, a novel class of rDNA repeats was found in somatic hybrids between distantly related Nicotiana and Atropa (Borisjuk et al., 1988). Later on, this observation let to the discovery of an “amplification promoting sequence” (APS) within the tobacco 35S IGS. The cloned APS element apparently increased the copy number of linked reporter genes in transgenic experiments resembling the origin of DNA replication (Borisjuk et al., 2000). It transpired that the structure of the Nicotiana 35S IGS is highly complex, bearing repetitive subregions which apparently account for species-specific differences in rDNA structure (Volkov et al., 1996; Borisjuk et al., 1997). Genomic analysis showed numerous SNPs (single nucleotide polymorphisms) in the tobacco 35S IGS, which indicated that the mutation rate in that region may be faster than that of coding regions, arguing for variable selection pressures acting on different parts of the rDNA unit (Lunerova et al., 2017).
The Fate of rDNA in Solanaceae Hybrids and Allopolypoids
The multigene families in both plants and animals reveal high levels of intra-species homogeneity and inter-species diversity. These features underlie the concept of concerted evolution put forward by geneticists in the second half of the last century (Brown et al., 1972; Zimmer et al., 1980; Dover, 1982). It has become clear that concerted evolution (i.e., homogenization) processes affect nearly all repeated families including non-coding satellites and rDNA. The tandemly arranged rDNA represent a textbook example of concerted evolution since their hundreds of units show little or no intragenomic variation (reviewed by (Eickbush and Eickbush, 2007; Nieto Feliner and Rossello, 2012). The presence of rDNA arrays that are homogeneous for different variants in interbreeding populations of Drosophila melanogaster indicated that there is little recombination between the arrays while there might be intensive recombination within the arrays, leading to their overall homogeneity (Schlotterer and Tautz, 1994).
Towards the end of the last century the VH group explored the Solanaceae family (nightshades) which includes many economically important crops such as tomato, potato and tobacco. Within the family, the Nicotiana genus, whose center of diversity is Latin America, contains at least 50 allopolyploids of different ages and genome compositions. Nicotiana tabacum (tobacco) is, perhaps, the most well-known allotetraploid (2n = 4x = 48, genome composition SSTT) and has long been a favorite model for plant genetic studies (Goodspeed, 1954) including transgenosis and chromosome evolution (Manoharlal et al., 2019; Dodsworth et al., 2020). It is a relatively recent (ca. 0.1 myrs old; Lim et al., 2007) allopolyploid originating from hybridization of progenitor species close to Nicotiana sylvestris (2n = 2x = 24, S genome) and Nicotiana tomentosiformis (2n = 2x = 24, T genome). Its parental S- and T-genomes are relatively intact with few intergenomic translocations (Lim et al., 2004a). The question was: What is the fate of parental 35S rDNAs in tobacco? Are they intact or have they been modified by allopolyploidy?
In order to clarify the fate of parental 35S rDNA in the genome of N. tabacum, NB and RV decided to sequence the 35S IGS regions. From 1990 to 1992, with the support of the Alexander v. Humboldt Foundation, NB worked in the group of VH, who by then was a well-known leader in rDNA research. Here he performed restriction mapping of numerous Solanum species and other Solanaceae (Borisjuk et al., 1994) and cloned and sequenced the 35S IGS of S. tuberosum (Borisjuk and Hemleben, 1993) and N. tabacum (Borisjuk et al., 1997). During this time VH’s lab used RFLP (restriction fragment length polymorphism) for characterizing artificial somatic hybrids of Solanum tuberosum and various wild Solanum species produced by protoplast fusion in the lab of Prof. Helga Ninnemann by Dr. Lieselotte Schilde-Rentschler, mainly to introduce pathogen resistant characters into the cultivated potato (Schweizer et al., 1993). Therefore, the Solanum research of NB and RV was very complementary to the hybrid identification research. Interestingly, the rDNA of one fusion partner disappeared very quickly (see below for Nicotiana).
RV obtained an Austrian exchange service Research Fellowship, and in 1993 he went to the lab of Prof. Dieter Schweizer (Department of Cytology and Genetics, University of Vienna), where the 35S IGS of Arabidopsis thaliana had recently been sequenced and characterized (Gruendler et al., 1989). In Dieter Schweizer’s laboratory, RV cloned and sequenced 35S IGS of N. sylvestris and N. tomentosiformis (Volkov et al., 1996, 1999b).
In 1996, RV received the AvH Research Fellowship, moved to Tübingen and joined the VH group in order to investigate further the rDNA in Solanaceae. Comparative analysis of the 35S IGS sequences of N. tabacum, N. tomentosiformis and N. sylvestris allowed the molecular evolution of parental rDNA in the genome of N. tabacum to be deduced. Strikingly, only units similar to the paternal N. tomentosiformis genome (T-genome) were cloned from the tobacco genome (Volkov et al., 1999b), while clones from the maternal parent were not recovered, indicating elimination of the S-genome rDNA. It was found that the rDNA repeats of N. tabacum originated from N. tomentosiformis, which involved reconstruction of subrepeated regions in the 35S IGS upstream and downstream of the transcription initiation site. These cloning results well-resonated with the non-additivity of tobacco 35S rDNA restriction fragments observed in previous Southern hybridization experiments (Borisjuk et al., 1989; Miroshnichenko et al., 1989; Volkov et al., 1991; Kovarik et al., 1996). Clearly, thousands of parental rDNA units were overwritten by novel hybrid-specific units in relatively short evolutionary time (<100 thousand years; Borisjuk et al., 1997; Volkov et al., 1999a,b). Molecular cytogenetics approaches carried out by Andrew Leitch’s group (University of London) revealed that the number of tobacco rDNA loci is additive, i.e., there is a single locus in the T-genome and three loci in the S-genome (Lim et al., 2000). Only a small number of unconverted and highly methylated S-genome units were detected in the tobacco genome by molecular methods. In 2017 Jana Lunerova (AK group) using locus-specific FISH probes addressed the question of chromosomal localization of these transcriptionally inactive rRNA genes (Lunerova et al., 2017). It appeared that the residual (about 8% of total rDNA) S-genome units are located on a small acrocentric chromosome S12 while active homogenized genes are located on chromosomes T3, S10, and S11. Of note, the S-genome rDNA loci were found in the variety SR-1 and wild tobacco collected by Sandy Knapp (Natural History Museum London) in Bolivia, but not in the variety 09555 (Kovarik et al., 2004). Thus, the process of cultivation and high inbreeding may potentially influence the behavior of rDNA in allopolyploids. The Leitch’s and AK’s groups further confirmed partial and complete homogenization of parental rDNAs in another two Nicotiana allotetraploids, Nicotiana rustica (Indian tobacco, 2n = 4x = 48; Matyasek et al., 2003) and Nicotiana arentsii (2n = 4x = 48), respectively (Kovarik et al., 2004). Hence genetic interactions of rDNA loci seem to be a rather general feature of rDNA evolution in Nicotiana allopolyploids.
Fulnecek et al. (2002) investigated the structure of 5S rDNA loci which occurs separately from 35S rDNA loci in most plant genomes (Hemleben and Grierson, 1978; Garcia et al., 2016). Locus-specific FISH together with pulsed-field gel electrophoresis mapping showed that parental 5S rDNA arrays remained relatively intact and were inherited at expected ratios in tobacco allotetraploid. Therefore, in contrast to 35S rDNA, the 5S rDNA loci do not genetically interact in tobacco allotetraploids. The reason for higher genetic stability of 5S rDNA compared to 35S rDNA is not fully understood. However, 5S rDNA is highly methylated (more than the genome average; Fulnecek et al., 1998), while 35S rDNA units contain many undermethylated sites particularly in intergenic spacers (Torres-Ruiz and Hemleben, 1994). Hypomethylated sites in 35S IGS were also observed in other species including Arabidopsis (Earley et al., 2006), cucumber (Torres-Ruiz and Hemleben, 1994), potato (Komarova et al., 2004) and wheat (Sardana et al., 1993). It is possible that apart from transcription regulation these undermethylated sites might be important for adopting chromatin conformation favorable to recombination processes (Kovarik et al., 2008).
Volkov et al. (2017) applied a combination of karyological and molecular methods to investigate chromosomal localization, molecular organization and evolution of 5S and 35S rDNA in Atropa belladonna (Solanaceae), one of the oldest known flowering plant allohexaploids. Intensive sequence homogenization between three pairs of 35S rDNA loci on separate chromosomes was found, presumably inherited from tetraploid and diploid ancestor species. Only four out of six 35S rDNA sites appeared transcriptionally active, demonstrating nucleolar dominance. For 5S rDNA, three size variants of repeats were detected, with the major class represented by repeats containing all functional 5S IGS elements required for transcription, whereas intermediate and short length repeats contained defects both in the spacer and coding sequences. The functional 5S rDNA variants are nearly identical at the sequence level, pointing to their origin from a single parental species. Localization of the 5S rRNA genes on two chromosome pairs further supports uniparental inheritance from the tetraploid progenitor. The data demonstrate complex evolutionary dynamics of rDNA loci in allohexaploid species of Atropa belladonna. The high level of sequence unification revealed in 5S and 35S rDNA loci of this ancient hybrid species have been seemingly achieved by different molecular mechanisms.
Nucleolar Dominance in Solanaceae Hybrids and Allopolyploids
Nucleolar dominance is an epigenetic phenomenon in which one parental array is inactivated in interspecific hybrids and allopolyploids. It was first described at the cytological level by Navashin (Navashin, 1934), who observed that in interspecific hybrids of Crepis (Asteraceae) only chromosomes of one crossing partner carried secondary constrictions at metaphase. This chromosomal region was not lost in hybrids but could be reactivated to produce normal nucleoli in hybrids with a different crossing partner. Experiments with epigenetic inhibitors performed in plants towards the end of the last century established that histone deacetylation and DNA methylation pathways interact in a self-reinforcing mechanism, maintaining silencing of partner rDNA units in hybrids (Chen and Pikaard, 1997a; Chen et al., 1988). The VH and AK groups investigated nucleolar dominance from different perspectives, asking questions about the influence of structural features of the 35S IGS, cytosine methylation of rDNA units and developmental stability of nucleolar dominance. To address these questions, they used well-defined natural and synthetic Nicotiana and Solanum (Solanaceae) allotetraploids. A comparison of 35S rDNA organization in several Solanum species revealed (Borisjuk and Hemleben, 1993; Borisjuk et al., 1994) that S. lycopersicum (tomato), S. tuberosum (potato) and wild species S. bulbocastanum possess 35S IGS of nearly identical length but contain different number of subrepeats up- and downstream of the TIS. Accordingly, VH and RV suggested using these species to elucidate the presumptive role of subrepeated elements in nucleolar dominance. In 1998, Nataliya Komarova from RV’s group moved from Ukraine to VH’s lab, where she studied expression of parental 35S rDNA in Solanum lycopersicum x S. tuberosum and S. tuberosum x S. bulbocastanum artificial somatic alloploids produced by protoplast fusion and back-crossed lines, which were kindly provided by E. Jacobsen and H.J. de Jong (Wageningen University, The Netherlands) and by L. Schilde-Rentschler and H. Ninnemann (University of Tübingen, Germany). It appeared that an expression hierarchy exists: In leaves, roots, and petals of the respective allopolyploids, rDNA of S. lycopersicum dominates over rDNA of S. tuberosum, whereas rDNA of S. tuberosum dominates over that of the wild species S. bulbocastanum. Also, in a monosomic addition line carrying only one NOR-bearing chromosome of tomato in a potato background, the dominance effect was maintained. These results demonstrated that there is possible correlation between transcriptional dominance and number of conserved elements downstream of the transcription start in the Solanum rDNA (Komarova et al., 2004). The authors proposed that this sequence motif could be a recognition site for DNA-interacting proteins involved in modulation of rDNA transcription (Borisjuk et al., 1997; Volkov et al., 2003). Remarkably, no correlation between the number of upstream subrepeats and differential transcription/silencing of 35S rDNA in Solanum allopolyploids was found. The latter contrasts with observations made in the allohexaploid wheat (AABBDD), where longer B-genome units containing more upstream subrepeats are active while the shorter units located in the D genome are usually inactive (Sardana et al., 1993). Units bearing longer upstream elements also seem to be dominant in recently (<100 years) formed Tragopogon allotetraploids (Matyasek et al., 2016). In contrast, sexual F1 hybrids resulting from crossing of N. sylvestris x N. tomentosiformis plants and a synthetic tobacco line show codominance (Dadejova et al., 2007), despite apparent differences in the 35S IGS structure of progenitor units. Thus, it seems that the role of 35S IGS repeat elements in regulation of rDNA expression varies from system to system. Certainly, a direct proof for an enhancer/silencing role of these elements, as shown in Xenopus laevis 35S IGS (Caudy and Pikaard, 2002), is missing in plants.
Developmental stability of nucleolar dominance was investigated in hybrids of Brassica and Solanum. Classical experiments in Brassica napus allotetraploids showed developmental lability of nucleolar dominance and partial reactivation of under-dominant genes in floral organs (Chen and Pikaard, 1997b). In the early 1990s RV in the VH group employed, perhaps for the first time, quantitative RT-PCR for the analysis of nucleolar dominance in plants. Using this method, they determined the levels of homoelogous ETS transcripts in different organs of tomato x potato hybrids showing a strong nucleolar dominance of tomato genes in leaf but not in anthers and calli (Komarova et al., 2004). Activation of partner units was apparently linked to changes in DNA methylation and chromatin organization. Indeed, profound changes in condensation of rDNA chromatin were observed between tobacco leaf and root (Koukalova et al., 2005).
Earlier cytogenetic data from N. tabacum indicated that unconverted parental units of N. sylvestris-origin were highly methylated, perhaps located at a locus on chromosome S12 (Lim et al., 2000) that does not show secondary constrictions at metaphase (a hallmark of genetic inactivity). More recently (Dadejova et al., 2007) used RT-PCR to investigate expression of rRNA genes in a number of synthetic Nicotiana hybrids (including reciprocal crosses) with a genomic composition similar to natural N. tabacum (SSTT), N. rustica (PPUU) and N. arentsii (UUWW) allotetraploids differing in age and genome donors. They found strong uniparental rDNA silencing of N. paniculata genes in N. paniculata × N. undulata F1 hybrids (genome composition corresponding to natural N. rustica), whereas N. sylvestris × N. tomentosiformis (N. tabacum) and N. undulata × N. wigandioides (N. arentsii) F1 hybrids showed little or no silencing (i.e., co-dominance). Based on these observations, Kovarik et al. (2008) proposed that nucleolar dominance, established early in allopolyploid formation including F1 hybrids, plays a significant role in further molecular evolution of rDNA. It has been suggested that epigenetic silencing of rDNA loci makes them less vulnerable to homogenization and more likely to be lost, perhaps thousands or millions of years later.
In 2003, AK visited the VH lab in Tübingen. At that time, both groups were fascinated by the dynamics of repetitive sequences, especially their species- and sometimes even population-specific features. As a result of fruitful discussions during a stroll around the old castle (whose walls remember the discovery of DNA by Friedrich Miescher) we wrote a review paper on the behavior of satellite DNA repeats in plant hybrids (Hemleben et al., 2007). The outcome was a productive collaborative research on allopolyploidy carried out in labs at the University of Tübingen, Queen Mary College of the University of London and the Czech Academy of Science. The findings are significant for our understanding of evolution of plant species since the world of angiosperms is largely dominated by allopolyploids.
Concluding Remarks
Looking back over 50 years of research on rDNA, it is amazing to see how many complex factors stimulated or supported this field of study. To start with: In the early 1960s, the interest in molecular biology was rising: Basic functions of cell organelles were clarified. Electron microscopy, ultracentrifugation, radioactive labelling, and gel electrophoresis and hybridization assays were the key experimental methods, revealing a complex and highly organized ribosome construction and assembly process. As in bacteria, but with variation in size in all eukaryotes, the 40S subunit were shown to contain 18S rRNA while the 60S subunit associates with 25/28S (plants/animals), 5.8S and a smaller 5S rRNA. Both subunits form a ribonucleoprotein complex which assembles into a functional 80S ribosome. Transcription of the 18S, 5.8S and 25S was found to occur by RNA polymerase I as a large polycistronic precursor (pre-rRNA) containing tandem repeat sequences of the 18S-5.8S-25S rDNA multigene family, which is subsequently processed into the mature rRNAs, whereas the 5S rRNA genes (5S rDNA) are transcribed by RNA polymerase III. Gene technologies, gene cloning and DNA sequencing showed that strongly conserved parts alternate with more variable regions, especially in the intergenic regions of these multigene families. Cooperation of various experts delivered further valuable information and opened up the broad field of plant molecular phylogeny, molecular evolution and molecular systematics, supported by the rapidly growing field of whole genome sequencing coupled with more and more sophisticated computer evaluation of the data obtained. New techniques of cytogenetics made genome evolution and species formation visible. Especially for plants, the process of homo- and allopolyploidy by natural hybridization could be followed. The phenomenon of nucleolar dominance helped us to understand the mechanisms of silencing of rDNA from one partner probably leading to elimination of rRNA genes in allopolyploids.
Author Contributions
VH conceived and designed the study. AK, DG, NB, RV, and VH contributed to the writing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
AK: The work was supported by the Czech Science Foundation (grant 19-03442S). VH: Our work was always supported by the German Science Foundation (DFG), some parts by the Federal Ministry of Research and Technology (BMFT), today called BMBF; the Alexander v. Humboldt Foundation (Bonn, Germany) greatly supported the research stay of NB and RV at Tübingen University which is gratefully acknowledged. DG is grateful to former UK governments for financial support during BSc studies at University of East Anglia and PhD (Science & Industry Award) at the University of Edinburgh and to the University of Nottingham. RA is grateful to the Ministry of Education and Science of Ukraine, the Alexander von Humboldt Foundation, German Academic Exchange Service (DAAD) and Austrian Exchange Service (OeAD) for the financial support provided to him and members of his research group. NB is grateful for the earlier support from the Alexander von Humboldt Foundation; he is currently supported by a personal grant from the Huaiyin Normal University (Huai’an, China).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Many Diploma and PhD students and Postdocs contributed to the rDNA research in VH’s lab, especially Birgit Lewecke, Harald Friedrich (†), Katrin Schiebel, Jutta Gerstner, Martin Ganal, Ramon Torres-Ruiz, Ulrike Zentgraf, Klaus King, Jürgen Jobst, Klaus Unfried, Günther Schweizer, Christine Zanke, Guido Grimm, Matthias Schlee, Thomas Grabe, which is highly acknowledged. DG is grateful to Ulrich Loening for his guidance and encouragement during the course of my PhD studies at the University of Edinburgh. Thanks also to the late Max Birnstiel for shining the light on nucleolar rDNA. VH and RV: Many Diploma and PhD students and Postdocs were involved in the study of rDNA in our labs. Especially, we would like to express our appreciation for the great contribution of Irina Panchuk, Nataliya Komarova, Yuri Tynkevich, Antonina Shelyfist, Olga Ishchenko and Kristina Bushyla. NB is grateful for the contribution made by Ljudmilla Borisjuk, Volodymyr Kolosha, Nikolai Friesen, Anton Stepanenko, Guimin Chen and Gregoriy Petjuch. NB and RV are grateful to Galina Miroshnichenko (Moscow, Russia) for her support/advice at early stage of our research careers. We are grateful to the members of The International Research Network (IRN) Polyploidy and Biodiversity (PolyDiv). AK wish to remember Yoong Lim (Queen Mary, University of London) and Ann Kenton (Kew Garden, Richmond, UK), both excellent cytogeneticists, who pioneered molecular cytogenetics of Nicotiana in early 90s, both of whom have sadly passed.
References
Adesnik, M., and Levinthal, C. (1969). Synthesis and maturation of ribosomal RNA in Escherichia coli. J. Mol. Biol. 46, 281–303. doi: 10.1016/0022-2836(69)90422-7
Appels, R., Moran, L. B., and Gustafson, J. P. (1986). Rye heterochromatin. 1. Studies on clusters of the major repeating sequence and the identification of a new dispersed repetitive sequence element. Can. J. Genet. Cytol. 28, 645–657. doi: 10.1139/g86-094
Appels, R., Wang, P., and Islam, S. (2021). Integrating wheat nucleolus structure and function: variation in the wheat ribosomal RNA and protein genes. Front. Plant Sci.
Bendich, A. J., and Anderson, R. S. (1974). Novel properties of satellite DNA from muskmelon. Proc. Natl. Acad. Sci. U. S. A. 71, 1511–1515. doi: 10.1073/pnas.71.4.1511
Bersaglieri, C., and Santoro, R. (2019). Genome organization in and around the nucleolus. Cell 8:579. doi: 10.3390/cells8060579
Birnstiel, M., Speirs, J., Purdom, I., Jones, K., and Loening, U. E. (1968). Properties and composition of isolated ribosomal DNA satellite of Xenopus laevis. Nature 219, 454–463. doi: 10.1038/219454a0
Birnstiel, M. L., Wallace, H., Stirlin, L., and Fischberger, M. (1966). Localization of the ribosomal DNA complements in the nucleolar organizer regions in Xenopus laevis. Natl. Cancer Inst. Monogr. 23, 431–447.
Borisjuk, N., Borisjuk, L., Komarnytsky, S., Timeva, S., Hemleben, V., Gleba, Y., et al. (2000). Tobacco ribosomal DNA spacer element stimulates amplification and expression of heterologous genes. Nat. Biotechnol. 18, 1303–1306. doi: 10.1038/82430
Borisjuk, N., Borisjuk, L., Petjuch, G., and Hemleben, V. (1994). Comparison of nuclear ribosomal RNA genes among Solanum species and other Solanaceae. Genome 37, 271–279. doi: 10.1139/g94-038
Borisjuk, N. V., Davidjuk, Y. M., Kostishin, S. S., Miroshnichenco, G. P., Velasco, R., Hemleben, V., et al. (1997). Structural analysis of rDNA in the genus Nicotiana. Plant Mol. Biol. 35, 655–660. doi: 10.1023/A:1005856618898
Borisjuk, N., and Hemleben, V. (1993). Nucleotide sequence of the potato rDNA intergenic spacer. Plant Mol. Biol. 21, 381–384. doi: 10.1007/Bf00019953
Borisjuk, N. V., Kostyshin, S. S., Volkov, R. A., and Miroshnichenko, G. P. (1989). Ribosomal RNA gene organization in higher plants from Nicotiana genus. Mol. Biol. 23, 1067–1074.
Borisjuk, N. V., and Miroshnichenko, G. P. (1989). Organization of ribosomal RNA genes in Brassica oleracea, B. campestris and their natural allotetraploid hybrid B. napus. Genetika 25, 417–424.
Borisjuk, N. V., Momot, V. P., and Gleba, Y. (1988). Novel class of rDNA repeat units in somatic hybrids between Nicotiana and Atropa. Theor. Appl. Genet. 76, 108–112. doi: 10.1007/Bf00288839
Brown, D. D., and Gurdon, J. B. (1964). Absence of ribosomal RNA synthesis in anucleolate mutant of Xenopus laevis. Proc. Natl. Acad. Sci. U. S. A. 51, 139–146. doi: 10.1073/pnas.51.1.139
Brown, J. W. S., and Shaw, P. J. (1998). Small nucleolar RNAs and pre-rRNA processing in plants. Plant Cell 10, 649–657. doi: 10.1105/tpc.10.5.649
Brown, D. D., Wensink, P. C., and Jordan, E. (1972). A comparison of ribosomal DNAs of Xenopus laevis and Xenopus mulleri: evolution of tandem genes. J. Mol. Biol. 63, 57–73. doi: 10.1016/0022-2836(72)90521-9
Cantara, W. A., Crain, P. F., Rozenski, J., McCloskey, J. A., Harris, K. A., Zhang, X. N., et al. (2011). The RNA modification database, RNAMDB: 2011 update. Nucl. Acids Res. 39, D195–D201. doi: 10.1093/nar/gkq1028
Capesius, I. (1997). Analysis of the ribosomal RNA gene repeat from the moss Funaria hygrometrica. Plant Mol. Biol. 33, 559–564. doi: 10.1023/a:1005740031313
Caudy, A. A., and Pikaard, C. S. (2002). Xenopus ribosomal RNA gene intergenic spacer elements conferring transcriptional enhancement and nucleolar dominance-like competition in oocytes. J. Biol. Chem. 277, 31577–31584. doi: 10.1074/jbc.M202737200
Chandrasekhara, C., Mohannath, G., Blevins, T., Pontvianne, F., and Pikaard, C. S. (2016). Chromosome-specific NOR inactivation explains selective rRNA gene silencing and dosage control in Arabidopsis. Genes Dev. 30, 177–190. doi: 10.1101/gad.273755.115
Chandrashekaran, M. (2006). Erwin Bünning (1906–1990): a centennial homage. J. Biosci. 31, 5–12. doi: 10.1007/BF02705230
Chen, Z. J., Comai, L., and Pikaard, C. S. (1988). Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc. Natl. Acad. Sci. USA 95, 14891–14896. doi: 10.1073/pnas.95.25.14891
Chen, Z. J., and Pikaard, C. S. (1997a). Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 11, 2124–2136. doi: 10.1101/gad.11.16.2124
Chen, Z. J., and Pikaard, C. S. (1997b). Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc. Natl. Acad. Sci. U. S. A. 94, 3442–3447. doi: 10.1073/pnas.94.7.3442
Chen, G., Stepanenko, A., and Borisjuk, N. (2021). Mosaic arrangement of the 5S rDNA in the aquatic plant Landoltia punctata (Lemnaceae). Front. Plant Sci. 12:678689. doi: 10.3389/fpls.2021.678689
Cox, B. J., and Turnock, G. (1973). Synthesis and processing of ribosomal RNA in cultured plant cells. Eur. J. Biochem. 37, 367–376. doi: 10.1111/j.1432-1033.1973
Dadejova, M., Lim, K. Y., Souckova-Skalicka, K., Matyasek, R., Grandbastien, M. A., Leitch, A., et al. (2007). Transcription activity of rRNA genes correlates with a tendency towards intergenomic homogenization in Nicotiana allotetraploids. New Phytol. 174, 658–668. doi: 10.1111/j.1469-8137.2007.02034.x
Dahlberg, A. E., and Peacock, A. C. (1971). Studies of 16 and 23 S ribosomal RNA of Escherichia coli using composite gel electrophoresis. J. Mol. Biol. 55, 61–74. doi: 10.1016/0022-2836(71)90281-6
Delcasso-Tremousaygue, D., Grellet, F., Panabieres, F., Ananiev, E. D., and Delseny, M. (1988). Structural and transcriptional characterization of the external spacer of a ribosomal RNA nuclear gene from a higher plant. Eur. J. Biochem. 172, 767–776. doi: 10.1111/j.1432-1033.1988.tb13956.x
Denk, T., and Grimm, G. (2010). The oaks of western Eurasia: traditional classifications and evidence from two nuclear markers. Taxon 59, 351–366. doi: 10.1002/tax.592002
Denk, T., Grimm, G. W., and Hemleben, V. (2005). Patterns of molecular and morphological differentiation in Fagus (Fagaceae): phylogenetic implications. Am. J. Bot. 92, 1006–1016. doi: 10.3732/ajb.92.6.1006
Dodsworth, S., Kovarik, A., Grandbastien, M.-A., Leitch, I. J., and Leitch, A. R. (2020). “Repetitive DNA dynamics and polyploidization in the genus Nicotiana (Solanaceae),” in The Tobacco Plant Genome. Compendium of Plant Genomes. eds. N. Ivanov, N. Sierro, and M. Peitsch (Cham: Springer).
Dover, G. A. (1982). Molecular drive: a cohesive mode of species evolution. Nature 299, 111–117. doi: 10.1038/299111a0
Earley, K., Lawrence, R. J., Pontes, O., Reuther, R., Enciso, A. J., Silva, M., et al. (2006). Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 20, 1283–1293. doi: 10.1101/gad.1417706
Eickbush, T. H., and Eickbush, D. G. (2007). Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics 175, 477–485. doi: 10.1534/genetics.107.071399
Ellis, T. H., Lee, D., Thomas, C. M., Simpson, P. R., Cleary, W. G., Newman, M. A., et al. (1988). 5S rRNA genes in Pisum: sequence, long range and chromosomal organization. Mol. Gen. Genet. 214, 333–342. doi: 10.1007/BF00337732
Flavell, R. B., O'Dell, M., and Thompson, W. F. (1988). Regulation of cytosine methylation in ribosomal DNA and nucleolus organizer expression in wheat. J. Mol. Biol. 204, 523–534. doi: 10.1016/0022-2836(88)90352-X
Fraser, R., and Loening, U. (1974). RNA synthesis during synchronous cell division in cultured explants of Jerusalem artichoke tuber. J. Exp. Bot. 25, 847–859. doi: 10.1093/jxb/25.5.847
Friedrich, H., Hemleben, V., Meagher, R. B., and Key, J. L. (1979). Purification and restriction endonuclease mapping of soybean 18 S and 25 S ribosomal RNA genes. Planta 146, 467–473. doi: 10.1007/Bf00380862
Fulnecek, J., Lim, K. Y., Leitch, A. R., Kovarik, A., and Matyasek, R. (2002). Evolution and structure of 5S rDNA loci in allotetraploid Nicotiana tabacum and its putative parental species. Heredity 88, 19–25. doi: 10.1038/sj.hdy.6800001
Fulnecek, J., Matyasek, R., Kovarik, A., and Bezdek, M. (1998). Mapping of 5-methylcytosine residues in Nicotiana tabacum 5S rRNA genes by genomic sequencing. Mol. Gen. Genet. 259, 133–141. doi: 10.1007/s004380050798
Galian, J. A., Rosato, M., and Rossello, J. A. (2012). Early evolutionary colocalization of the nuclear ribosomal 5S and 45S gene families in seed plants: evidence from the living fossil gymnosperm Ginkgo biloba. Heredity 108, 640–646. doi: 10.1038/hdy.2012.2
Gall, J. G. (1981). Chromosome structure and the C-value paradox. J. Cell Biol. 91, 3s–14s. doi: 10.1083/jcb.91.3.3s
Ganal, M., and Hemleben, V. (1986). Comparison of the ribosomal-RNA genes in 4 closely related Cucurbitaceae. Plant Syst. Evol. 154, 63–77. doi: 10.1007/Bf00984868
Ganal, M., Torres, R., and Hemleben, V. (1988). Complex structure of the DNA ribosomal spacer of Cucumis sativus (cucumber). Mol. Gen. Genet. 212, 548–554. doi: 10.1007/BF00330863
Garcia, S., Borowska-Zuchowska, N., Wendel, J. F., Ainouche, M., Kuderova, A., and Kovarik, A. (2020). The utility of graph clustering of 5S ribosomal DNA homoeologs in plant allopolyploids, homoploid hybrids and cryptic introgressants. Front. Plant Sci. 11:41. doi: 10.3389/fpls.2020.00041
Garcia, S., Garnatje, T., and Kovarik, A. (2012). Plant rDNA database: ribosomal DNA loci information goes online. Chromosoma 121, 389–394. doi: 10.1007/s00412-012-0368-7
Garcia, S., and Kovarik, A. (2013). Dancing together and separate again: gymnosperms exhibit frequent changes of fundamental 5S and 35S rRNA gene (rDNA) organisation. Heredity 111, 23–33. doi: 10.1038/hdy.2013.11
Garcia, S., Kovarik, A., Leitch, A. R., and Garnatje, T. (2016). Cytogenetic features of rRNA genes across land plants: analysis of the plant rDNA database. Plant J. 89, 1020–1030. doi: 10.1111/tpj.13442
Garcia, S., Lim, K. Y., Chester, M., Garnatje, T., Pellicer, J., Valles, J., et al. (2009). Linkage of 35S and 5S rRNA genes in Artemisia (family Asteraceae): first evidence from angiosperms. Chromosoma 118, 85–97. doi: 10.1007/s00412-008-0179-z
Garcia, S., Panero, J. L., Siroky, J., and Kovarik, A. (2010). Repeated reunions and splits feature the highly dynamic evolution of 5S and 35S ribosomal RNA genes (rDNA) in the Asteraceae family. BMC Plant Biol. 10:176. doi: 10.1186/1471-2229-10-176
Gerstner, J., Schiebel, K., von Waldburg, G., and Hemleben, V. (1988). Complex organization of the length heterogeneous 5' external spacer of mung bean (Vigna radiata) ribosomal DNA. Genome 30, 723–733. doi: 10.1139/g88-120
Gleba, Y. Y., Hinnisdaels, S., Sidorov, V. A., Kaleda, V. A., Parokonny, A. S., Boryshuk, N. V., et al. (1988). Intergeneric asymmetric hybrids between Nicotiana plumbaginifolia and Atropa belladonna obtained by "gamma-fusion". Theor. Appl. Genet. 76, 760–766. doi: 10.1007/BF00303523
Goldberg, R. B., Bemis, W. P., and Siegel, A. (1972). Nucleic acid hybridization studies within the genus Cucurbitaceae. Genetics 72, 253–266. doi: 10.1093/genetics/72.2.253
Gray, M. W. (2017). Lynn Margulis and the endosymbiont hypothesis: 50 years later. Mol. Biol. Cell 28, 1285–1287. doi: 10.1091/mbc.E16-07-0509
Grebenstein, B., Röser, M., Sauer, W., and Hemleben, V. (1998). Molecular phylogenetic relationships in Aveneae (Poaceae) species and other grasses as inferred from ITS1 and ITS2 rDNA sequences. Plant Syst. Evol. 213, 233–250. doi: 10.1007/Bf00985203
Grierson, D. (1972). The Synthesis of rRNA in Developing Primary Leaves of Phaseolus aureus. PhD, Edinburgh, UK.
Grierson, D. (1984). “Structure and expression of nuclear genes,” in Plant Molecular Biology. eds. D. Grierson and S. N. Covey (Glasgow: Blackie).
Grierson, D., and Hemleben, V. (1977). Ribonucleic acid from higher plant Matthiola incana. Molecular weight measurements and DNA RNA hybridization studies. Biochim. Biophys. Acta 475, 424–436. doi: 10.1016/0005-2787(77)90058-2
Grierson, D., and Loening, U. E. (1972). Distinct transcription products of ribosomal genes in 2 different tissues. Nature New Biol. 235, 80–82. doi: 10.1038/newbio235080a0
Grierson, D., and Loening, U. (1974). Ribosomal RNA precursors and synthesis of chloroplast and cytoplasmic ribosomal acid in leaves of Phaseolus aureus. Eur. J. Biochem. 44, 501–507. doi: 10.1111/j.1432-1033.1974.tb03508.x
Grierson, D., Rogers, M. E., Sartiran, M., and Loening, U. E. (1970). The synthesis of ribosomal RNA in different organisms: structure and evolution of rRNA precursor. Cold Spring Harb. Symp. Quant. Biol. 35, 589–598. doi: 10.1101/Sqb.1970.035.01.074
Grierson, D., and Smith, H. (1973). The synthesis and stability of ribosomal RNA in blue-green algae. Eur. J. Biochem. 36, 280–285. doi: 10.1111/j.1432-1033.1973.tb02911.x
Grimm, G. W., Schlee, M., Komarova, N. Y., Volkov, R. A., and Hemleben, V. (2005). “Low-level taxonomy and intrageneric evolutionary trends in higher plants,” in From Plant Taxonomy to Evolutionary Biology. Nova Acta Leopoldina NF, Vol. 92, No. 342. eds. P. K. Endress, U. Lüttge, and B. Parthier (Stuttgart: Wissenschaftl. Verlagsges. mbH), 129–145.
Grossmann, K., Friedrich, H., and Seitz, U. (1980). Purification and characterization of chromatin-bound DNA-dependent RNA polymerase-I from parsley (Petroselinum crispum) - influence of nucleoside triphosphates. Biochem. J. 191, 165–171. doi: 10.1042/bj1910165
Grossmann, K., Seitz, U., and Seitz, H. U. (1979). Transcription and release of RNA in isolated nuclei from parsley cells. Z. Naturforsch. C. J. Biosci. 34, 431–435. doi: 10.1515/znc-1979-5-619
Gruendler, P., Unfried, I., Pointner, R., and Schweizer, D. (1989). Nucleotide sequence of the 25S-18S ribosomal gene spacer from Arabidopsis thaliana. Nucl. Acids Res. 17, 6395–6396. doi: 10.1093/nar/17.15.6395
Grummt, I., Sorbaz, H., Hofmann, A., and Roth, E. (1985). Spacer sequences downstream of the 28S RNA coding region are part of the mouse rDNA transcription unit. Nucl. Acids Res. 13, 2293–2304. doi: 10.1093/nar/13.7.2293
Handa, H., Kanamori, H., Tanaka, T., Murata, K., Kobayashi, F., Robinson, S. J., et al. (2018). Structural features of two major nucleolar organizer regions (NORs), nor-B1 and nor-B2, and chromosome-specific rRNA gene expression in wheat. Plant J. 96, 1148–1159. doi: 10.1111/tpj.14094
Hartley, M. R., and Ellis, R. J. (1973). Ribonucleic acid synthesis in chloroplasts. Biochem. J. 134, 249–262. doi: 10.1042/bj1340249
Hemleben, V. (1972). Untersuchungen zur Biosynthese und Funktion von Nucleinsäuren in höheren Pflanzen. Habilitation, University of Tübingen.
Hemleben, V., Ermisch, N., Kimmich, D., Leber, B., and Peter, G. (1975). Studies on fate of homologous DNA applied to seedlings of Matthiola incana. Eur. J. Biochem. 56, 403–411. doi: 10.1111/j.1432-1033.1975.tb02246.x
Hemleben, V., and Grierson, D. (1978). Evidence that in higher plants the 25S and 18S rRNA genes are not interspersed with genes for 5S rRNA. Chromosoma 65, 353–358. doi: 10.1007/Bf00286414
Hemleben, V., Grierson, D., and Dertmann, H. (1977). Use of equilibrium centrifugation in actinomycin cesium chloride for purification of ribosomal DNA. Plant Sci. Lett. 9, 129–135. doi: 10.1016/0304-4211(77)90090-6
Hemleben, V., Kovarik, A., Torres, R. A., Volkov, R. A., and Beridze, T. (2007). Plant highly repeated satellite DNA: molecular evolution, distribution and use for identification of hybrids. Syst. Biodivers. 5, 277–289. doi: 10.1017/S147720000700240X
Hemleben, V., and Werts, D. (1988). Sequence organization and putative regulatory elements in the 5S rRNA genes of 2 higher plants (Vigna radiata and Matthiola incana). Gene 62, 165–169. doi: 10.1016/0378-1119(88)90591-4
Hemleben, V., and Zentgraf, U. (1994). “Structural organisation and regulation of transcription by RNA polymerase I of plant nuclear ribosomal genes,” in Plant Promoters and Transcription Factors. ed. L. Nover (Berlin/Heidelberg: Springer-Verlag), 3–24.
Hemleben-Vielhaben, V. (1966). Characterization of rapidly labelled nucleic acids in tissues of plant seedlings. Z. Naturforsch. Pt. B. 21, 983–992. doi: 10.1515/znb-1966-1016
Hoang, P. T. N., Fiebig, A., Novák, P., Macas, J., Cao, H. X., Stepanenko, A., et al. (2020). Chromosome-scale genome assembly for the duckweed Spirodela intermedia, integrating cytogenetic maps, PacBio and Oxford Nanopore libraries. Sci. Rep. 10:19230. doi: 10.1038/s41598-020-75728-9
Ingle, J., Timmis, J. N., and Sinclair, J. (1975). The relationship between satellite deoxyribonucleic acid, ribosomal ribonucleic acid gene redundancy, and genome size in plants. Plant Physiol. 55, 496–501. doi: 10.1104/pp.55.3.496
Ishchenko, O. O., Bednarska, O. I., and Panchuk, I. I. (2021). Application of 5S ribosomal DNA for molecular taxonomy of subtribe Loliinae (Poaceae). Cytol. Genet. 55, 10–18. doi: 10.3103/S0095452721010096
Ishchenko, O. O., Mel’nyk, V. М., Parnikoza, І. Y., Budzhak, V. V., Panchuk, І. І., Kunakh, V. А., et al. (2020). Molecular organization of 5S ribosomal DNA and taxonomic status of Avenella flexuosa (L.) Drejer (Poaceae). Cytol. Genet. 54, 505–513. doi: 10.3103/S0095452720060055
Ishchenko, O. O., Panchuk, І. І., Andreev, І. O., Kunakh, V. A., and Volkov, R. A. (2018). Molecular organization of 5S ribosomal DNА of Deschampsia antarctica. Cytol. Genet. 52, 416–421. doi: 10.3103/S0095452718060105
Jo, S. H., Koo, D. H., Kim, J. F., Hu, C.-G., Lee, S., Yang, T. J., et al. (2009). Evolution of ribosomal DNA-derived satellite repeat in tomato genome. BMC Plant Biol. 9:42. doi: 10.1186/1471-2229-9-42
Jobst, J., King, K., and Hemleben, V. (1998). Molecular evolution of the internal transcribed spacers (ITS1 and ITS2) and phylogenetic relationships among species of the family Cucurbitaceae. Mol. Phylogenetics Evol. 9, 204–219. doi: 10.1006/mpev.1997.0465
Jung, H., Winefield, C., Bombarelym, A., Prentis, P., and Waterhouse, P. (2019). Tools and strategies for long-read sequencing and de novo assembly of plant genomes. Trends Plant Sci. 24, 700–724. doi: 10.1016/j.tplants.2019.05.003
Kalendar, R., Tanskanen, J., Chang, W., Antonius, K., Sela, H., Peleg, O., et al. (2008). Cassandra retrotransposons carry independently transcribed 5S RNA. Proc. Natl. Acad. Sci. U. S. A. 105, 5833–5838. doi: 10.1073/pnas.0709698105
King, K., Torres, R. A., Zentgraf, U., and Hemleben, V. (1993). Molecular evolution of the intergenic spacer in the nuclear ribosoma RNA genes of Cucurbitaceae. J. Mol. Evol. 36, 144–152. doi: 10.1007/Bf00166250
Kirov, I., Gilyok, M., Knyazev, A., and Fesenko, I. (2018). Pilot satellitome analysis of the model plant, Physcomitrella patens, revealed a transcribed and high-copy IGS related tandem repeat. Comp. Cytogenet. 12, 493–513. doi: 10.3897/CompCytogen.v12i4.31015
Komarova, N. Y., Grabe, T., Huigen, D. J., Hemleben, V., and Volkov, R. A. (2004). Organization, differential expression and methylation of rDNA in artificial Solanum allopolyploids. Plant Mol. Biol. 56, 439–463. doi: 10.1007/s11103-004-4678-x
Komarova, N. Y., Grimm, G. W., Hemleben, V., and Volkov, R. A. (2008). Molecular evolution of 35S rDNA and taxonomic status of Lycopersicon within Solanum sect. Petota. Plant Syst. Evol. 276, 59–71. doi: 10.1007/s00606-008-0091-2
Koukalova, B., Fojtova, M., Lim, K. Y., Fulnecek, J., Leitch, A. R., and Kovarik, A. (2005). Dedifferentiation of tobacco cells is associated with ribosomal RNA gene hypomethylation, increased transcription, and chromatin alterations. Plant Physiol. 139, 275–286. doi: 10.1104/pp.105.061788
Kovarik, A., Dadejova, M., Lim, Y. K., Chase, M. W., Clarkson, J. J., Knapp, S., et al. (2008). Evolution of rDNA in Nicotiana allopolyploids: a potential link between rDNA homogenization and epigenetics. Ann. Bot. 101, 815–823. doi: 10.1093/aob/mcn019
Kovarik, A., Fajkus, J., Koukalova, B., and Bezdek, M. (1996). Species-specific evolution of telomeric and rDNA repeats in the tobacco composite genome. Theor. Appl. Genet. 92, 1108–1111. doi: 10.1007/BF00224057
Kovarik, A., Matyasek, R., Lim, K. Y., Skalicka, K., Koukalova, B., Knapp, S., et al. (2004). Concerted evolution of 18-5.8-26S rDNA repeats in Nicotiana allotetraploids. Biol. J. Linn. Soc. 82, 615–625. doi: 10.1111/j.1095-8312.2004.00345.x
Layat, E., Sáez-Vásquez, J., and Tourmente, S. (2012). Regulation of pol I-transcribed 45S rDNA and pol III-transcribed 5S rDNA in Arabidopsis. Plant Cell Physiol. 53, 267–276. doi: 10.1093/pcp/pcr177
Leaver, C. J., and Key, J. L. (1970). Ribosomal RNA synthesis in plants. J. Mol. Biol. 49, 671–680. doi: 10.1016/0022-2836(70)90290-1
Leber, B., and Hemleben, V. (1979). Structure of plant nuclear and ribosomal DNA containing chromatin. Nucl. Acids Res. 7, 1263–1282. doi: 10.1093/nar/7.5.1263
Leweke, B., and Hemleben, V. (1982). Organization of rDNA in chromatin: plants. Cell Nucleus 11, 225–253.
Lim, K. Y., Kovarik, A., Matyasek, R., Bezdek, M., Lichtenstein, C. P., and Leitch, A. R. (2000). Gene conversion of ribosomal DNA in Nicotiana tabacum is associated with undermethylated, decondensed and probably active gene units. Chromosoma 109, 161–172. doi: 10.1007/s004120050424
Lim, K. Y., Kovarik, A., Matyasek, R., Chase, M. W., Clarkson, J. J., Grandbastien, M. A., et al. (2007). Sequence of events leading to near complete genome turnover in allopolyploid Nicotiana within five million years. New Phytol. 175, 756–763. doi: 10.1111/j.1469-8137.2007.02121.x
Lim, K. Y., Matyasek, R., Kovarik, A., and Leitch, A. (2004a). Genome evolution in allotetraploid Nicotiana. Biol. J. Linn. Soc. 82, 599–606. doi: 10.1111/j.1095-8312.2004.00344.x
Lim, K. Y., Skalicka, K., Koukalova, B., Volkov, R. A., Matyasek, R., Hemleben, V., et al. (2004b). Dynamic changes in the distribution of a satellite homologous to intergenic 26-18S rDNA spacer in the evolution of Nicotiana. Genetics 166, 1935–1946. doi: 10.1534/genetics.166.4.1935
Lin, C. Y., Chen, Y. M., Guilfoyle, T. J., and Key, J. L. (1976). Selective modulation of RNA polymerase I activity during growth transitions in the soybean seedling. Plant Physiol. 58, 614–617. doi: 10.1104/pp.58.5.614
Loening, U. E. (1969). The determination of molecular weight of ribonucleic acid by polyacrylamide-gel electrophoresis - effects of changes in conformation. Biochem. J. 113, 131–138. doi: 10.1042/bj1130131
Lunerova, J., Renny-Byfield, S., Matyasek, R., Leitch, A., and Kovarik, A. (2017). Concerted evolution rapidly eliminates sequence variation in rDNA coding regions but not in intergenic spacers in Nicotiana tabacum allotetraploid. Plant Syst. Evol. 303, 1043–1060. doi: 10.1007/s00606-017-1442-7
Macas, J., Navratilova, A., and Meszaros, T. (2003). Sequence subfamilies of satellite repeats related to rDNA intergenic spacer are differentially amplified on Vicia sativa chromosomes. Chromosoma 112, 152–158. doi: 10.1007/s00412-003-0255-3
Manoharlal, R., Saiprasad, G. V. S., and Kovarik, A. (2019). “Smoking and tobacco use,” in New Research on Tobacco. ed. U. Dreher (New York: Nova Science Publishers Inc), 23–82.
Marazia, T., Barsanti, P., and Maggini, F. (1980). Individual quantitative rDNA variation in 3 species of the Cucurbitaceae family. Biochem. Genet. 18, 509–517. doi: 10.1007/Bf00484398
Matsuda, K., and Siegel, A. (1967). Hybridization of plant ribosomal RNA to DNA - isolation of a DNA component rich in ribosomal RNA cistrons. Proc. Natl. Acad. Sci. U. S. A. 58, 673–680. doi: 10.1073/pnas.58.2.673
Matyasek, R., Dobesova, E., Huska, D., Jezkova, I., Soltis, P. S., Soltis, D. E., et al. (2016). Interpopulation hybridization generates meiotically stable rDNA epigenetic variants in allotetraploid Tragopogon mirus. Plant J. 85, 362–377. doi: 10.1111/tpj.13110
Matyasek, R., Lim, K. Y., Kovarik, A., and Leitch, A. R. (2003). Ribosomal DNA evolution and gene conversion in Nicotiana rustica. Heredity 91, 268–275. doi: 10.1038/sj.hdy.6800333
McClintock, B. (1934). The relationship of a particular crhomosomal element to the development of the nucleoli in Zea mays. Z. Zellforsch Microsk. 21, 294–398. doi: 10.1007/BF00374060
Michael, T., Bryant, D., Gutierrez, R., Borisjuk, N., Chu, P., Zhang, H., et al. (2017). Comprehensive definitions of genome features in Spirodela polyrhiza by high-depth physical mapping and short-read DNA sequencing strategies. Plant J. 89, 617–635. doi: 10.1111/tpj.13400
Michael, T. P., and Van Buren, R. (2020). Building near-complete plant genomes. Curr. Opin. Plant Biol. 54, 26–33. doi: 10.1016/j.pbi.2019.12.009
Miroshnichenko, G. P., Borisjuk, N. V., and Volkov, R. A. (1989). Organization of rDNA repeat units in the Solanaceae sexual and parasexual hybrids. Biochemistry 54, 669–675.
Mohannath, G., Pontvianne, F., and Pikaard, C. S. (2016). Chromosome-specific selective nucleolus organizer inactivation in Arabisopsis is a chromosome position-effect phenomenon. Proc. Natl. Acad. Sci. U. S. A. 113, 13426–13431. doi: 10.1073/pnas.1608140113
Moss, T. (2004). At the crossroads of growth control: making ribosomal RNA. Curr. Opin. Genet. Dev. 14, 210–217. doi: 10.1016/j.gde.2004.02.005
Nagl, W., Ehrendorfer, F., and Hemleben, V. (1979). “Genome and Chromatin: Organization, Evolution and Function,” in Plant Systematics and Evolution. 2nd Edn. eds. W. Nagl, F. Ehrendorfer, and V. Hemleben (Wien: Springer Verlag).
Navashin, M. (1934). Chromosomal alterations caused by hybridisation and their bearing upon certain genetic problems. Cytologia 5, 169–203.
Nieto Feliner, G., and Rossello, J. A. (2012). “Concerted evolution of multigene families and homeologous recombination,” in Plant Genome Diversity. ed. J. F. Wendel (Wien: Springer-Verlag), 171–194.
Noller, H. F., Kop, J., Wheaton, V., Brosius, J., Gutell, R. R., Kopylov, A. M., et al. (1981). Secondary structure model for 23S ribosomal RNA. Nucl. Acids Res. 9, 6167–6189. doi: 10.1093/nar/9.22.6167
Pikaard, C. S., and Reeder, R. H. (1988). Sequence elements essential for function of the Xenopus laevis ribosomal DNA enhancers. Mol. Cell. Biol. 8, 4282–4288. doi: 10.1128/Mcb.8.10.4282
Poczai, P., and Hyvonen, J. (2010). Nuclear ribosomal spacer regions in plant phylogenetics: problems and prospects. Mol. Biol. Rep. 37, 1897–1912. doi: 10.1007/s11033-009-9630-3
Pontvianne, F., Abou-Ellail, M., Douet, J., Comella, P., Matia, I., Chandrasekhara, C., et al. (2010). Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet. 6:e1001225. doi: 10.1371/journal.pgen.1001225
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rabanal, F. A., Mandáková, T., Soto-Jiménez, L. M., Greenhalgh, R., Parrott, D. L., Lutzmayer, S., et al. (2017). Epistatic and allelic interactions control expression of ribosomal RNA gene clusters in Arabidopsis thaliana. Genome Biol. 18:75. doi: 10.1186/s13059-017-1209-z
Rathgeber, J., and Capesius, I. (1990). Nucleotide sequence of the intergenic spacer and the 18S ribosomal-RNA gene from mustard (Sinapis alba). Nucl. Acids Res. 18, 1288–1288. doi: 10.1093/nar/18.5.1288
Ribeiro, T., Dos Santos, K. G., Richard, M. M., Sévignac, M., Thareau, V., Geffroy, V., et al. (2017). Evolutionary dynamics of satellite DNA repeats from Phaseolus beans. Protoplasma 254, 791–801. doi: 10.1007/s00709-016-0993-8
Robicheau, B. M., Susko, E., Harrigan, A. M., and Snyder, M. (2017). Ribosomal RNA genes contribute to the formation of pseudogenes and junk DNA in the human genome. Genome Biol. Evol. 9, 380–397. doi: 10.1093/gbe/evw307
Rogers, S. O., and Bendich, A. J. (1987). Ribosomal RNA genes in plants: variability in copy number and in the intergenic spacer. Plant Mol. Biol. 9, 509–520. doi: 10.1007/BF00015882
Rogers, S. O., Honda, S., and Bendich, A. J. (1986). Variation in the ribosomal RNA genes among individuals of Vicia faba. Plant Mol. Biol. 6, 339–345. doi: 10.1007/Bf00034941
Röser, M., Winterfeld, G., Grebenstein, B., and Hemleben, V. (2001). Molecular diversity and physical mapping of 5S rDNA in wild, and cultivated oat grasses (Poaceae: Aveneae). Mol. Phylogenet. Evol. 21, 198–217. doi: 10.1006/mpev.2001.1003
Sáez-Vásquez, J., and Delseny, M. (2019). Ribosome biogenesis in plants: from functional 45S ribosomal DNA organization to ribosome assembly factors. Plant Cell 31, 1945–1967. doi: 10.1105/tpc.18.00874
Sáez-Vásquez, J., and Pikaard, C. S. (2000). RNA polymerase I holoenzyme-promoter interactions. J. Biol. Chem. 275, 37173–37180. doi: 10.1074/jbc.M006057200
Salim, D., Bradford, W. D., Freeland, A., Cady, G., Wang, J., Pruitt, S. C., et al. (2017). DNA replication stress restricts ribosomal DNA copy number. PLoS Genet. 13:e1007006. doi: 10.1371/journal.pgen.1007006
Sanger, F., Nicklen, S., and Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74, 5463–5467. doi: 10.1073/pnas.74.12.5463
Sardana, R., Odell, M., and Flavell, R. (1993). Correlation between the size of the intergenic regulatory region, the status of cytosine methylation of ribosomal-RNA genes and nucleolar expression in wheat. Mol. Gen. Genet. 236, 155–162. doi: 10.1007/Bf00277107
Schiebel, K., and Hemleben, V. (1989). Nucleotide sequence of the 18S-25S spacer region from rDNA of mung bean. Nucl. Acids Res. 17, 2852–2852. doi: 10.1093/nar/17.7.2852
Schlee, M., Göker, M., Grimm, G. W., and Hemleben, V. (2011). Genetic patterns in the Lathyrus pannonicus complex (Fabaceae) reflect ecological differentiation rather than biogeography and traditional subspecific division. Biol. J. Linn. Soc. 165, 402–421. doi: 10.1111/j.1095-8339.2011.01125.x
Schlotterer, C., and Tautz, D. (1994). Chromosomal homogeneity of Drosophila ribosomal DNA arrays suggests intrachromosomal exchanges drive concerted evolution. Curr. Biol. 4, 777–783. doi: 10.1016/S0960-9822(00)00175-5
Schweizer, G., Borisjuk, N., Borisjuk, L., Stadler, M., Stelzer, T., Schilde, L., et al. (1993). Molecular analysis of highly repeated genome fractions in Solanum and their use as markers for the characterization of species and cultivars. Theor. Appl. Genet. 85-85, 801–808. doi: 10.1007/Bf00225022
Scoles, C. J., Gill, B. S. Z., Xin, Y., Clarke, B. C., McIntyre, C. L., Chapman, C., et al. (1988). Frequent duplication and deletion events in the 5S RNA genes and the associated spacer regions of the Triticeae. Plant Syst. Evol. 160, 105–122.
Seitz, U., and Seitz, U. (1973). Biosynthetic pathway of ribosomal-RNA in blue-green algae (Anacystis nidulans). Archiv fur Mikrobiologie. 90, 213–222. doi: 10.1007/Bf00424973
Seitz, U., and Seitz, U. (1979). Molecular weight of rRNA precursor molecules and their processing in higher plant cells. Z. Naturforsch. C. J. Biosci. 34, 253–258. doi: 10.1515/znc-1979-3-416
Selig, C., Wolf, M., Müller, T., Dandekar, T., and Schultz, J. (2008). The ITS2 database II: homology modelling RNA structure for molecular systematics. Nucl. Acids Res. 36, D377–D380. doi: 10.1093/nar/gkm827
Sims, J., Sestini, G., Elgert, C., von Haeseler, A., and Schlögelhofer, P. (2021). Sequencing of the Arabidopsis NOR2 reveals its distinct organization and tissue-specific rRNA ribosomal variants. Nat. Commun. 12, 387. doi: 10.1038/s41467-020-20728-6
Soltis, D. E., and Soltis, P. S. (2016). Mobilizing and integrating big data in studies of spatial and phylogenetic patterns of biodiversity. Plant Divers. 38, 264–270. doi: 10.1016/0022-2836(88)90353-1
Souza, G., Marques, A., Ribeiro, T., Dantas, L. G., Speranza, P., Guerra, M., et al. (2019). Allopolyploidy and extensive rDNA site variation underlie rapid karyotype evolution in Nothoscordum section Nothoscordum (Amaryllidaceae). Bot. J. Linn. Soc. 190, 215–228. doi: 10.1093/botlinnean/boz008
Szymanski, M., Specht, T., Barciszewska, M. Z., Barciszewski, J., and Erdmann, V. A. (1998). 5S rRNA Data Bank. Nucl. Acids Res. 26, 156–159. doi: 10.1093/nar/26.1.156
Thompson, W. F., and Flavell, R. B. (1988). DNAse-I sensitivity of ribosomal RNA genes in chromatin and nucleolar dominance in wheat. J. Mol. Biol. 204, 535–548. doi: 10.1016/0022-2836(88)90353-1
Tomecki, R., Sikorski, P. J., and Zakrzewska-Placzek, M. (2017). Comparison of preribosomal RNA processing pathways in yeast, plant and human cells - focus on coordinated action of endo- and exoribonucleases. FEBS Lett. 591, 1801–1850. doi: 10.1002/1873-3468.12682
Torres, R. A., Zentgraf, U., and Hemleben, V. (1989). Species and genus specificity of the intergenic spacer (IGS) in the ribosoma RNA genes of Cucurbitaceae. Z. Naturforsch. C. J. Biosci. 44, 1029–1034. doi: 10.1515/znc-1989-11-1224
Torres-Ruiz, R. A., and Hemleben, V. (1994). Pattern and degree of methylation in ribosomal RNA genes of Cucurbita pepo L. Plant Mol. Biol. 26, 1167–1179. doi: 10.1007/Bf00040697
Tulpová, Y. O., Kovařík, A., Toegelová, H., Navrátilová, P., Kapustová, V., Hřibová, E., et al. (2020). Anatomy, transcription dynamics and evolution of wheat ribosomal RNA loci deciphered by a multi-omics approach. BioRxriv [Preprint]. doi: 10.1101/2020.08.29.273623
Tynkevich, Y. O., and Volkov, R. A. (2014). Structural organization of 5S ribosomal DNA in Rosa rugosa. Cytol. Genet. 48, 1–6. doi: 10.3103/S0095452714010095
Tynkevich, Y. O., and Volkov, R. A. (2019). 5S ribosomal DNA of distantly related Quercus species: molecular organization and taxonomic application. Cytol. Genet. 53, 459–466. doi: 10.3103/S0095452719060100
Unfried, K., Schiebel, K., and Hemleben, V. (1991). Subrepeats of rDNA intergenic spacer present as prominent independent satellite DNA in Vigna radiata but not in Vigna angularis. Gene 99, 63–68. doi: 10.1016/0378-1119(91)90034-9
Volkov, R. A., Bachmair, A., Panchuk, I. I., Kostyshyn, S. S., and Schweizer, D. (1999a). 25S-18S rDNA intergenic spacer of Nicotiana sylvestris (Solanaceae): primary and secondary structure analysis. Plant Syst. Evol. 218, 89–97. doi: 10.1007/bf01087037
Volkov, R. A., Borisjuk, N. V., Kostishin, S. S., and Panchuk, I. I. (1991). Variability of rRNA genes in Nicotiana correlates with the chromosome reconstruction. Mol. Biol. 25, 442–450.
Volkov, R. A., Borisjuk, N. V., Panchuk, I. I., Schweizer, D., and Hemleben, V. (1999b). Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Mol. Biol. Evol. 16, 311–320. doi: 10.1093/oxfordjournals.molbev.a026112
Volkov, R. A., Komarova, N. Y., and Hemleben, V. (2007). Ribosomal DNA in plant hybrids: inheritance, rearrangement, expression. Syst. Biodivers. 5, 261–276. doi: 10.1017/S1477200007002447
Volkov, R. A., Komarova, N. Y., Panchuk, I. I., and Hemleben, V. (2003). Molecular evolution of rDNA external transcribed spacer and phylogeny of sect. Petota (genus Solanum). Mol. Phylogenetics Evol. 29, 187–202. doi: 10.1016/s1055-7903(03)00092-7
Volkov, R., Kostishin, S., Ehrendorfer, E., and Schweizer, D. (1996). Molecular organization and evolution of the external transcribed rDNA spacer region in two diploid relatives of Nicotiana tabacum (Solanaceae). Plant Syst. Evol. 201, 117–129. doi: 10.1007/Bf00989055
Volkov, R. A., Kozeretska, I. A., Kyryachenko, S. S., Andreev, I. O., Maidanyuk, D. N., Parnikoza, I. Y., et al. (2010). Molecular evolution and variability of ITS1–ITS2 in populations of Deschampsia antarctica from two regions of the maritime Antarctic. Pol. Sci. 4, 469–478. doi: 10.1016/j.polar.2010.04.011
Volkov, R. A., Medina, F. J., Zentgraf, U., and Hemleben, V. (2004). “Molecular cell biology: organization and molecular evolution of rDNA, nucleolar dominance and nucleolus structure,” in Progress in Botany. Vol. 65. eds. K. Esser, U. Lüttge, W. Beyschlag, and J. Murata (Berlin, Heidelberg, New York: Springer Verlag), 106–146.
Volkov, R. A., Panchuk, I. I., Borisjuk, N. V., Maluszynska, J., and Hemleben, V. (2017). Evolutional dynamics of 45S and 5S ribosomal DNA in ancient allohexaploid Atropa belladonna. BMC Plant Biol. 17, 21–24. doi: 10.1186/s12870-017-0978-6
Volkov, R. A., Zanke, C., Panchuk, I. I., and Hemleben, V. (2001). Molecular evolution of 5S rDNA of Solanum species (sect. Petota): application for molecular phylogeny and breeding. Theor. Appl. Genet. 103, 1273–1282. doi: 10.1007/s001220100670
Wang, W., Wan, T., Becher, H., Kuderova, A., Leitch, I. J., Garcia, S., et al. (2019). Remarkable variation of ribosomal DNA organization and copy number in gnetophytes, a distinct lineage of gymnosperms. Ann. Bot. 123, 767–781. doi: 10.1093/aob/mcy172
Weis, B. L., Kovacevic, J., Missbach, S., and Schleiff, E. (2015). Plant-specific features of ribosome biogenesis. Trends Plant Sci. 20, 729–740. doi: 10.1016/j.tplants.2015.07.003
Wenzel, W., and Hemleben, V. (1982a). A comparative study of genomes in angiosperms. Plant Syst. Evol. 139, 209–227. doi: 10.1007/Bf00989326
Wenzel, W., and Hemleben, V. (1982b). DNA-sequence organization and RNA complexity in Matthiola incana (Brassicaceae). Plant Syst. Evol. 140, 75–86. doi: 10.1007/Bf02409898
Wicke, S., Costa, A., Munoz, J., and Quandt, D. (2011). Restless 5S: the re-arrangement(s) and evolution of the nuclear ribosomal DNA in land plants. Mol. Phylogenet. Evol. 61, 321–332. doi: 10.1016/j.ympev.2011.06.023
Yan, Q., Zhu, C. M., Guang, S. H., and Feng, X. Z. (2019). The functions of non-coding RNAs in rRNA regulation. Front. Genet. 10:290. doi: 10.3389/fgene.2019.00290
Yokota, Y., Kawata, T., Iida, Y., Kato, A., and Tanifuji, S. (1989). Nucleotide sequences of the 5.8S rRNA gene and internal transcribed spacer regions in carrot and broad bean ribosomal DNA. J. Mol. Evol. 29, 294–301. doi: 10.1007/Bf02103617
Zakrzewska-Placzek, M., Souret, F. F., Sobczyk, G. J., Green, P. J., and Kufel, J. (2010). Arabidopsis thaliana XRN2 is required for primary cleavage in the pre-ribosomal RNA. Nucl. Acids Res. 38, 4487–4502. doi: 10.1093/nar/gkq172
Zentgraf, U., and Hemleben, V. (1992). Complex-formation of nuclear proteins with the RNA polymerase-I promoter and repeated elements in the external transcribed spacer of Cucumis sativus ribosomal DNA. Nucl. Acids Res. 20, 3685–3691. doi: 10.1093/nar/20.14.3685
Zentgraf, U., and Hemleben, V. (1993). Nuclear proteins interact with RNA polymerase-I promoter and repeated elements of the 5' external transcribed spacer of the rDNA of cucumber in a single-stranded stage. Plant Mol. Biol. 22, 1153–1156. doi: 10.1007/Bf00028984
Keywords: rDNA research history, rRNA precursor, rRNA processing, molecular evolution, epigenetics, polyploidy, hybridization, nucleolar dominance
Citation: Hemleben V, Grierson D, Borisjuk N, Volkov RA and Kovarik A (2021) Personal Perspectives on Plant Ribosomal RNA Genes Research: From Precursor-rRNA to Molecular Evolution. Front. Plant Sci. 12:797348. doi: 10.3389/fpls.2021.797348
Edited by:
Wellington Ronildo Clarindo, Universidade Federal de Viçosa, BrazilReviewed by:
Andrea Pedrosa-Harand, Federal University of Pernambuco, BrazilMilene Miranda Praça-Fontes, Universidade Federal do Espírito Santo, Brazil
Copyright © 2021 Hemleben, Grierson, Borisjuk, Volkov and Kovarik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vera Hemleben, vera.hemleben@uni-tuebingen.de; Ales Kovarik, kovarik@ibp.cz
†ORCID: Vera Hemleben, orcid.org/0000-0002-5171-472
Donald Grierson, orcid.org/0000-0002-2238-8072
Nikolai Borisjuk, orcid.org/0000-0001-5250-9771
Roman A. Volkov, orcid.org/0000-0003-0673-2598
Ales Kovarik, orcid.org/0000-0003-2896-0698
 Vera Hemleben1*†
Vera Hemleben1*† Donald Grierson
Donald Grierson Nikolai Borisjuk
Nikolai Borisjuk Roman A. Volkov
Roman A. Volkov Ales Kovarik
Ales Kovarik