- 1College of Agronomy, Northwest A&F University, Xianyang, China
- 2College of Tobacco, Henan Agricultural University, Zhengzhou, China
Drought is the main abiotic stress factor limiting the growth and yield of wheat (Triticum aestivum L.). Therefore, improving wheat tolerance to drought stress is essential for maintaining yield. Previous studies have reported on the important role of TaNRX1 in conferring drought stress tolerance. Therefore, to elucidate the regulation mechanism by which TaNRX1 confers drought resistance in wheat, we generated TaNRX1 overexpression (OE) and RNA interference (RNAi) wheat lines. The results showed that the tolerance of the OE lines to drought stress were significantly enhanced. The survival rate, leaf chlorophyll, proline, soluble sugar content, and activities of the antioxidant enzymes (catalase, superoxide dismutase, and peroxidase) of the OE lines were higher than those of the wild type (WT); however, the relative electrical conductivity and malondialdehyde, hydrogen peroxide, and superoxide anion levels of the OE lines were lower than those of the WT; the RNAi lines showed the opposite results. RNA-seq results showed that the common differentially expressed genes of TaNRX1 OE and RNAi lines, before and after drought stress, were mainly distributed in the plant–pathogen interaction, plant hormone signal transduction, phenylpropane biosynthesis, starch and sucrose metabolism, and carbon metabolism pathways and were related to the transcription factors, including WRKY, MYB, and bHLH families. This study suggests that TaNRX1 positively regulates drought stress tolerance in wheat.
Introduction
Thioredoxin (TRX) is a class of small proteins with a molecular weight of approximately 12 kDa widely found in animals, plants, fungi, and microorganisms (Meyer et al., 2012). TRX contains a conserved active site domain, WCG/PPC, and two cysteine (Cys) residues in the active center that participate in the redox reaction through the reversible conversion of thiol–disulfide to maintain the stability of the cell environment (Powis and Montfort, 2001; Marx et al., 2003). TRX plays an irreplaceable role in maintaining the balance of the intracellular redox state and regulating the spatial structure and activity of the proteins. It is also involved in regulating plant growth and development and the response process in resisting drought, high temperature, and other adverse stresses (Marx et al., 2003). TRX participates in a series of physiological and biochemical processes in cells by reducing disulfide bonds in the target proteins (Wong et al., 2004; Joudrier et al., 2005; Traverso et al., 2007; Schurman and Buchanan, 2008). Nucleoredoxin (NRX) is a member of the TRX family, and its molecular weight is considered to be larger than that of TRX.
Nucleoredoxin in plants was first isolated from maize (Zea mays L.) by Laughner et al. (1998). Based on the number of TRX-like domains and its amino acid sequence, the plant NRX can be classified into three types. Type I NRX contains three TRX-like domains; the first and third domains contain typical WCG/PPC redox active sites, and the second domain is an atypical TRX-like domain. Type II NRX contains two TRX-like domains and the atypical TRX active sites (WYP/AK/PC and W/R/HCL/A/V/RPC/G). Type III NRX contains two TRX-like domains, including the highly conserved WCRPC redox active site and the typical TRX active site WCPPC/F/S (Marchal et al., 2014). Laughner et al. (1998) found that the maize NRX consists of three TRX-like domains, of which the first and third domains contain the active site WCPPC of the typical TRX-like domain, and the third domain exhibits a disulfide bond reduction ability in vitro. Li et al. (2016) also found that the first and third domains of the cotton (Gossypium barbadense L.) GbNRX1 contain the active site WCG/CPC, which is a typical TRX-like domain; GbNRX1 also has an obvious disulfide bond reduction activity. Both Arabidopsis thaliana NRX type I (AtNRX1) and type II (AtNRX2) exhibit disulfide bond reduction abilities in vitro (Marchal et al., 2014). Cheng (2021) showed that the three domains of wheat (Triticum aestivum L.) TaNRX1 exhibit insulin disulfide bond reduction activity in vitro; among these domains, the third domain has the strongest reduction activity and the second domain has the weakest activity.
Funato et al. (2013) found that NRX directly interacts with phosphofructokinase 1 (PFK1) to regulate the activity of PFK1, thereby maintaining the balance between glycolysis and pentose phosphate pathways. Zhao (2016) showed that AtNRX1 is involved in the regulation of reactive oxygen species (ROS) levels in Arabidopsis seedlings. Kneeshaw et al. (2017) showed that NRX1 targets enzymes in the hydrogen peroxide (H2O2) clearance pathway, including catalase (CAT); an NRX1 mutant showed reduced CAT activity and was extremely sensitive to oxidative stress. NRX1 regulates CAT activity, enhances the detoxification ability of CAT, and protects antioxidant enzymes from oxidative stress induced by ROS, thereby protecting plant cells from oxidative stress (Kneeshaw et al., 2017). Evidently, NRX plays an important role in plants, and an understanding of NRX and its target proteins will help elucidate the molecular mechanism of NRX in plant resistance to adverse stresses.
Wheat is one of the most important food crops worldwide. Wheat production is often adversely affected by biotic and abiotic stresses (Tuan et al., 2019), among which drought is the main abiotic stress factor limiting global wheat production (Valluru et al., 2016). It severely affects the growth and development of wheat plants by causing various physiological and biochemical damage, resulting in serious wheat yield declines (Dey et al., 2016; Zhou et al., 2016; Ma J. et al., 2017; Saradadevi et al., 2017; Yadav et al., 2019). Therefore, a comprehensive understanding of the molecular mechanisms of drought resistance in wheat is important for increasing wheat yield and ensuring food security.
Previously, in our laboratory, Zhang et al. (2014) cloned a TaNRX1 gene in common wheat and reported that TaNRX1 is associated with drought resistance; however, the molecular mechanisms underlying drought resistance in wheat require further elucidation. In this study, TaNRX1 overexpression (OE) and RNA interference (RNAi) transgenic wheat plants were subjected to drought stress treatment to elucidate the drought resistance mechanism of TaNRX1. This study identified an excellent candidate gene TaNRX1 for the production of drought-resistance wheat plants, selection and promotion of drought-resistant wheat varieties, and improvement in wheat yield and food security.
Materials and Methods
Plant Materials
The cDNA sequence of TaNRX1 (NCBI ID: KC890769) was cloned from Jinmai 47, a drought-tolerant common wheat variety. The wheat variety JW1 and expression vector pLGY-02 were provided by Professor Genying Li (Crop Research Institute, Shandong Academy of Agricultural Sciences, China). The vector pLGY-02 used has been presented in Liang et al. (2020).
Vector Construction and Generation of Transgenic Wheat
To generate transgenic plants overexpressing TaNRX1, the full-length coding sequence (CDS) of TaNRX1 was cloned into the KpnI-HF and SpeI-HF (Supplementary Table 1) (New England Biolabs, Beijing, China) sites of pLGY-02, a plant transformation vector driven by the maize ubiquitin promoter. The RNAi vector was constructed to silence TaNRX1 in wheat, and two 223-bp truncated fragments (forward and reverse fragments) of TaNRX1 CDS (ΔTaNRX1) were amplified and inserted into the pLGY-02 vector to form a hairpin structure (ΔTaNRX1-intron-ΔTaNRX1). The truncated fragments contained KpnI and SacI restriction sites. The constructed vector was then transformed into wheat (JW1) immature embryos via Agrobacterium tumefaciens-mediated transformation (Cheng et al., 1997).
Screening of Homogeneous Transgenic Wheat Lines
In this experiment, specific primers HygF1 and HygR1 (Supplementary Table 1) of the transgenic vector hygromycin resistance gene were used to detect different lines of transgenic wheat in the T1 generation and to screen the seeds of the transgenic wheat with a resistance: sensitivity ratio of 3:1 to identify single-copy transgenic lines. Genomic DNA was isolated from transgenic wheat leaves using the cetyltrimethylammonium bromide method (Porebski et al., 1997). The DNA was then used for polymerase chain reaction (PCR) assays, and PCR-amplified products were separated by 1.0% agarose gel electrophoresis. The T2 generation of the transgenic wheat was detected by PCR, and the T3 generation was detected by PCR and RT-qPCR.
Reverse Transcription Quantitative PCR
Total RNA was extracted from the leaves of the transgenic wheat using TRNzol Universal Reagent DP424 (TianGen Biotech, Beijing, China). cDNA was synthesized using EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix AE311-02 (TransGen Biotech, Beijing, China). To determine the transgenic wheat lines and drought stress periods for RNA-seq, RT-qPCR was performed using the TB Green Premix Ex Taq (Takara, Beijing, China). Each reaction was performed in triplicate. In addition, RT-qPCR was used to determine the relative expression levels of TaNRX1 in WT and transgenic wheat lines (TaNRX1-OE-1, TaNRX1-OE-3, and TaNRX1-OE-6 and TaNRX1-RNAi-6, TaNRX1-RNAi-8, and TaNRX1-RNAi-9) during different drought stress periods. All primers used in the experiments are listed in Supplementary Table 1. The actin gene was used as an endogenous control for template cDNA normalization, and the 2–ΔΔ CT method (Livak and Schmittgen, 2001) was used to calculate the relative expression levels.
Drought Stress Treatment
To determine the effect of drought stress on roots during the seedling stage, the germinated seeds of WT, OE, and RNAi wheat lines were cultivated with 1/2 Hoagland nutrient solution and 1/2 Hoagland nutrient solution with 20% (M/V) polyethylene glycol (PEG6000). Plants were cultivated in a light incubator at 20–25°C, with a 16-h-light/8-h-dark photoperiod, with humidity being approximately 70% and light intensity being 18,000 Lux. After 10 days of seed germination, morphological and physiological parameters of plant roots were measured once the final age of seedlings was 14 days.
To analyze the drought tolerance of wheat plants at the seedling stage, WT and TaNRX1 transgenic wheat seeds were germinated in petri dishes and transplanted into pots containing the same soil volumes (10 plants per pot) in a greenhouse at 15–20°C, with a 16-h-light/8-h-dark photoperiod for 4 weeks. Control plants were watered normally, and drought stress in plants was induced by withholding watering for 10 days when the relative water content of the soil was approximately 60%; the physiological indexes of the WT and transgenic wheat lines under control and drought stress conditions were measured. For 4-week-old WT, OE, and RNAi wheat plants, watering was withheld for 14 days when the soil relative water content was below 50%; rewatering was resumed after 3 days, and the phenotype and survival rates were recorded.
To analyze the drought resistance of the wheat at the heading stage, WT and TaNRX1 transgenic wheat lines were grown in pots with the same soil volumes (six plants per pot) in a greenhouse at 15–20°C, with a 16-h-light/8-h-dark photoperiod. The water supply of 45-day-old plants was stopped when the tip of the main spike emerged from the flag leaf sheath (Zhang et al., 2017). The soil relative water content (RWC) of the drought-stressed plants was maintained at 35–40%, and the control plants were watered normally. At 21 days after drought treatment, the phenotype was observed and chlorophyll content and photosynthesis-related indicators were measured. All drought stress treated plants were homogenous lines of the transgenic wheat T3 generation.
Measurement of Drought-Relevant Physiological Indicators
Wild type, OE, and RNAi wheat plants were used to measure the water loss rate according to Srivastava’s method (Srivastava et al., 2017). RWC was measured according to the method described by Chen et al. (2016). The content of proline and soluble sugar were measured according to the method described by Wang N. et al. (2017). Malondialdehyde (MDA) content was measured using the thiobarbituric acid method (Zhang et al., 2020). Relative electrical conductivity (REC) was determined according to the method described by Ma Y. et al. (2017).
Hydrogen peroxide (H2O2) and superoxide anions (O2–) were detected using 3,3-diamino-benzidinestaining (DAB) and nitroblue tetrazolium (NBT) staining, respectively. For DAB staining, the wheat control and drought stress leaves were immersed in DAB staining solution (0.4 mg/mL) for 4 h and then decolorized in an ethanol:acetic acid:glycerol solution at a ratio of 3:1:1 (V/V/V). For NBT staining, the wheat leaves were immersed in NBT staining solution (0.1 mg/mL) for 6 h and then decolorized according to the NBT staining method (Wang N. et al., 2017). Images were obtained using an Olympus stereo microscope (Olympus, Tokyo, Japan). The H2O2 content was measured using a H2O2 test box (Suzhou Keming Biotechnology Co., Ltd., Suzhou, China). The O2– level was measured according to the method described by Wang K. et al. (2017). The superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD) contents were measured using an SOD, CAT, and POD test box (Suzhou Keming Biotechnology Co., Ltd., Suzhou, China). The photosynthesis-related indicators at the heading stage were measured using a portable photosynthetic system (LI-6400XT, United States). In this study, at least three replicates were performed for each experiment.
RNA Sequencing
T3 homozygous lines of transgenic wheat and the WT were used for RNA-seq. Wheat seedlings were cultured in the same pot with 1/2 Hoagland nutrient solution for 2 weeks and then transferred into fresh 1/2 Hoagland nutrient solution containing 20% PEG6000. Leaves were collected at 0 and 6 h following drought treatment, and three biological replicates were used for RNA-seq.
RNA extraction and RNA-seq analysis were performed by Biomarker Biotech Company (Beijing, China). The RNA integrity number, with ≥7 samples, was selected for subsequent analysis. RNA-seq was performed using Illumina Novaseq 6000 system with paired-end sequencing and 150 bp sequencing reads. The Pearson correlation coefficient between each pair of biological replicates was calculated to confirm the reliability of the RNA-seq data. Gene expression levels were measured based on fragments per kilobase of transcript per million fragments mapped values. For significant differentially expressed genes (DEGs), log2[fold change] ≥ 2 and a false discovery rate < 0.05 were used as the screening criteria. Gene Ontology (GO) enrichment analysis was used to determine the main biological functions of DEGs, and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was used to determine the main physiological and biochemical metabolic pathways of DEGs. All RNA-seq data were analyzed using the Biomark Cloud Platform (BMKCloud1). To verify the reliability of the RNA-seq data, we randomly selected seven DEGs to determine their relative expression levels by RT-qPCR. RT-qPCR was performed as described in Section “Reverse Transcription Quantitative PCR.”
Statistical Analysis
All experiments were independently repeated at least three times to obtain reliable data for statistical analysis. Statistical analysis was performed using SPSS 20.0 (SPSS, Chicago, IL, United States). The parametric tests one-way analysis of variance was used for data analysis, and the least significant difference post hoc was used to determine significant differences between means (*, statistically significant at P < 0.05; **, statistically significant at P < 0.01).
Results
Generation and Identification of TaNRX1 Transgenic Wheat Lines
To study the drought resistance and molecular mechanism of TaNRX1 in wheat, we used TaNRX1 OE and RNAi-silenced wheat lines. A total of 140 embryos were infected; among these, after screening and regeneration, 16 OE and 15 RNAi plants were obtained.
To identify homozygous transgenic wheat lines, T1–T3 generations were confirmed by PCR. The vector pLGY-02 contained hygromycin resistance, and the 950 bp DNA segment was amplified in the transgenic wheat lines; however, no amplification was detected in WT plants (Supplementary Figure 1). RT-qPCR analysis demonstrated that the relative expression levels of TaNRX1 in the three OE lines (TaNRX1-OE-1, TaNRX1-OE-3, and TaNRX1-OE-6) were significantly (P < 0.05) higher than those in the WT (Figure 1A). Similarly, the relative expression levels of TaNRX1 in three RNAi lines (TaNRX1-RNAi-6, TaNRX1-RNAi-8, and TaNRX1-RNAi-9) decreased by approximately 76, 44, and 66%, respectively (Figure 1B). These results confirmed that TaNRX1 was integrated and expressed in the wheat genome; the OE (TaNRX1-OE-1, TaNRX1-OE-3, and TaNRX1-OE-6) and RNAi (TaNRX1-RNAi-6, TaNRX1-RNAi-8, and TaNRX1-RNAi-9) lines were selected for subsequent experiments. To understand whether other TRX or NRX members were down-regulated by off-targeting, we compared expression levels of TRX and NRX members in WT and RNAi lines that were not subjected to drought stress (drought stress 0 h) in RNA-seq data. According to whether the protein has at least one Thioredoxin domain, a total of 109 TRX and NRX members were identified. Results showed that compared with expression levels of TRX and NRX members in WT, those of other TRX and NRX members in the RNAi lines were not down-regulated (Supplementary Table 2), indicating no off-targeting, and therefore the RNAi lines were used for subsequent experiments.
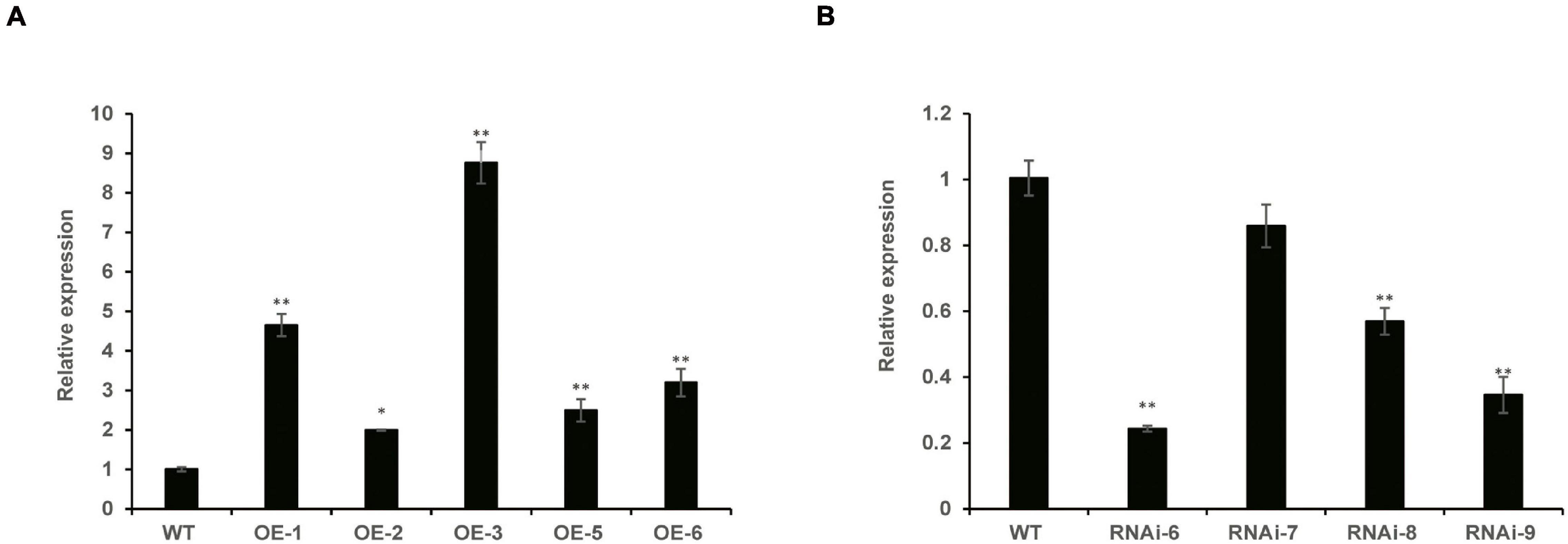
Figure 1. The relative expression of TaNRX1 in transgenic wheat. (A) The relative expression of TaNRX1 in overexpression lines (n = 3). (B) The relative expression of TaNRX1 in RNA interference lines (n = 3). Data represent the mean ± SD. WT, wild type; OE-1-OE-6, TaNRX1 overexpression T3 homogeneous lines; RNAi-6–RNAi-9, TaNRX1 RNA interference T3 homogeneous lines. *P < 0.05, **P < 0.01 represent significant difference between the transgenic line and the WT, respectively.
Drought Resistance of TaNRX1 Transgenic Wheat at the Seedling and Heading Stages
To examine drought tolerance of transgenic wheat at different growth stages, we determined the drought resistance function of TaNRX1 by subjecting transgenic wheat to drought stress at the seedling and heading stages. Under control conditions, root morphology between WT and transgenic wheat at the seedling stage were not differed (Figure 2A). After 10 days of drought stress, the average longest root length of the OE lines [TaNRX1-OE-1 (12.5 cm), TaNRX1-OE-3 (12.5 cm), and TaNRX1-OE-6 (13.0 cm)] was significantly (P < 0.01) longer than that of the WT (9.17 cm), whereas that of the RNAi lines was significantly (P < 0.05) shorter than that of the WT (Figure 2B). After drought stress, the 2, 3, 5-triphenyltetrazolium chloride staining of the OE lines was darker and the root activity was higher than those of the WT; RNAi lines were light and had low root activity (Figures 2C,D). The root dry weight of the OE lines was significantly (P < 0.01) greater than that of the WT (Figure 2E); there was no significant difference of lateral root numbers between the WT and transgenic wheat lines (Figure 2F). The results show that TaNRX1 OE enhanced root growth and activity under drought stress.
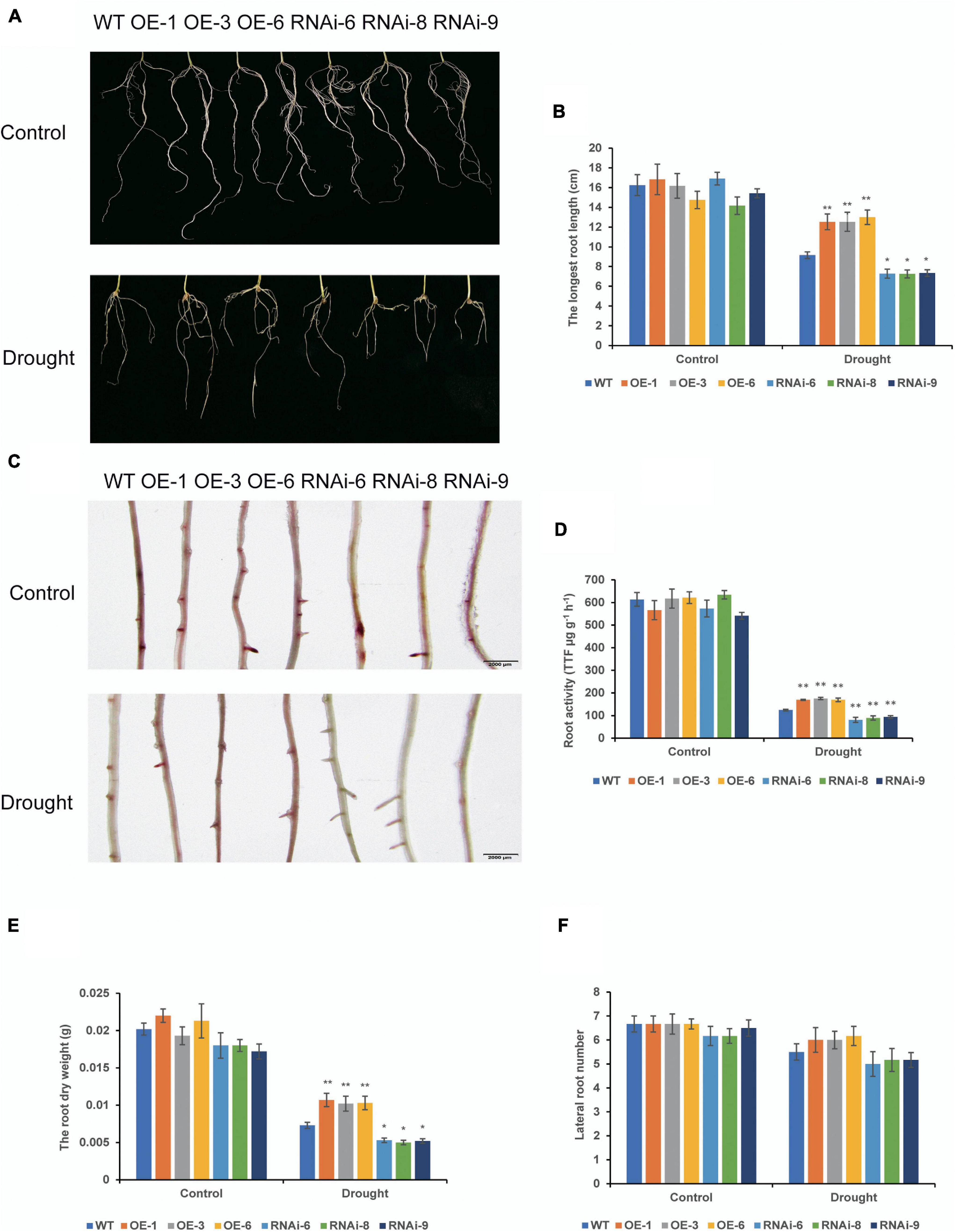
Figure 2. Effect of drought stress on the root system of WT and transgenic wheat at the seedling stage. (A) The roots phenotypes of WT and TaNRX1 transgenic wheat were cultured in 1/2 Hoagland nutrient solution and 1/2 Hoagland nutrient solution with 20% (M/V) polyethylene glycol (PEG6000) for 10 days. (B) The longest root length (n = 6). (C) 2, 3, 5-triphenyltetrazolium chloride (TTC) staining of roots. Scale bar = 2 mm. (D) Root activity (n = 3). (E) Root dry weight (n = 6). (F) Lateral root numbers (n = 6). Data represent the mean ± SD. WT, wild type; OE-1–OE-6, TaNRX1 overexpression T3 homogeneous lines; RNAi-6–RNAi-9, TaNRX1 RNA interference T3 homogeneous lines. *P < 0.05, **P < 0.01 represent the significant difference between the transgenic line and the WT, respectively.
Before drought stress, there was no visibly discernible difference in phenotype between the WT and transgenic wheat plants at the seedling stage (Figure 3A). After 14 days of drought stress, both WT and transgenic wheat plants exhibited different wilting and yellowing. The difference between OE and WT was not obvious, but the wilting in RNAi lines were more severe than that in the WT (Figure 3A). The survival rates of the WT and transgenic wheat lines after rewatering for 3 days were significantly (P < 0.05) different. The survival rates of the OE lines [TaNRX1-OE-1 (93.3%), TaNRX1-OE-3 (86.7%), and TaNRX1-OE-6 (80.0%)] were the highest, followed by the WT (63.3%); the survival rates of the RNAi lines [TaNRX1-RNAi-6 (30.0%), TaNRX1-RNAi-8 (33.3%), and TaNRX1-RNAi-9 (26.7%)] were the lowest (Figure 3B). The relative expression of TaNRX1 in WT, OE lines and RNAi lines under drought stress for 10 and 14 days were determined by qRT-PCR. After 10 and 14 days of drought stress, the expression levels of TaNRX1 in OE lines were significantly (P < 0.01) higher than those of WT and RNAi lines, while the expression levels of TaNRX1 in RNAi lines were still lower than WT (Supplementary Figure 2). These results indicate that TaNRX1 OE enhanced wheat drought tolerance at the seedling stage, whereas RNAi of the TaNRX1 gene significantly reduced drought resistance.
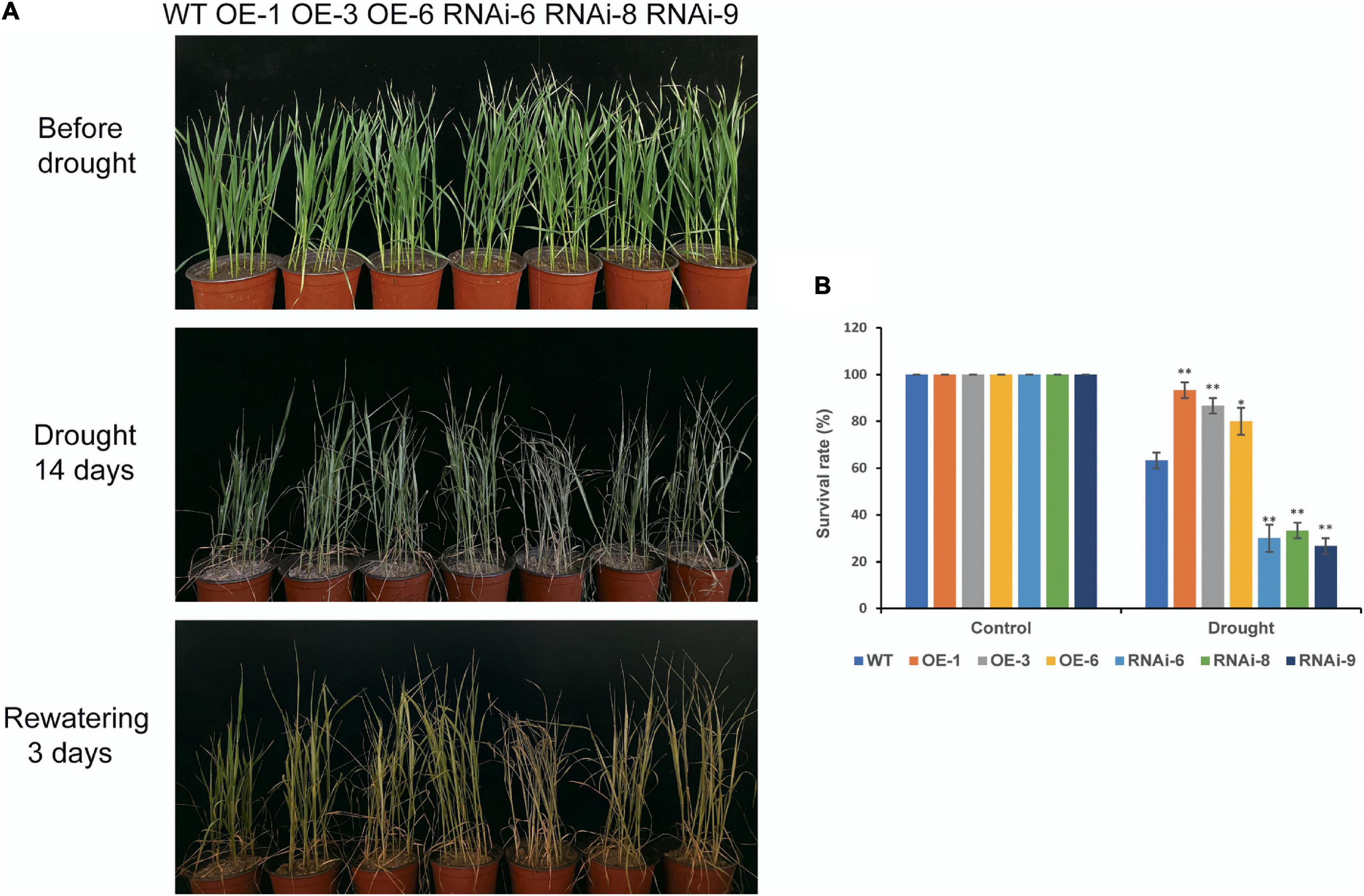
Figure 3. Phenotype and survival rate of WT and TaNRX1 transgenic wheat under drought stress at the seedling stage. (A) WT and TaNRX1 transgenic wheat phenotype at 4 weeks, after 14 days drought treatment, and 3 days rewatering. (B) Survival rate (n = 6). Data represent the mean ± SD. WT, wild type; OE-1–OE-6, TaNRX1 overexpression T3 homogeneous lines; RNAi-6–RNAi-9, TaNRX1 RNA interference T3 homogeneous lines. *P < 0.05, **P < 0.01 represent the significant difference between the transgenic line and the WT, respectively.
There were no major growth defects in OE and RNAi compared to those in WT under normal conditions, OE lines were denser than WT, while RNAi lines were less dense than WT at the heading stage (Figure 4A). After drought stress treatment, leaves of both WT and transgenic wheat lines turned yellow and wilted. Leaf wilt of OE lines were less severe, while that of WT and RNAi lines were more severe (Figure 4A). Under normal conditions, there was no significant difference in the total chlorophyll content between the WT and transgenic wheat lines. After 21 days of drought stress, the total chlorophyll content of the RNAi lines was significantly (P < 0.01) lower than that of the WT and OE lines (Figure 4B). There was no distinct difference in the net photosynthetic rate (Pn) and other photosynthetic indicators between the WT and transgenic wheat flag leaves under control conditions. After drought stress controlled the soil RWC at 35–40% for 21 days, the Pn of the OE lines was significantly (P < 0.01) higher than that of the WT (Figure 4C). The stomatal conductance and transpiration rate were similar to those of Pn, while those of OE lines were higher than those of WT and RNAi lines. The OE-3 was significantly different (P < 0.01), and the rest of the lines were not significantly different (P > 0.05) (Figures 4D,E). There was no significant difference in intercellular CO2 concentration (Figure 4F). These results suggest that TaNRX1 OE can improve the photosynthetic performance of wheat under drought stress.
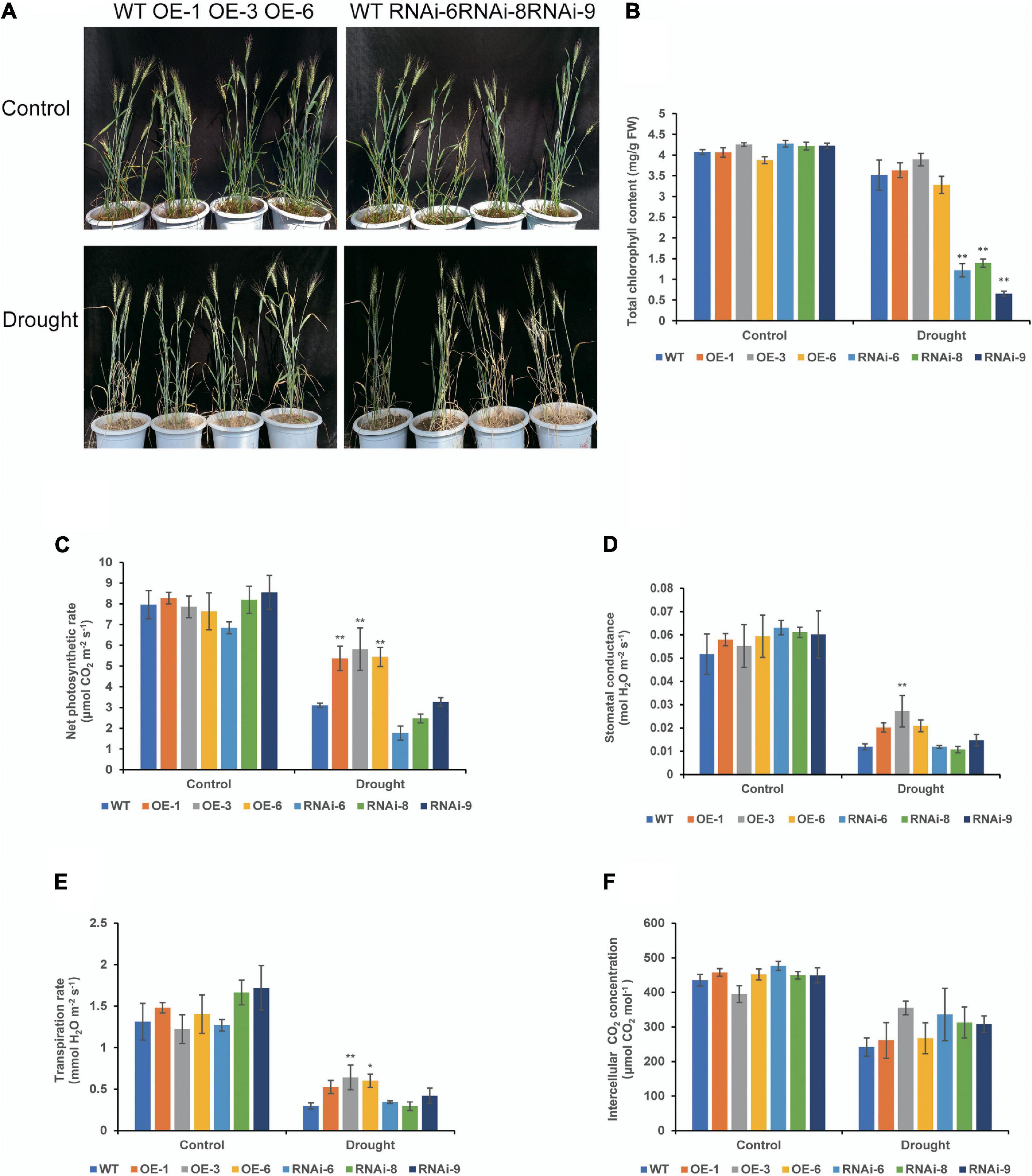
Figure 4. Phenotype and photosynthetic parameters of WT and TaNRX1 transgenic wheat under drought stress at the heading stage. (A) Phenotype of 45-day-old WT and TaNRX1 transgenic wheat in normal water supply and 21 days of drought stress (the soil relative water content was 35–40%). (B) Total chlorophyll content (n = 4). (C) Net photosynthetic rate (n = 6). (D) Stomatal conductance (n = 6). (E) Transpiration rate (n = 6). (F) Intercellular CO2 concentration (n = 6). Data represent the mean ± SD. WT, wild type; OE-1–OE-6, TaNRX1 overexpression T3 homogeneous lines; RNAi-6–RNAi-9, TaNRX1 RNA interference T3 homogeneous lines. *P < 0.05, **P < 0.01 represent the significant difference between the transgenic line and the WT, respectively.
Effects of Drought Stress on the Water Retention of Transgenic Wheat
The leaf water loss rate of 4-week-old WT, OE, and RNAi wheat lines increased gradually with the prolonging of leaf detachment time; the leaf water loss rate of the OE lines was significantly (P < 0.05) lower than that of the WT for 0.5–6 h in vitro, the RNAi lines was higher than that of the WT, but the difference was not significant (P > 0.05) (Figure 5A). There was no significant difference in RWC between the WT and transgenic wheat lines under the control conditions. After drought stress, the RWC of both the WT and transgenic wheat showed a downward trend. The RWC of the OE lines was the highest and significantly (P < 0.01) increased by 14.23% compared with that of the WT, whereas the RWC of the RNAi lines was the lowest and significantly (P < 0.05) decreased by 9.16% compared with that of the WT (Figure 5B).
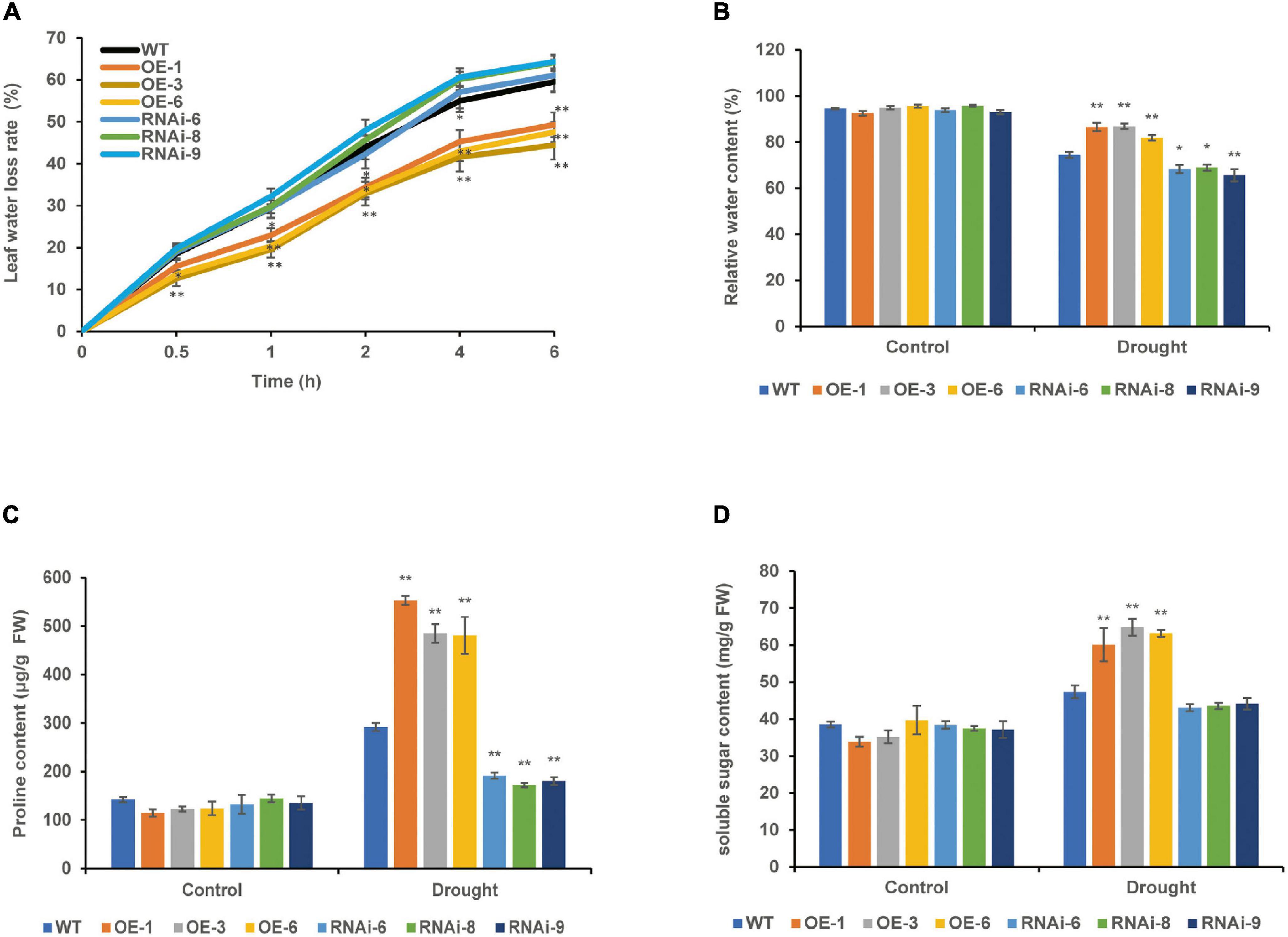
Figure 5. Effect of drought stress on the water status of WT and transgenic wheat. (A) Leaf water loss rate of 4 weeks WT and TaNRX1 transgenic wheat (n = 15). (B) Relative water content of 4 weeks WT and TaNRX1 transgenic wheat in normal water supply and 10 days of drought stress (n = 6). (C) Proline content (n = 4). (D) Soluble sugar content (n = 4). Data represent the mean ± SD. WT, wild type; OE-1–OE-6, TaNRX1 overexpression T3 homogeneous lines; RNAi-6–RNAi-9, TaNRX1 RNA interference T3 homogeneous lines. *P < 0.05, **P < 0.01 represent the significant difference between the transgenic line and the WT, respectively.
Under control conditions, the proline and soluble sugar contents were not significantly different between the WT and transgenic wheat lines. After 10 days of drought stress, the proline content of the OE lines [TaNRX1-OE-1 (553.3 μg/g), TaNRX1-OE-3 (485.0 μg/g), and TaNRX1-OE-6 (480.9 μg/g)] was significantly (P < 0.05) higher than that of the WT (292.0 μg/g), whereas the proline content of the RNAi lines [TaNRX1-RNAi-6 (191.2 μg/g), TaNRX1-RNAi-8 (172.2 μg/g), and TaNRX1-RNAi-9 (180.1 μg/g)] was significantly (P < 0.05) lower than that of the WT (291.7 μg/g) (Figure 5C). The soluble sugar in leaves also showed the same result under drought stress; the soluble sugar content of the OE lines [TaNRX1-OE-1 (60.1 mg/g), TaNRX1-OE-3 (64.8 mg/g), and TaNRX1-OE-6 (63.2 mg/g)] was significantly (P < 0.05) higher than that of the WT (47.3 mg/g) (Figure 5D). Thus, under drought stress, TaNRX1 OE in transgenic wheat was associated with the accumulation of osmotic adjustment substances, such as proline and soluble sugar, and enhances water retention ability, thereby reducing water loss after drought stress and enhancing the water retention capacity of wheat.
Effects of Drought Stress on the Reactive Oxygen Species Levels of Transgenic Wheat
To clarify the effect of TaNRX1 OE or RNAi on wheat ROS levels, we measured H2O2, O2–, and antioxidant enzyme activity in WT, OE, and RNAi wheat. Under normal conditions, DAB and NBT staining results between the WT and transgenic wheat lines were not varied. After 10 days of drought stress, DAB staining of the RNAi lines was the darkest, followed by the WT, and that of the OE lines was the lightest (Figure 6A). The quantitative determination of H2O2 also showed that the accumulation of H2O2 in the leaves of the OE lines [TaNRX1-OE-1 (1.3 μmol/g), TaNRX1-OE-3 (1.4 μmol/g), and TaNRX1-OE-6 (1.3 μmol/g)] was significantly (P < 0.05) lower than that in the leaves of the WT (2.5 μmol/g) (Figure 6B). The results of the NBT staining of leaves was similar to those of DAB staining (Figure 6C), and the accumulation of O2– in the leaves of the OE lines [TaNRX1-OE-1 (83.9 nmol/g), TaNRX1-OE-3 (80.6 nmol/g), and TaNRX1-OE-6 (85.6 nmol/g)] was significantly (P < 0.01) lower than that in the leaves of the WT (103.6 nmol/g); however, the RNAi lines showed the opposite results (Figure 6D).
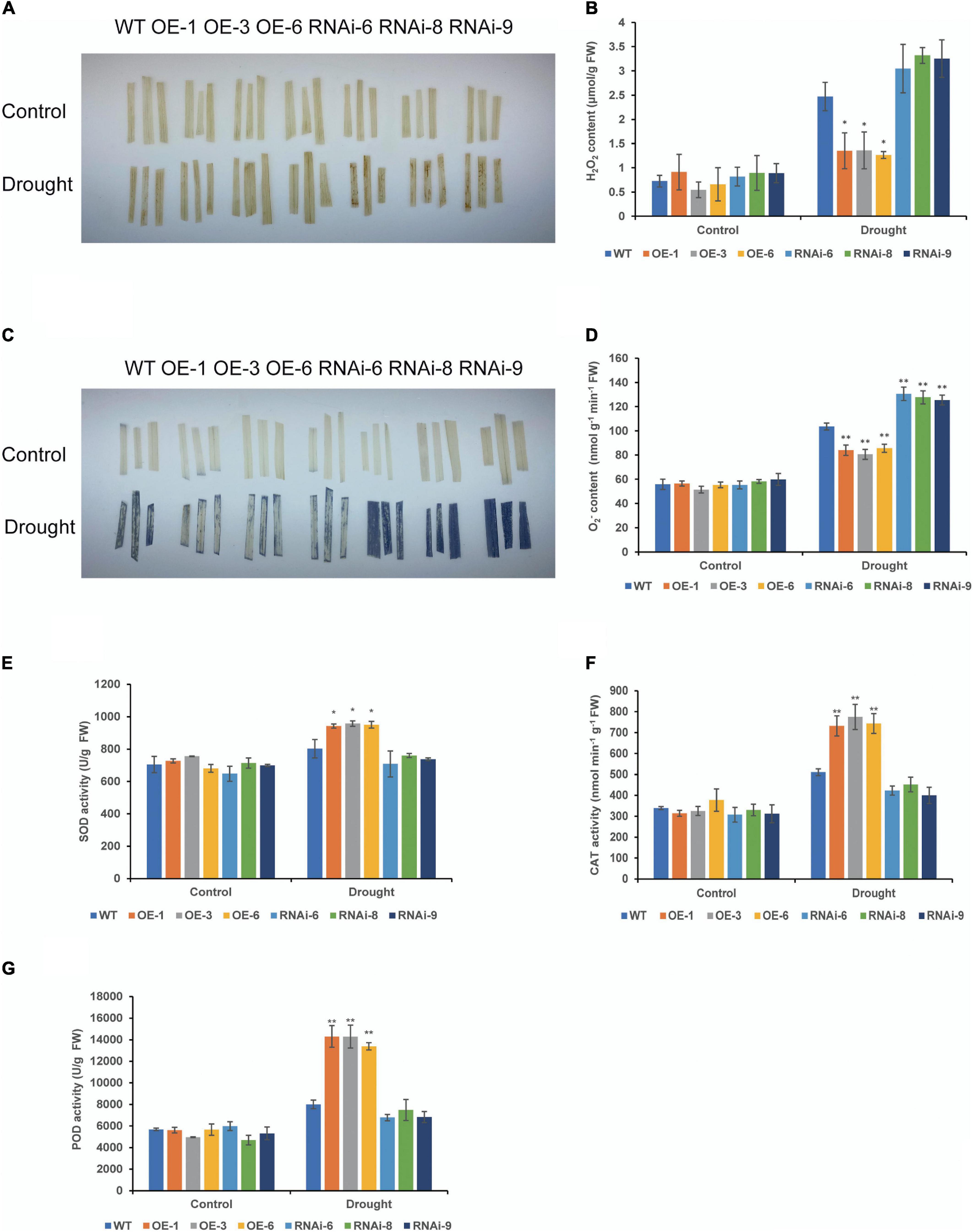
Figure 6. Effect of drought stress on the ROS of WT and transgenic wheat. (A) Histochemical staining was used to detect H2O2 by DAB staining with 4 weeks WT and TaNRX1 transgenic wheat in normal water supply and 10 days of drought stress. (B) H2O2 content (n = 3). (C) Histochemical staining was used to detect O2– by NBT staining. (D) O2– content (n = 4). (E) Superoxide dismutase (SOD) content (n = 4). (F) Catalase (CAT) content (n = 3). (G) Peroxidase (POD) content (n = 4). Data represent the mean ± SD. WT, wild type; OE-1–OE-6, TaNRX1 overexpression T3 homogeneous lines; RNAi-6–RNAi-9, TaNRX1 RNA interference T3 homogeneous lines. *P < 0.05, **P < 0.01 represent the significant difference between the transgenic line and the WT, respectively.
Superoxide dismutase, CAT, and POD are important antioxidant enzymes in plants and are vital for scavenging ROS. There were no significant differences in SOD, CAT, and POD activities under normal conditions in wheat leaves. After 10 days of drought stress, the activities of SOD, CAT, and POD enzymes were significantly (P < 0.05) higher than those in the WT, and activities of SOD, CAT, and POD enzymes in RNAi lines were lower than that of WT, but the difference was not significant (Figures 6E–G). Thus, TaNRX1 OE was related to the increase of antioxidant enzymes activity and reduced the accumulation of ROS under drought stress.
Under normal conditions, there were no significant differences in the MDA content and REC between the WT and transgenic wheat leaves. After 10 days of drought stress, the MDA content of the RNAi lines [TaNRX1-RNAi-6 (24.3 μmol/g), TaNRX1-RNAi-8 (20.8 μmol/g), and TaNRX1-RNAi-9 (24.4 μmol/g)] was the highest, followed by the WT (14.0 μmol/g). The MDA content of the OE lines [TaNRX1-OE-1 (9.3 μmol/g), TaNRX1-OE-3 (7.9 μmol/g), and TaNRX1-OE-6 (8.7 μmol/g)] was the lowest, with significant differences in the WT and transgenic wheat lines. Changes in REC and MDA content showed similar results (Figures 7A,B).
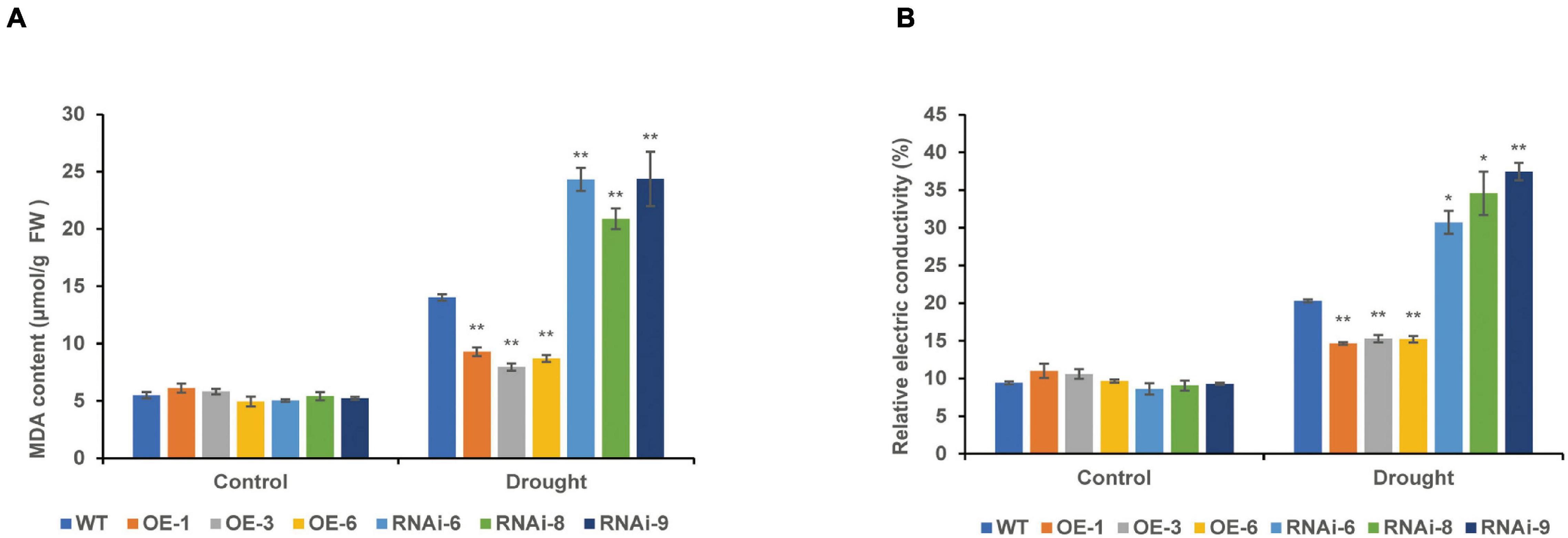
Figure 7. Effect of drought stress on the malondialdehyde and relative electrical conductivity of WT and transgenic wheat. (A) Malondialdehyde content of 4 weeks WT and TaNRX1 transgenic wheat in normal water supply and 10 days of drought stress (n = 4). (B) Relative electrical conductivity (n = 4). Data represent the mean ± SD. WT, wild type; OE-1–OE-6, TaNRX1 overexpression T3 homogeneous lines; RNAi-6–RNAi-9, TaNRX1 RNA interference T3 homogeneous lines. *P < 0.05, **P < 0.01 represent the significant difference between the transgenic line and the WT, respectively.
RNA-Seq
To identify DEGs after TaNRX1 OE or RNAi in wheat, we performed RNA-seq. RT-qPCR results showed that the OE (TaNRX1-OE-3) and RNAi (TaNRX1-RNAi-6) lines had the highest and lowest relative expression levels of TaNRX1 after 6 h of drought stress, respectively (Figure 8). Therefore, 2-week-old WT, TaNRX1-OE-3, and TaNRX1-RNAi-6 plants under drought stress (0 and 6 h) were selected for RNA-seq.
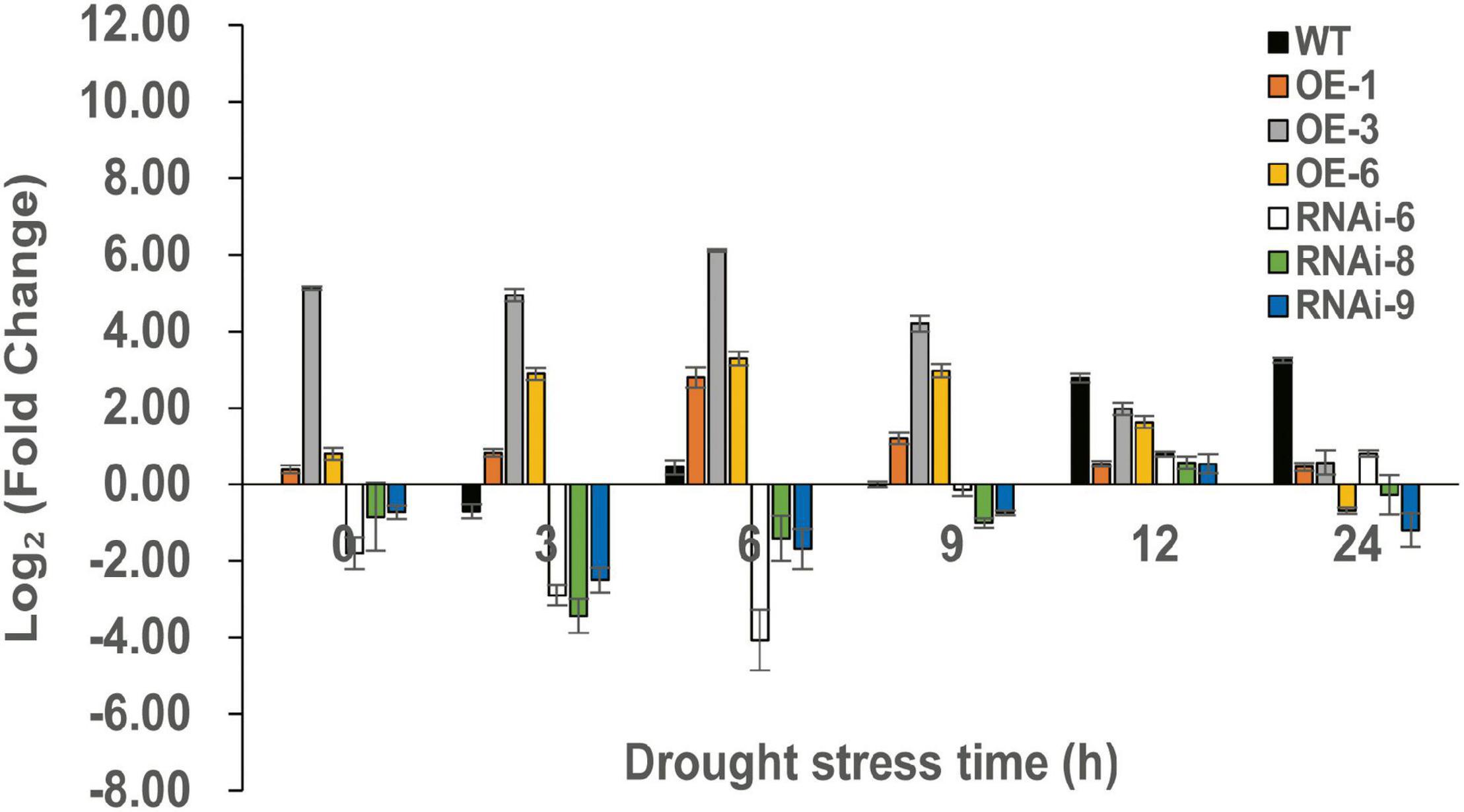
Figure 8. Relative expression of TaNRX1 in different lines of transgenic wheat under different drought stress times. Data represent the mean ± SD (n = 3). WT, wild type; OE-1–OE-6, TaNRX1 overexpression T3 homogeneous lines; RNAi-6–RNAi-9, TaNRX1 RNA interference T3 homogeneous lines.
The RNA-seq analysis of 18 samples of WT and transgenic wheat produced 211.93 Gb of clean data, before and after drought stress; the clean data of each sample reached 10.03 Gb, and the percentage of the Q30 base was 93.74% or above. The sequences of the raw reads have been deposited in the NCBI Sequence Read Archive (SRA) under accession numbers SRR16077951, SRR16077950, SRR16077959, SRR16077958, SRR16077957, SRR16077956, SRR16077955, SRR16077954, SRR16077953, SRR16077952, SRR16077949, SRR16077948, SRR16077965, SRR16077964, SRR16077963, SRR16077962, SRR16077961, and SRR16077960 at the SRA (Sequence Read Achieve) of NCBI. Clean reads of each sample were mapped to a reference genome, and the alignment efficiency ranged from 91.09 to 93.95%. The GC content in all samples exceeded 50%, indicating that the RNA-seq data were highly reliable and could be used for subsequent experimental analysis.
Pearson’s correlation coefficient (r) was used as an evaluation index for biological repeat correlation. The closer r2 is to 1, the stronger the correlation between the two duplicated samples. Sample correlation results in this experiment are shown in Supplementary Figure 3. One of the three biological replicates of each sample showed a low correlation. After removing replicates with a low correlation, DEGs were identified according to the differing expression levels of genes in different samples, and functional annotation and enrichment analyses were conducted.
The number of DEGs identified in different RNA-seq comparisons is shown in Supplementary Figure 4. In total, 11,692 genes were differentially expressed, of which 7,858 were upregulated and 3,834 were downregulated in the WT after drought stress compared with those under pre-drought stress. In total, 9,140 genes were differentially expressed, with 5,765 being upregulated and 3,375 downregulated in the OE lines. The RNAi lines had a total of 9,725 DEGs, with 4,342 being upregulated and 5,383 downregulated. Compared with the WT before drought stress, the OE lines had a total of 7,458 DEGs, with 3,770 being upregulated and 3,688 downregulated. Compared with the WT, the RNAi lines had a total of 8,942 genes that were differentially expressed, with 5,789 being upregulated and 3,153 downregulated under control conditions. Compared with the WT after drought stress, the OE lines had 427 DEGs, of which 203 were upregulated and 224 downregulated. Compared with the WT, the RNAi lines had 3,275 genes that were differentially expressed, with 586 being upregulated and 2,689 downregulated after drought stress.
In order to identify the DEGs specifically affected by TaNRX1, the DEGs in WT, TaNRX1 OE, and RNAi wheat were compared before and after drought stress. We removed the DEGs shared with the WT, and a total of 1,991 common DEGs were identified in the transgenic wheat (Supplementary Figure 5). DEGs were verified using RT-qPCR analysis. To this end, we randomly selected seven DEGs, including triphosphate isomerase, heat shock protein 70, and chlorophyll a/b binding protein. The gene expression trend was consistent with the RNA-seq results (Supplementary Figure 6), which strongly indicated the reliability of the RNA-seq data for use in subsequent analysis.
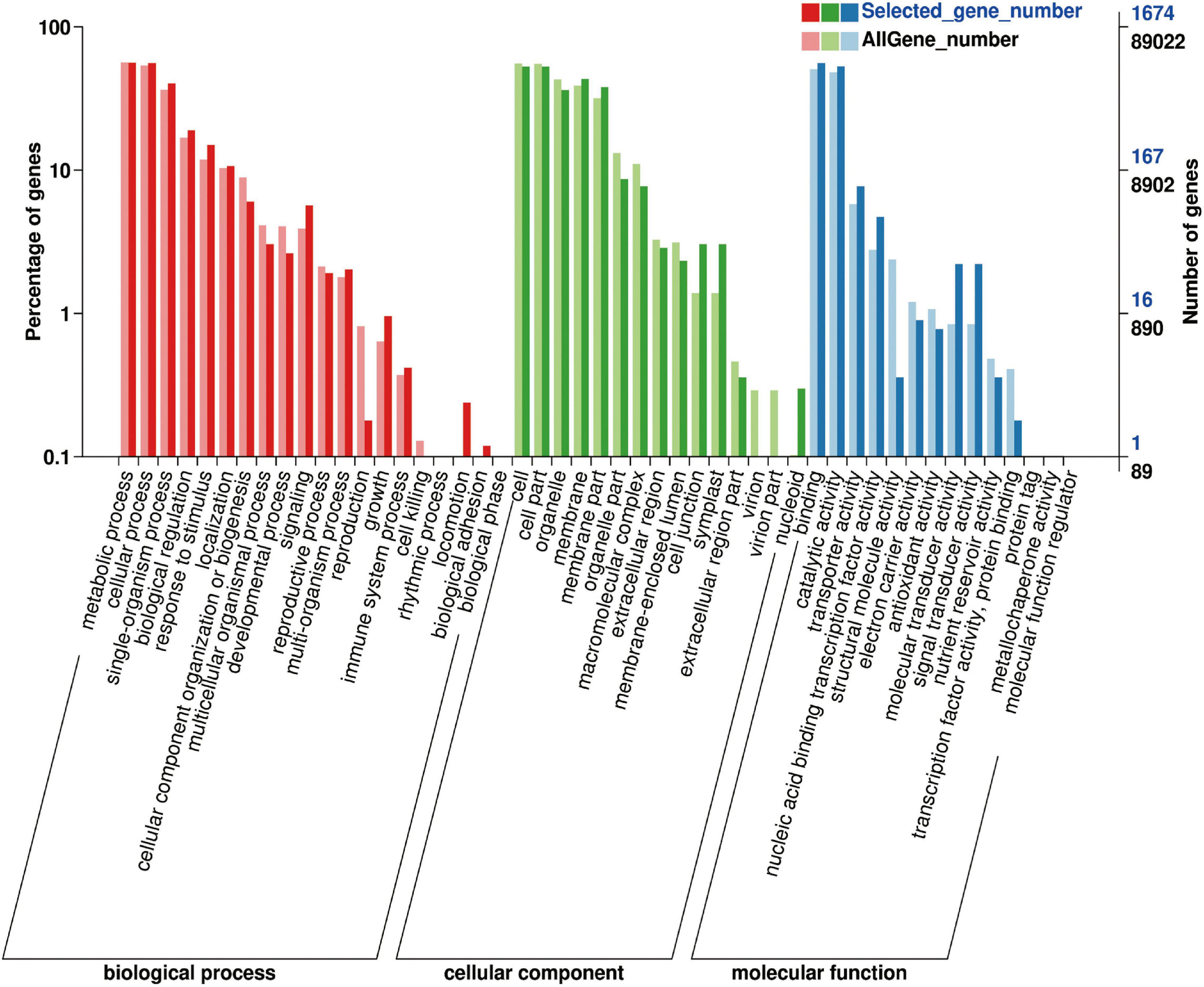
The 1,991 DEGs were further analyzed using GO and KEGG. GO analysis described the biological components, cellular components, and molecular functions of the genes. Among the biological processes, DEGs were mainly concentrated in metabolic, cellular, single-organism processes; biological regulation; response to stimulus; and localization. Among the cell components, DEGs were mainly distributed in cells, membranes, and organelles. The molecular functions of the DEGs included binding ability, catalytic activity, transporter activity, nucleotide binding transcription factor activity, molecular transducer activity, and signal transducer activity (Figure 9).
To further explore the metabolic pathways involved in the 1,991 DEGs, the DEGs were assigned to 71 KEGG pathways. The DEGs were mainly distributed in the plant–pathogen interaction (45 genes), plant hormone signal transduction (40 genes), phenylpropanoid biosynthesis (31 genes), starch and sucrose metabolism (23 genes), carbon metabolism (22 genes), glycolysis and gluconeogenesis (19 genes), amino sugar and nucleotide sugar metabolism (18 genes), and biosynthesis of amino acids (18 genes) pathways (Figure 10).
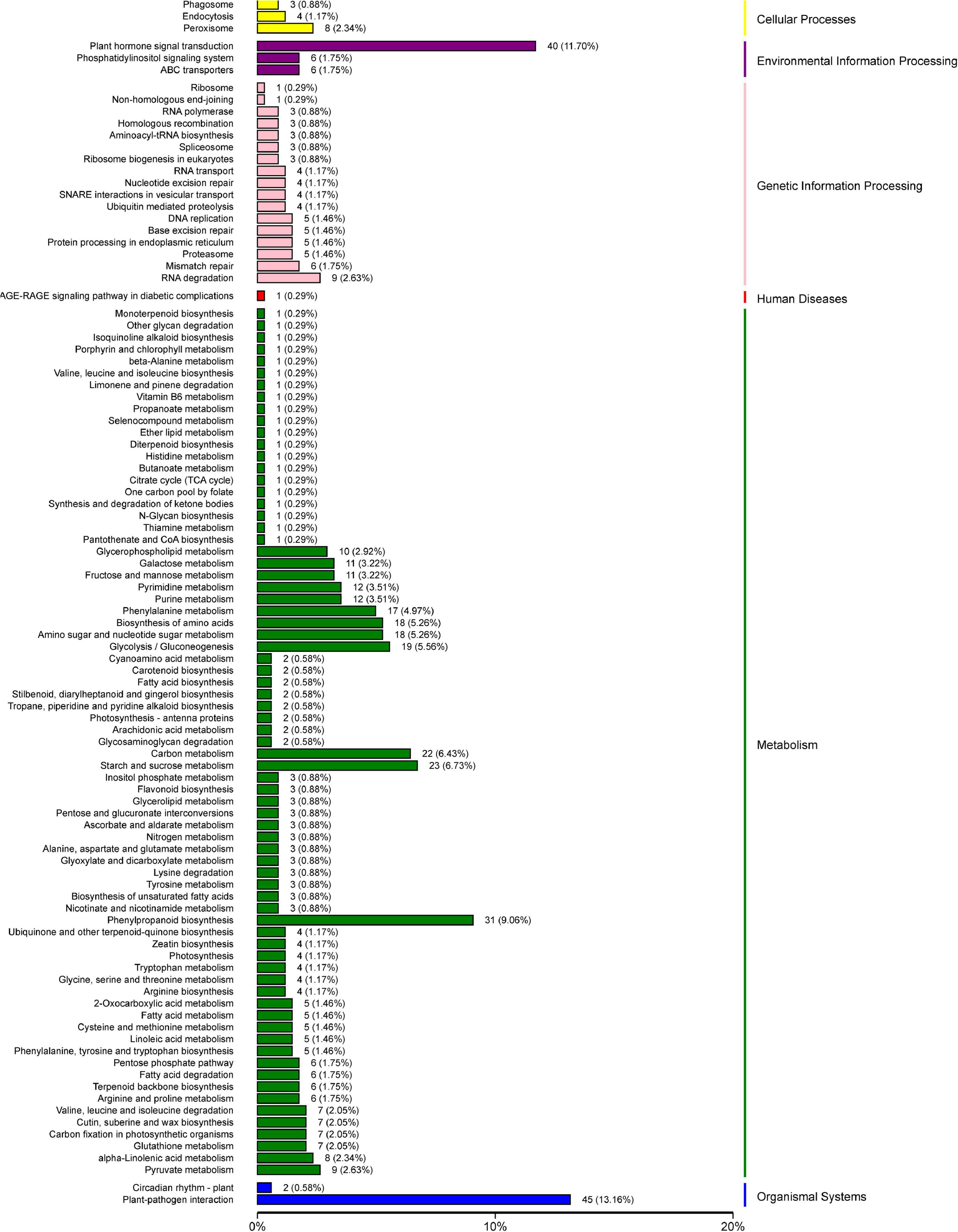
Figure 10. Physiological and biochemical metabolic pathway KEGG enrichment analysis of 1,991 differentially expressed genes.
Discussion
TaNRX1 Positively Regulates Drought Tolerance of Wheat
Nucleoredoxin belongs to the TRX superfamily, which is widely distributed in organisms. It performs the function of electron transfer, which is involved in a variety of life activities (Montrichard et al., 2009; Urbainsky et al., 2018). To date, many studies have been conducted on the biological function of the NRX gene, but information on the specific mechanism of how the gene participates in the response to abiotic stress in wheat is still very limited.
In this study, we selected three wheat lines with TaNRX1 OE (TaNRX1-OE-1, TaNRX1-OE-3, and TaNRX1-OE-6) and three wheat lines with TaNRX1 knockdown (TaNRX1-RNAi-6, TaNRX1-RNAi-8, and TaNRX1-RNAi-9) to study the function of this gene in the drought tolerance of wheat. The results showed that the TaNRX1-OE lines had relatively better growth and development than those of the WT and TaNRX1-RNAi lines at both the seedling and heading stages under drought stress conditions (Figures 3A, 4A), and that the survival rate of TaNRX1-OE lines was higher after rehydration after drought stress at the seedling stage (Figure 3B). Compared with WT, TaNRX1-OE lines had more dry matter content of roots, longer root length and stronger root activity (Figures 2B–E), suggesting that TaNRX1-OE lines might have stronger water absorption capacity. Drought stress can change the chlorophyll content in plant leaves. It is generally believed that when the degree of drought stress approaches the critical state of plants, chlorophyll begins to degrade and thus its content decreases (Tohru et al., 2001). In this study, following drought stress experiments, the chlorophyll content of TaNRX1-OE lines decreased by the same level or at a slightly lower level than that in the WT (Figure 4B), suggesting that TaNRX1-OE lines can tolerate a higher degree of drought stress than the WT. In addition, as photosynthesis is key to the growth and development of crops as well as yield and quality, a change in water status becomes an important limiting factor affecting the photosynthetic activities of crops (Guo et al., 2013). In this study, TaNRX1-OE lines exhibited better photosynthetic performance (Figures 4C–F). Under drought treatment, the Pn content of leaves decreased slightly, suggesting that TaNRX1 OE improved the physiological activity intensity in wheat, enhanced the ability to capture external CO2, and led to an increase in photosynthetic intensity. In TaNRX1-OE lines, the Gs in leaves was relatively high, which could decrease CO2 diffusion resistance, facilitate CO2 exchange between the stomata and the outside world, and promote an increase in Pn content. The Tr of leaves in TaNRX1-OE lines was small, suggesting that TaNRX1 OE reduced transpiration and water loss in leaves to a certain extent to maintain the normal physiological activities of plants. These results suggest that under drought stress, the carbon assimilation capacity of TaNRX1-OE lines was significantly improved, whereas that of TaNRX1-RNAi lines was contrary, suggesting that TaNRX1 positively regulates the drought tolerance of transgenic wheat.
Plant–Pathogen Interaction Pathway Mediates the Resistance of TaNRX1 Transgenic Wheat to Drought Stress
The DEGs in the plant–pathogen interaction pathway mainly included Ca2+ signal transduction-related genes and NBS-LRR genes. As sessile organisms, plants often suffer from biotic (necrotrophic and biotrophic pathogens) and abiotic (drought, salt, heat, or cold) stresses in the natural environment. Several studies have shown that the defense mechanisms of these two forms of stress in plants are related; that is, some stress resistance genes can respond to both biotic and abiotic stresses simultaneously (Atkinson and Urwin, 2012; Ku et al., 2018). Among the many signal transduction pathways in plants, Ca2+ is a multifunctional second messenger. The change in Ca2+ concentration inside and outside the cell is an important part of the signal transduction pathway of plants in response to growth, development, and environmental stimuli. Three types of Ca2+ signaling systems have been identified, which include calmodulin, calmodulin-like protein (CML), calcium-dependent protein kinase, and calcineurin B-like protein. CMLs are involved in various plant growth and development processes, hormone-regulated cell activities, and related defense mechanisms induced by pathogens and inducers. They are induced by various stresses (Bender and Snedden, 2013). CMLs are sensitive to abiotic stresses such as abscisic acid (ABA), methyl jasmonate, and drought and salt stresses (Mccormack et al., 2005). Magnan et al. (2008) found that water shortage treatment induced a rapid and transient increase in the expression level of AtCML9. The expression level increased rapidly by seven times within 10 min, which suggests that AtCML9 is associated with the improvement of water deficit tolerance. CML20 is a negative regulator of ABA-induced stomatal movement in Arabidopsis. The loss of CML20 resulted in a smaller stomatal aperture and less water loss in the leaves of the CML20 gene deletion mutant than in the WT (Wu et al., 2017). Compared with WT tomato plants, tomato plants overexpressing ShCML44 have lower accumulation of MDA and a lower degree of membrane damage under cold and drought stress. The activities of antioxidant enzymes, gas exchange, and water retention were enhanced. The loss of CML20 also showed a decrease in ROS levels and an increase in the RWC (Munir et al., 2016).
NBS-LRR proteins containing nucleotide binding sites (NBSs) and leucine-rich repeats (LRRs) are the main immune receptors in plants. In total, 413 NBS resistance genes of orchardgrass (Dactylis glomerata L.) were studied; among these, 11 were differentially expressed under waterlogging stress, 5 were differentially expressed under waterlogging and drought stress, and 1 was differentially expressed under waterlogging and heat stress, suggesting that some NBS-LRR genes also respond to abiotic stress while defending against pathogen infection (Ren et al., 2020).
In the RNA-seq results of this study, the 1,991 DEGs before and after drought stress treatment mainly included TaCML10, TaCML25, TaCML26, TaCML27, TaCML30, TaCML31, and the NBS-LRR protein gene TaRPM1 in the plant–pathogen interaction pathway. ROS accumulate under abiotic stress conditions and are considered an important index of plant tolerance to abiotic stress (You and Chan, 2015; Xu et al., 2018). According to the results of previous studies, it was speculated that after the overexpression of TaNRX1, the changes in the expression levels of these DEGs improve the drought resistance of transgenic wheat by mediating the changes in ABA content and the removal of ROS. The DAB and NBT staining results of the leaves of TaNRX1-OE and TaNRX1-RNAi lines showed that the ROS in wheat lines overexpressing TaNRX1 were effectively removed (Figures 6A,C). The higher leaf RWC and the lower water loss rate of isolated leaves in TaNRX1-OE lines suggested that the stomatal closure of wheat leaves increased after TaNRX1 OE, whereas TaNRX1-RNAi lines showed the opposite result (Figures 5A,B). The results of these physiological indices were consistent with our prediction above.
The Plant Hormone Signal Transduction Pathway Affects the Resistance of Transgenic Wheat With TaNRX1 Gene to Drought Stress
The DEGs in plant hormone signal transduction pathways mainly include genes related to ABA hormone and cytokinin (CTK) hormone signal transduction. Protein phosphatase 2C (PP2C) is a monomer serine/threonine protein phosphatase dependent on Mg2+ or Mn2+ (Cohen, 1989; Stern et al., 2007). PP2C is widely involved in various ABA signaling pathways in higher plants, including ABA-induced seed germination and dormancy, regulation of guard cells and ion channels, stomatal closure, and biological and abiotic stress. Liu et al. (2012) reported an Arabidopsis gene AtPP2CG1, which encodes a protein belonging to the G subgroup of the PP2C family and regulates the response of Arabidopsis to salt stress, with its regulation being ABA dependent. The RNA-seq results in this study showed that the expression levels of TaPP2C37, TaPP2C50, and TaPP2C68 were all increased in TaNRX1-OE, TaNRX1-RNAi, and WT lines after drought treatment compared with those at pre-drought treatment, and the increased expression was higher in TaNRX1-OE lines.
Cytokinins also play an important role in plant resistance to abiotic stress. The CTK signal is mediated by the CTK receptor histidine kinase (HK), histidine-containing phosphotransfer (HPT), and a complex two-component system (TCS) consisting of an effector response regulator (RR) (Hwang and Sheen, 2001; Hutchison and Kieber, 2002). Jain et al. (2006) systematically studied the expression of A-type RR genes in different organs under different exogenous hormone treatments and under different environmental stresses by quantitative PCR. The results showed that most RR genes were induced by CTK expression. In addition, the expression level of OsRR6 was significantly increased under high salinity, water shortage, and low temperature stresses. The RNA-seq results in this study showed that the expression levels of TaHPT4, TaHPT5, and TaR22 were all increased in the three lines after drought treatment compared with those at pre-drought treatment, and the extent of upregulation was higher in TaNRX1-OE lines. These results suggest that TaNRX1 could improve the drought resistance of TaNRX1-OE lines by mediating the expression of TCS-related genes and regulating the CTK signaling pathway. In this study, the chlorophyll content of TaNRX1-OE lines decreased by the same level or at a slightly lower level than that in the WT after drought stress (Figure 4B). However, CTKs could inhibit the decomposition of chlorophyll (Bajguz and Piotrowska-Niczyporuk, 2014; Dobránszki and Mendler-Drienyovszki, 2014), which confirmed that TaNRX1 mediates the CTK signaling pathway.
Phenylalanine Metabolism Pathway Is Associated With the Resistance of Transgenic Wheat With TaNRX1 Gene to Drought Stress
Under different abiotic stresses, plants maintain their performance using various methods. One is to improve secondary metabolism, as the accumulation of secondary metabolites helps plants to alleviate the damage caused by various stresses (Sharma et al., 2012; Shahzad et al., 2018). Phenylpropane-like metabolism is a typical representative of plant secondary metabolism. Under drought and other adverse conditions, the metabolic processes of phenylpropanes actively participate in the stress resistance of plants. The metabolites in the reaction process not only stabilize cell osmotic potential and cell structure or avoid cell collapse and metabolic disorders caused by cell dehydration, they also eliminate oxidative damage to plants by promptly removing the ROS produced during the stress resistance process.
Isoprene production in transgenic tobacco maintains plant photosynthesis by affecting phenylpropane metabolism and protects plants from the effects of drought stress (Tattini et al., 2014). Transcriptome analysis revealed that the metabolism of hormones and ROS as well as cell wall biosynthesis, especially phenylpropane-like metabolism, promotes root growth in wheat under drought conditions (Dalal et al., 2018). Herrero et al. (2013) showed that AtPOD72 is involved in lignin formation, and Fernandez-Pérez et al. (2015) showed that inhibition of the last step of lignin biosynthesis affects whole phenylpropanol biosynthesis. The RNA-seq results of this study showed that the expression levels of Taβ-Glu12, TaPOD5, TaPOD11, and TaPOD55 in TaNRX1-OE, TaNRX1-RNAi, and WT lines were all increased after drought treatment, and the extent of upregulation was greater in TaNRX1-OE lines. These results suggest that TaNRX1 may enhance drought resistance in wheat by mediating phenylpropane metabolism.
Multiple Transcription Factors Are Related to the Resistance of Transgenic Wheat With TaNRX1 Gene to Drought Stress
Transcription factors are a class of important regulatory proteins that play an important and unique role in regulating plant growth and development and are involved in most life processes in cells. Transcription factors regulate gene transcription or termination through specific binding with nucleotide sequences in the promoter region of target genes, and act as important “switches” for downstream target genes, thus generating a defense response. In addition, they realize their biological functions by interacting with a variety of proteins (Brunelle and Chua, 1993). In this study, 1,991 DEGs contained a large number of transcription factor genes, among which the frequency of WRKY, MYB, and bHLH genes was the highest.
WRKY proteins are involved in plant abiotic stress responses through complex signal transduction pathways and play an important regulatory role. Drought-tolerant plants can improve their drought resistance by accumulating large amounts of sucrose, tetrose, and a series of raffinose family oligosaccharides. In particular, WRKY proteins are involved in drought responses in plants through glucose metabolism pathways (Wu et al., 2009) and can directly regulate the expression of drought resistance genes to improve drought resistance in plants. For example, WRKY transcription factors can regulate the expression of ABA-related genes and participate in ABA-mediated drought response pathways. Under drought stress conditions, ABA levels in plants usually increase and the stomata of the leaves are closed to maintain water in the cells (Sakamoto, 2004). In addition to directly regulating the expression patterns of target genes, some WRKY proteins indirectly regulate the expression levels of functional genes by interacting with other transcription factors (Wei et al., 2008). AtWRKY53 OE in Arabidopsis inhibits stomatal closure by reducing the hydrogen peroxide content in guard cells and accelerating starch metabolism, thereby negatively regulating drought (Sun and Yu, 2015). Cotton GhWRKY41 positively regulates plant salt tolerance and drought resistance by regulating stomatal closure and expression of antioxidant-related genes (Chu et al., 2015). AtWRKY46, AtWRKY54, and AtWRKY70 positively regulated growth and development and negatively regulated the drought stress response in A. thaliana (Chen et al., 2017). In this study, the RNA-seq results showed that the expression levels of TaWRKY53 and TaWRKY70-like were decreased in the TaNRX1-OE lines after drought stress treatment compared with that at pre-drought stress, whereas the increase in expression levels of TaWRKY41 was higher in the TaNRX1-OE lines. These findings are similar to those of the previous studies on A. thaliana and cotton, as described above.
The MYB transcription factor family is named by its conserved MYB domain and is present in all eukaryotes. MYB transcription factors not only respond to drought and other abiotic stresses by inducing stress-related gene expression and stomatal opening and closing, but also regulate phenylpropane-like metabolic pathways to regulate plant growth, development, and stress tolerance. The wheat MYB transcription factor TaODORANT1 positively regulates the response of transgenic tobacco to drought and salt stress (Wei et al., 2017). In this study, RNA-seq results showed that the expression of wheat MYB transcription factor gene TaODORANT1 was higher in TaNRX1-OE lines after drought stress than under pre-drought stress.
Basic/helix-loop-helix (bHLH) is a type of transcription factor that contains a basic/helix-loop-helix structure, is widely present in eukaryotes, and comprises many members. The expression of bHLH transcription factor OsbHLH148 in rice is significantly increased under drought conditions. OsbHLH148 participates in regulating the expression of genes related to the jasmonic acid signaling pathway, thereby increasing the drought tolerance of plants. In addition, another bHLH transcription factor RERT1 (OsbHLH006) in rice is also involved in mediating the jasmonic acid signal pathway and drought stress response (Seo et al., 2011). NtbHLH123 overexpression in plants shows lower electrolyte leakage under cold stress, reducing the content of malondialdehyde and the accumulation of reactive oxygen species (ROS), thereby alleviating the cold stress-induced oxidative damage of cell membranes (Zhao et al., 2018). The heterologous expression of the gene AabHLH35 of Anthurium andraeanum in Arabidopsis can improve the cold and drought tolerance of transgenic Arabidopsis species (Jiang et al., 2019). In this study, RNA-seq results showed that the expression of wheat bHLH transcription factor gene TabHLH35 was higher in TaNRX1-OE lines after drought stress than under pre-drought stress.
bZIP-type transcription factors recognize cis-acting elements whose core sequence is ACGT, including CACGTG (G box), GACGTC (C box), and TACGTA (A box), and the promoter regions of some genes induced by light or ABA containing these elements (Nakagawa et al., 1996). ABA and abiotic stress induce ABF/AREB expression, and ABA induces AREB1/2 phosphorylation. This phosphorylation is necessary for downstream gene induction by ABRE1/2 and could occur at the phosphorylation site of casein kinase II in the conserved domain. Kinase activates the bZIP protein through phosphorylation so that it can bind to the ABRE element in the promoter region of the gene, resulting in the expression of downstream genes regulated by ABA (Schlögl et al., 2008). Zong et al. (2016) found that OsbZIP23 can positively regulate the expression of OsNCED4 by directly binding to the promoter of OsNCED4. It can then positively regulate the content of ABA in rice, thus further activating ABA signal transduction. The RNA-seq results in this study showed that the expression levels of wheat bZIP transcription factor genes TabZIP23 and TaTRAB1 were higher in TaNRX1-OE lines after drought stress than under pre-drought stress. It was speculated that TaNRX1 OE in wheat mediated ABA signal transduction by affecting the expression of wheat bZIP transcription factor genes, thereby enhancing the drought resistance of wheat.
Some Proteins Encoded by Differentially Expressed Genes Are Potential Target Proteins of TaNRX1
Thioredoxin family members are molecular redox switches in organisms. They reduce the target protein through the transfer of electrons, thereby changing the catalytic activity or binding ability of the target protein, which in turn affects more biological functions (Collin et al., 2003). Montrichard et al. (2009) reviewed approximately 500 established and potential target proteins of TRX in terrestrial plants and aerobic microorganisms. Urbainsky et al. (2018) used a differential thiol-labeling technique and quantitative mass spectrometry to identify 609 NRX target proteins in the neuronal cells of mice. The biological processes and molecular functions of these interacting proteins were analyzed, and the results show that NRXs have multiple functions in the redox regulation of metabolic pathways, cell morphology, and signal transduction. Compared with previous studies, we found that some of the proteins encoded by the 1,991 common DEGs identified in this study, such as enzymes involved in the glycolysis/gluconeogenesis pathway and aldehyde dehydrogenases (ALDHs) were also target proteins of TRX or NRX. Glycolysis uses many enzymes to convert glucose into pyruvate and produce ATP, and the activation of genes related to glycolysis and gluconeogenesis may play a role in the drought response of plants. When plants are subjected to abiotic stresses, such as drought, the ATP-generating reaction catalyzed by pyruvate kinase is inhibited. As a glycolytic enzyme, pyruvate, phosphate dikinase (PPDK) can bypass the reaction catalyzed by pyruvate kinase and produce two ATP equivalents, which is very important for plants to cope with stress (Plaxton and Tran, 2011). Sucrose synthase (SUS) is a key enzyme involved in sucrose metabolism. As SUS requires less energy to catalyze sucrose decomposition, plants tend to rely more on SUS to provide reducing sugar when under stress and with limited energy supply to maintain normal growth and development (Fu and Park, 1995; Ricard et al., 1998; Biemelt et al., 1999). Soluble sugars play an important role in stress resistance in plants. They facilitate the plant to absorb water better by maintaining the osmotic pressure balance in the cell so that the cell can resist the adverse environment. The results of this study show that after drought stress, TaNRX1-OE lines had higher contents of proline and soluble sugar in the leaves compared with those in the WT and TaNRX1-RNAi lines, which resulted in higher leaf RWC and a lower leaf water loss rate in vitro. Water retention and water holding capacity of transgenic wheat leaves with the TaNRX1 gene were enhanced under drought stress. The RNA-seq results showed that genes involved in the glycolysis/gluconeogenesis pathway, including TaACS, TaPGM, TaPFP, TaSUS, and TaTPI, which are the encoding genes of acetyl-CoA synthase, phosphoglucose transgenic enzyme, pyrophosphate: fructose 6-phosphate 1-phosphate transferase, sucrose synthase and propanones phosphate isomerase, respectively, were all more upregulated in TaNRX1-OE lines than in the WT and TaNRX1-RNAi lines, which is consistent with the results of the above physiological indicators.
Reactive oxygen species can promote the peroxidation of membrane lipids to cause chemical reactions that generate corresponding aldehydes and cause the accumulation of aldehydes in the organism. Excessive aldehydes react with proteins and nucleic acids, destroying the normal structure and function of proteins and nucleic acids. The total amount of MDA determines the level of lipid peroxidation in plant cells (Perozich et al., 2010). ALDHs are a large class of NAD(P)+-dependent catalytic enzymes that can combine NAD+ or NADP+ to catalyze the oxidative dehydrogenation of endogenous or exogenous aldehydes to generate corresponding carboxylic acids (Rodrigues et al., 2006), thereby reducing the accumulation of aldehydes in plants and achieving detoxification. Thus, ALDHs can alleviate the toxicity of plants under abiotic stress conditions and improve the ability of plants to tolerate adversity. In this study, the MDA content and REC of leaves of TaNRX1-OE lines were significantly lower than those of the WT and RNAi lines after drought stress, which is consistent with the changes in aldehyde dehydrogenase gene expression before and after drought stress, as indicated by the RNA-seq results.
Conclusion
We demonstrated that TaNRX1 positively regulates drought resistance in wheat. TaNRX1 affected the expression of genes related to drought stress through transcriptomics, with these DEGs being mainly distributed in plant–pathogen interactions, plant hormone signal transduction, phenylpropane biosynthesis, starch and sucrose metabolism, and carbon metabolism pathways. This study could provide a new sight for the analysis of drought resistance mechanism in wheat and a valuable target for the production of drought-resistant wheat varieties.
Data Availability Statement
The sequences of the raw reads have been deposited in the NCBI Sequence Read Archive (SRA) under accession numbers SRR16077951, SRR16077950, SRR16077959, SRR16077958, SRR16077957, SRR16077956, SRR16077955, SRR16077954, SRR16077953, SRR16077952, SRR16077949, SRR16077948, SRR16077965, SRR16077964, SRR16077963, SRR16077962, SRR16077961, and SRR16077960.
Author Contributions
XZ designed the experiments with the help of GX. YZ, and JZ performed the experiments and wrote the manuscript. FW, TS, YY, MY, QF, and YNY provided assistance with the experiments. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the National Natural Science Foundation of China (31671693), National Key Research and Development Program of China (2016YFD0101802), and Key Research and Development Project of Shaanxi Province (2019ZDLNY04-05).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Jie Yang for the construction of the overexpression vector of transgenic wheat, Genying Li (Shandong Academy of Agricultural Sciences) for the transgenic wheat RNA interference vector construction and genetic transformation, Ruibo Li for the screening of homogeneous transgenic wheat lines, and Yangyang Guo and Wusa Qin for transgenic wheat breeding.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.756338/full#supplementary-material
Supplementary Figure 1 | The PCR detection for T1–T3 generations of transgenic wheat. WT, wild type; 1–11, transgenic wheat T1–T3 independent lines; M, 2,000 bp marker.
Supplementary Figure 2 | The relative expression of TaNRX1 in transgenic wheat after drought 10 and 14 days (n = 3). Data represent the mean ± SD. WT, wild type; OE-1–OE-6, TaNRX1 overexpression T3 homogeneous lines; RNAi-6–RNAi-9, TaNRX1 RNA interference T3 homogeneous lines. *P < 0.05, **P < 0.01 represent significant difference between the transgenic line and the WT, respectively. The 2–ΔΔ CT method was used to calculate the relative expression levels of genes.
Supplementary Figure 3 | Heat map of correlation between samples. W01–W03, wild type drought stress 0 h three biological duplicates; W61–W63, wild type drought stress 6 h three biological duplicates; E01–E03, TaNRX1-OE-3 drought stress 0 h three biological duplicates; E61–E63, TaNRX1-OE-3 drought stress 6 h three biological duplicates; R01–R03, TaNRX1-RNAi-6 drought stress 0 h three biological duplicates; R61–R63, TaNRX1-RNAi-6 drought stress 6 h three biological duplicates.
Supplementary Figure 4 | The bar graph of WT, TaNRX1 overexpression and RNA interference wheat lines differentially expressed genes after PEG6000 treatment 0 and 6 h. DEG set, the name of the differentially expressed genes set; DEG number, the number of differentially expressed genes; up-regulated, the number of up-regulated genes; down-regulated, the number of down-regulated genes. W0, wild type drought stress 0 h; W6, wild type drought stress 6 h; E0, TaNRX1-OE-3 drought stress 0 h; E6, TaNRX1-OE-3 drought stress 6 h; R0, TaNRX1-RNAi-6 drought stress 0 h; R6, TaNRX1-RNAi-6 drought stress 6 h.
Supplementary Figure 5 | Venn diagrams of differentially expressed genes in WT and transgenic wheat lines before and after drought stress. W0, wild type drought stress 0 h; W6, wild type drought stress 6 h; E0, TaNRX1-OE-3 drought stress 0 h; E6, TaNRX1-OE-3 drought stress 6 h; R0, TaNRX1-RNAi-6 drought stress 0 h; R6, TaNRX1-RNAi-6 drought stress 6 h.
Supplementary Figure 6 | RT-qPCR verification of RNA-seq results. (A) Relative expression of 70 kDa heat shock protein. (B) Relative expression of WPI6 gene. (C) Relative expression of receptor-like protein kinase. (D) Relative expression of triosephosphate isomerase. (E) Relative expression of chloroplast a/b binding protein. (F) Relative expression of protein phosphatase 2C. (G) Relative expression of UDP-glucose-6-dehydrogenase. Data represent the mean ± SD (n = 3). E0, TaNRX1-OE-3 drought stress 0 h; E6, TaNRX1-OE-3 drought stress 6 h; R0, TaNRX1-RNAi-6 drought stress 0 h; R6, TaNRX1-RNAi-6 drought stress 6 h. The 2–ΔΔ CT method was used to calculate the relative expression levels of genes.
Supplementary Table 1 | All primers used in the experiment.
Supplementary Table 2 | Nucleoredoxin (NRX) and TRX members off-target RNA-seq data. W01–W02, wild type drought stress 0 h two biological duplicates; R01–R02, TaNRX1-RNAi-6 drought stress 0 h two biological duplicates.
Footnotes
References
Atkinson, N. J., and Urwin, P. E. (2012). The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63, 3523–3543. doi: 10.1093/jxb/ers100
Bajguz, A., and Piotrowska-Niczyporuk, A. (2014). Interactive effect of brassinosteroids and cytokinins on growth, chlorophyll, monosaccharide and protein content in the green alga chlorella vulgaris (trebouxiophyceae). Plant Physiol. Biochem. 80, 176–183. doi: 10.1016/j.plaphy.2014.04.009
Bender, K. W., and Snedden, W. A. (2013). Calmodulin-related proteins step out from the shadow of their namesake. Plant Physiol. 163, 486–495. doi: 10.1104/pp.113.221069
Biemelt, S., Hajirezaei, M. R., Melzer, M., Albrecht, G., and Sonnewald, U. (1999). Sucrose synthase activity does not restrict glycolysis in roots of transgenic potato plants under hypoxic conditions. Planta 210, 41–49. doi: 10.1007/s004250050652
Brunelle, A. N., and Chua, N. H. (1993). Transcription regulatory proteins in higher plants. Curr. Opin. Genet. 3, 254–258. doi: 10.1016/0959-437X(93)90031-J
Chen, D., Wang, S., Cao, B., Cao, D., Leng, G., Li, H., et al. (2016). Genotypic variation in growth and physiological response to drought stress and re-watering reveals the critical role of recovery in drought adaptation in maize seedlings. Front. Plant Sci. 6:1241. doi: 10.3389/fpls.2015.01241
Chen, J., Nolan, T., Ye, H., Zhang, M., Tong, H., Xin, P., et al. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29, 1425–1439. doi: 10.1105/tpc.17.00364
Cheng, J. (2021). Effects of different domains on activity of wheat (Triticum aestivum) TaNRX1-D. J. Agric. Biotech. 29, 443–452. doi: 10.3969/j.issn.1674-7968.2021.03.004
Cheng, M., Fry, J. E., Pang, S. Z., Zhou, H. P., and Wan, Y. C. (1997). Genetic transformation of wheat mediated by agrobacterium tumefaciens. Plant Physiol. 115, 971–980. doi: 10.1104/pp.115.3.971
Chu, X., Chen, W., Chen, X., Lu, W., Han, L., Wang, X., et al. (2015). The cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic nicotiana benthamiana. PLoS One 10:e0143022. doi: 10.1371/journal.pone.0143022
Cohen, P. (1989). The structure and regulation of protein phosphatases. Annu. Rev. Biochem. 58, 453–508. doi: 10.1146/ANNUREV.BI.58.070189.002321
Collin, V., Issakidis-Bourguet, E., Marchand, C., Hirasawa, M., Lancelin, J.-M., Knaff, D. B., et al. (2003). The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J. Biol. Chem. 278, 23747–23752. doi: 10.1074/jbc.M302077200
Dalal, M., Sahu, S., Tiwari, S., Rao, A. R., and Gaikwad, K. (2018). Transcriptome analysis reveals interplay between hormones, ros metabolism and cell wall biosynthesis for drought-induced root growth in wheat. Plant Physiol. Biochem. 130, 482–492. doi: 10.1016/j.plaphy.2018.07.035
Dey, A., Samanta, M. K., Gayen, S., Sen, S. K., and Maiti, M. K. (2016). Enhanced gene expression rather than natural polymorphism in coding sequence of the Osbzip23 determines drought tolerance and yield improvement in rice genotypes. PLoS One 11:e0187172. doi: 10.1371/journal.pone.0150763
Dobránszki, J., and Mendler-Drienyovszki, N. (2014). Cytokinin-induced changes in the chlorophyll content and fluorescence of in vitro apple leaves. J. Plant Physiol. 171, 1472–1478. doi: 10.1016/j.jplph.2014.06.015
Fernandez-Pérez, F., Pomar, F., and Pedreño, M. Á, and Novo-Uzal, E. (2015). Suppression of Arabidopsis peroxidase 72 alters cell wall and phenylpropanoid metabolism. Plant Sci. 239, 192–199. doi: 10.1016/j.plantsci.2015.08.001
Fu, H. Y., and Park, W. D. (1995). Sink-and vascular-associated sucrose synthase functions are encoded by different gene classes in potato. Plant Cell 7, 1369–1385. doi: 10.2307/3870128
Funato, Y., Hayashi, T., Irino, Y., Takenawa, T., and Miki, H. (2013). Nucleoredoxin regulates glucose metabolism via phosphofructokinase 1. Biochem. Biophys. Res. Commun. 440, 737–742. doi: 10.1016/j.bbrc.2013.09.138
Guo, Q., Li, W. W., Liu, D. D., Wu, W., Liu, Y., Wen, X. X., et al. (2013). Seasonal characteristics of co2 fluxes in a rain-fed wheat field ecosystem at the loess plateau. Span. J. Agric. Res. 11, 980–988. doi: 10.5424/sjar/2013114-4373
Herrero, J. N., Fernández-Pérez, F., Yebra, T., Novo-Uzal, E., Pomar, F., and Pedreño, M. Á, et al. (2013). Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta 237, 1599–1612. doi: 10.1007/s00425-013-1865-5
Hutchison, C. E., and Kieber, J. J. (2002). Cytokinin signaling in Arabidopsis. Plant Cell 14, S47–S59. doi: 10.1105/tpc.010444
Hwang, I., and Sheen, J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389. doi: 10.1038/35096500
Jain, M., Tyagi, A. K., and Khurana, J. P. (2006). Molecular characterization and differential expression of cytokinin-responsive type-a response regulators in rice (oryza sativa). BMC Plant Biol. 6:1. doi: 10.1186/1471-2229-6-1
Jiang, L., Tian, X. K., Li, S. T., Fu, Y. X., Xu, J. J., and Wang, G. D. (2019). The AabHLH35 transcription factor identified from Anthurium andraeanum is involved in cold and drought tolerance. Plants 8:216. doi: 10.3390/plants8070216
Joudrier, P., Gautier, M. F., Lamotte, F. D., and Kobrehel, K. (2005). The thioredoxin h system: potential applications. Biotechnol. Adv. 23, 81–85. doi: 10.1016/j.biotechadv.2004.09.003
Kneeshaw, S., Keyani, R., Delorme-Hinoux, V., Imrie, L., and Spoel, S. H. (2017). Nucleoredoxin guards against oxidative stress by protecting antioxidant enzymes. Proc. Natl. Acad. Sci. U S A. 114, 8414–8419. doi: 10.1073/pnas.1703344114
Ku, Y. S., Sintaha, M., Cheung, M. Y., and Lam, H. M. (2018). Plant Hormone Signaling Crosstalks between Biotic and Abiotic Stress Responses. Int. J. Mol. Sci. 19:3206. doi: 10.3390/ijms19103206
Laughner, B. J., Sehnke, P. C., and Ferl, R. J. (1998). A novel nuclear member of the thioredoxin superfamily. Plant Physiol. 118, 987–996.
Li, Y. B., Han, L. B., Wang, H. Y., Zhang, J., Sun, S. T., Feng, D. Q., et al. (2016). The thioredoxin gbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to verticillium dahliae infection in cotton. Plant Physiol. 2016, 2392–2406. doi: 10.1104/pp.15.01930
Liang, F., Cui, Z. C., Wang, H. Y., and Liu, D. Q. (2020). Generation of TaTLP1 transgenic wheat (Triticum aestivum) and analysis of its resistance to leaf rust fungus. J. Agric. Biotech. 28, 963–973. doi: 10.3969/j.issn.1674-7968.2020.06.002
Liu, X., Zhu, Y., Zhai, H., Cai, H., Ji, W., Luo, X., et al. (2012). AtPP2CG1, a protein phosphatase 2C, positively regulates salt tolerance of Arabidopsis in abscisic acid-dependent manner. Biochem. Biophys. Res. Commun. 422, 710–715. doi: 10.1016/j.bbrc.2012.05.064
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, J., Li, R., Wang, H., Li, D., Wang, X., Zhang, Y., et al. (2017). Transcriptomics analyses reveal wheat responses to drought stress during reproductive stages under field conditions. Front. Plant Sci. 8:592. doi: 10.3389/fpls.2017.00592
Ma, Y., Yang, C., He, Y., Tian, Z., and Li, J. (2017). Rice OVATE family protein 6 regulates plant development and confers resistance to drought and cold stresses. J. Exp. Bot. 68, 4885–4898. doi: 10.1093/jxb/erx309
Magnan, F., Ranty, B., Charpenteau, M., Sotta, B., Galaud, J. P., and Aldon, D. (2008). Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 56, 575–589. doi: 10.1111/j.1365-313X.2008.03622.x
Marchal, C., Delorme-Hinoux, V., Bariat, L., Siala, W., Belin, C., Saez-Vasquez, J., et al. (2014). NTR/NRX define a new thioredoxin system in the nucleus of Arabidopsis thaliana cells. Mol. Plant 7, 30–44. doi: 10.1093/mp/sst162
Marx, C., Wong, J. H., and Buchanan, B. B. (2003). Thioredoxin and germinating barley: targets and protein redox changes. Planta 216, 454–460. doi: 10.1007/s00425-002-0857-7
Mccormack, E., Tsai, Y. C., and Braam, J. (2005). Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10, 383–389. doi: 10.1016/j.tplants.2005.07.001
Meyer, Y., Buchanan, B. B., Vignols, F., and Reichheld, J. P. (2012). Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu. Rev. Genet. 43, 335–367. doi: 10.1146/annurev-genet-102108-134201
Montrichard, F., Alkhalfioui, F., Yano, H., Vensel, W. H., Hurkman, W. J., and Buchanan, B. B. (2009). Thioredoxin targets in plants: the first 30 years. J. Proteom. 72, 452–474. doi: 10.1016/j.jprot.2008.12.002
Munir, S., Liu, H., Xing, Y., Hussain, S., Ouyang, B., Zhang, Y., et al. (2016). Overexpression of calmodulin-like (shcml44) stress-responsive gene from solanum habrochaites enhances tolerance to multiple abiotic stresses. Sci. Rep. 6:31772. doi: 10.1038/srep31772
Nakagawa, H., Ohmiya, K., and Hattori, T. (1996). A rice bzip protein, designated osbz8, is rapidly induced by abscisic acid. Plant J. 9, 217–227. doi: 10.1046/j.1365-313x.1996.09020217.x
Perozich, J., Nicholas, H., Wang, B. C., Lindahl, R., and Hempel, J. (2010). Relationships within the aldehyde dehydrogenase extended family. Protein Sci. 8, 137–146. doi: 10.1110/ps.8.1.137
Plaxton, W. C., and Tran, H. T. (2011). Metabolic adaptations of phosphate-starved plants. Plant Physiol. 156, 1006–1015. doi: 10.1104/pp.111.175281
Porebski, S., Bailey, L. G., and Baum, B. R. (1997). Modification of a ctab dna extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15, 8–15. doi: 10.1007/BF02772108
Powis, G., and Montfort, W. R. (2001). Properties and biological activities of thioredoxin. Annu. Rev. Pharmacol. Toxicol. 41, 261–295. doi: 10.1146/annurev.pharmtox.41.1.261
Ren, S. P., Sun, M., Yan, H. D., Wu, B. C., Jing, T. T., Huang, L. K., et al. (2020). Identification and distribution of NBS-encoding resistance genes of Dactylis glomerata L. and its expression under abiotic and biotic stress. Biochem. Genet. 58, 824–847. doi: 10.1007/s10528-020-09977-8
Ricard, B., Toai, T. V., Chourey, P., and Saglio, P. (1998). Evidence for the critical role of sucrose synthase for anoxic tolerance of maize roots using a double mutant. Plant Physiol. 116, 1323–1331. doi: 10.1104/pp.116.4.1323
Rodrigues, S. M., Andrade, M. O., Gomes, A., Damatta, F. M., Baracat-Pereira, M. C., Fontes, A. E. P. B., et al. (2006). Arabidopsis and tobacco plants ectopically expressing the soybean antiquitin-like aldh7 gene display enhanced tolerance to drought, salinity, and oxidative stress. J. Exp. Bot. 57, 1909–1918. doi: 10.1093/jxb/erj132
Sakamoto, H. (2004). Arabidopsis cys2/his2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 136, 2734–2746. doi: 10.1104/pp.104.046599
Saradadevi, R., Palta, J. A., and Siddique, K. H. M. (2017). ABA-mediated stomatal response in regulating water use during the development of terminal drought in wheat. Front. Plant Sci. 8:1251. doi: 10.3389/fpls.2017.01251
Schlögl, P. S., Nogueira, F. T., Drummond, R., Felix, J. M., De Rosa, V. E. Jr., Vicentini, R., et al. (2008). Identification of new ABA- and MEJA-activated sugarcane bZIP genes by data mining in the SUCEST database. Plant Cell Rep. 27, 335–345. doi: 10.1007/s00299-007-0468-7
Schurman, P., and Buchanan, B. B. (2008). The ferredoxin/thioredoxin system of oxygenic photosynthesis (1). Antioxid. Redox Signal. 10, 1235–1274. doi: 10.1089/ars.2007.1931
Seo, J. S., Joo, J., Kim, M. J., Kim, Y. K., Nahm, B. H., Song, S. I., et al. (2011). OsbHLH148, a basic helix-loop-helix protein, interacts with osjaz proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 65, 907–921. doi: 10.1111/j.1365-313X.2010.04477.x
Shahzad, B., Cheema, S. A., Farooq, M., Cheema, Z. A., Rehman, A., and Abbas, T. (2018). Growth stimulating influence of foliage applied brassica water extracts on morphological and yield attributes of bread wheat under different fertilizer regimes. Planta Daninha 36:e018178331. doi: 10.1590/s0100-83582018360100117
Sharma, P., Jha, A. B., Dubey, R. S., and Pessarakli, M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 1–26. doi: 10.1155/2012/217037
Srivastava, A. K., Zhang, C., Caine, R. S., Gray, J., and Sadanandom, A. (2017). Rice sumo protease overly tolerant to salt 1 targets the transcription factor, osbzip23 to promote drought tolerance in rice. Plant J. 92, 1031–1043. doi: 10.1111/tpj.13739
Stern, A., Privman, E., Rasis, M., Lavi, S., and Pupko, T. (2007). Evolution of the metazoan protein phosphatase 2c superfamily. J. Mol. Evol. 64, 61–70. doi: 10.1007/s00239-006-0033-y
Sun, Y. D., and Yu, D. Q. (2015). Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Rep. 34, 1295–1306. doi: 10.1007/s00299-015-1787-8
Tattini, M., Velikova, V., Vickers, C., Brunetti, C., Ferdinando, M. D., Trivellini, A., et al. (2014). Isoprene production in transgenic tobacco alters isoprenoid, non−structural carbohydrate and phenylpropanoid metabolism, and protects photosynthesis from drought stress. Plant Cell Environ. 37, 1950–1964. doi: 10.1111/pce.12350
Tohru, T., Hiroyuki, O., and Ken-ichiro, T. (2001). How do plants degrade the green color of their leaves current state of the study on chlorophyll degradation. Kagaku Seibutsu 39, 580–587. doi: 10.1271/kagakutoseibutsu1962.39.580
Traverso, J. A., Vignols, F., Cazalis, R., Pulido, A., Sahrawy, M., Cejudo, F. J., et al. (2007). PsTrxh1 and PsTrxh2 are both pea h-type thioredoxins with antagonistic behavior in redox imbalances. Plant Physiol. 143, 300–311. doi: 10.1104/pp.106.089524
Tuan, P. A., Yamasaki, Y., Kanno, Y., Seo, M., and Ayele, B. T. (2019). Transcriptomics of cytokinin and auxin metabolism and signaling genes during seed maturation in dormant and non-dormant wheat genotypes. Sci. Rep. 9:3983. doi: 10.1038/s41598-019-40657-9
Urbainsky, C., Nölker, R., Imber, M., Lübken, A., Mostertz, J., Hochgräfe, F., et al. (2018). Nucleoredoxin-dependent targets and processes in neuronal cells. Oxid. Med. Cell. Longev. 2018, 1–11. doi: 10.1155/2018/4829872
Valluru, R., Davies, W. J., Reynolds, M. P., and Dodd, I. C. (2016). Foliar abscisic acid-to-ethylene accumulation and response regulate shoot growth sensitivity to mild drought in wheat. Front. Plant Sci. 7:461. doi: 10.3389/fpls.2016.00461
Wang, K., Zhong, M., Wu, Y. H., Bai, Z. Y., Liang, Q. Y., Liu, Q. L., et al. (2017). Overexpression of a chrysanthemum transcription factor gene dgnac1 improves the salinity tolerance in chrysanthemum. Plant Cell Rep. 36, 571–581. doi: 10.1007/s00299-017-2103-6
Wang, N., Zhang, W., Qin, M., Li, S., Qiao, M., Liu, Z., et al. (2017). Drought tolerance conferred in Soybean (Glycine max. L) by GmMYB84, a novel R2R3-MYB transcription factor. Plant Cell Physiol. 58, 1764–1776. doi: 10.1093/pcp/pcx111
Wei, Q., Luo, Q., Wang, R., Fan, Z., Yuan, H., Yang, Z., et al. (2017). A wheat r2r3-type myb transcription factor taodorant1 positively regulates drought and salt stress responses in transgenic tobacco plants. Front. Plant Sci. 8:1374. doi: 10.3389/fpls.2017.01374
Wei, W., Zhang, Y., Lu, H., Guan, Z., and Chai, T. (2008). A novel WRKY transcriptional factor from thlaspi caerulescens negatively regulates the osmotic stress tolerance of transgenic tobacco. Plant Cell Rep. 27, 795–803. doi: 10.1007/s00299-007-0499-0
Wong, J. H., Cai, N., Balmer, Y., Tanaka, C. K., Vensel, W. H., Hurkman, W. J., et al. (2004). Thioredoxin targets of developing wheat seeds identified by complementary proteomic approaches. Phytochemistry 65, 1629–1640. doi: 10.1016/j.phytochem.2004.05.010
Wu, X., Shiroto, Y., Kishitani, S., Ito, Y., and Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing oswrky11 under the control of hsp101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Wu, X., Zhu, Q., Liu, H., Acharya, B. R., Li, C., and Wei, Z. (2017). CML20, an Arabidopsis Calmodulin-like protein, negatively regulates guard cell aba signaling and drought stress tolerance. Front. Plant Sci. 8:824. doi: 10.3389/fpls.2017.00824
Xu, N., Chu, Y., Chen, H., Li, X., Wu, Q., Jin, L., et al. (2018). Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA-mediated regulatory pathway and ROS scavenging. PLoS Genet. 14:e1007662. doi: 10.1371/journal.pgen.1007662
Yadav, A. K., Carroll, A. J., Estavillo, G. M., Rebetzke, G. J., and Pogson, B. J. (2019). Wheat drought tolerance in the field is predicted by amino acid responses to glasshouse-imposed drought. J. Exp. Bot. 70, 4931–4948. doi: 10.1093/jxb/erz224
You, J., and Chan, Z. (2015). ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 6:1092. doi: 10.3389/fpls.2015.01092
Zhang, F., Jiang, L., Ju, L. P., Jin, X. F., Wang, X., Zhang, X. K., et al. (2014). Cloning a novel gene TaNRX of Trx superfamily and developing its molecular markers related to drought resistance in common wheat. Acta Agron. Sin. 40, 29–36. doi: 10.3724/SP.J.1006.2014.00029
Zhang, N., Yin, Y., Liu, X., Tong, S., Xing, J., Zhang, Y., et al. (2017). The E3 Ligase TaSAP5 Alters Drought Stress Responses by Promoting the Degradation of DRIP Proteins. Plant Physiol. 175, 1878–1892. doi: 10.1104/pp.17.01319
Zhang, S. J., Li, Y. L., Song, G. Q., Gao, J., Zhang, R. Z., Li, W., et al. (2020). Heterologous expression of the ThIPK2 gene enhances drought resistance of common wheat. J. Integrat. Agricult. 19, 941–952. doi: 10.1016/s2095-3119(19)62714-0
Zhao, Q., Xiang, X., Liu, D., Yang, A., and Wang, Y. (2018). Tobacco transcription factor NtbHLH123 confers tolerance to cold stress by regulating the NtCBF pathway and reactive oxygen species homeostasis. Front. Plant Sci. 9:381. doi: 10.3389/fpls.2018.00381
Zhao, T. T. (2016). Thioredoxin regulates the pollen germination on the stigma in Arabidopsis. Ph. D. thesis. Shandong: University of Shandong Agricultural.
Zhou, J., Ma, C., Zhen, S., Cao, M., Zeller, F. J., Hsam, S. L. K., et al. (2016). Identification of drought stress related proteins from 1S(l)(1B) chromosome substitution line of wheat variety Chinese spring. Bot. Stud. 57:20. doi: 10.1186/s40529-016-0134-x
Keywords: wheat, nucleoredoxin, TaNRX1, drought, RNA-seq
Citation: Zhang Y, Zhou J, Wei F, Song T, Yu Y, Yu M, Fan Q, Yang Y, Xue G and Zhang X (2021) Nucleoredoxin Gene TaNRX1 Positively Regulates Drought Tolerance in Transgenic Wheat (Triticum aestivum L.). Front. Plant Sci. 12:756338. doi: 10.3389/fpls.2021.756338
Received: 10 August 2021; Accepted: 18 October 2021;
Published: 11 November 2021.
Edited by:
Yuri Shavrukov, Flinders University, AustraliaReviewed by:
Dhruv Lavania, University of Alberta, CanadaMarcelo Nogueira Do Amaral, Federal University of Pelotas, Brazil
Copyright © 2021 Zhang, Zhou, Wei, Song, Yu, Yu, Fan, Yang, Xue and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Xue, xuegangyan2009@sina.com; Xiaoke Zhang, zhangxiaoke66@126.com
†These authors have contributed equally to this work
 Yunrui Zhang1†
Yunrui Zhang1† Jianfei Zhou
Jianfei Zhou Tianqi Song
Tianqi Song Yang Yu
Yang Yu Ming Yu
Ming Yu