- 1State Key Laboratory of North China Crop Improvement and Regulation, Hebei Sub-center for National Maize Improvement Center, College of Agronomy, Hebei Agricultural University, Baoding, China
- 2Key Laboratory of Crop Physiology and Ecology, Ministry of Agriculture and Rural Affairs, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 3State Key Laboratory of Agrobiotechnology, National Maize Improvement Center, Department of Plant Genetics and Breeding, China Agricultural University, Beijing, China
- 4Institute of Botany, Chinese Academy of Sciences, Beijing, China
The vascular bundle plays an important role in nutrient transportation in plants and exerts great influence on crop yield. Maize is widely used for food, feed, and fuel, producing the largest yield in the world. However, genes and molecular mechanism controlling vascular bundle-related traits in maize have largely remained undiscovered. In this study, a natural population containing 248 diverse maize inbred lines genotyped with high-throughput SNP markers was used for genome-wide association study. The results showed that broad variations existed for the vascular bundle-related traits which are subject to genetic structure and it was suitable for association analysis. In this study, we identified 15, 13, 2, 1, and 5 SNPs significantly associated with number of small vascular bundle, number of large vascular bundle, average area of single small vascular bundle, average area of single large vascular bundle, and cross-sectional area, respectively. The 210 candidate genes in the confidence interval can be classified into ten biological processes, three cellular components, and eight molecular functions. As for the Kyoto Encyclopedia of Genes and Genomes analysis of the candidate genes, a total of six pathways were identified. Finally, we found five genes related to vascular development, three genes related to cell wall, and two genes related to the mechanical strength of the stalk. Our results provide the further understanding of the genetic foundation of vascular bundle-related traits in maize stalk.
Introduction
Maize is the most widely planted crop in the world and provides a large part of food for animals and human as well as materials for deep processing and energy. According to the forecast of the United Nations, the world’s population will exceed nine billion and global demand for maize will double (Ray et al., 2013) by 2050. In order to provide living space for the growing population, the cultivated land area has shrunk year by year worldwide. Thus, it is of great significance to increase the yield per unit area of maize.
Vascular bundles consist of xylem and phloem, two differentiated conductive tissues, and undifferentiated cambial or procambial stem cells. Vascular bundles provide mechanical support for plants. In maize, appropriate strength and tenacity would help the plant stand against winds and decrease the loss caused by lodging. Moreover, vascular bundles function as “flow” in the “source-flow-sink” system and transport photoassimilates produced by leaves (“source”) to the fruit (“sink”) and transfer water and salt from roots to the whole plants. Previous studies have observed significant correlation between the vascular bundle system and maize yield (Housley and Peterson, 1982; Nátrová, 1991).
Many studies have been carried out on the genes related to vascular bundle system in plants. In Arabidopsis, genes involved in vascular bundle patterning have been identified, such as MP, PHB, PHV, AtHB15, and REV (Hardtke and Berleth, 1998; McConnell et al., 2001; Zhong and Ye, 2004; Du and Wang, 2015). In crops, quantitative trait loci (QTL) for vascular bundle-related traits have been identified in tomato (Coaker et al., 2002), wheat (Sang et al., 2010), and rice (Bai et al., 2012; Zhai et al., 2018; Fei et al., 2019). Notably, genes that affect the vascular bundle system in rice have been reported, such as APO1, ABV, DEP1, and NAL1 (Qi et al., 2008; Terao et al., 2010; Fujita et al., 2013; Fei et al., 2019). However, the functional genes have not been fully discovered and the molecular mechanism of how the vascular bundle system influences the crop yield has remained largely unknown.
Progresses have been made in the study of maize vascular bundles system. Sakaguchi and Fukuda (2008) used the basal leaves to observe the continuous process of vascular bundle cell development and accordingly divided the longitudinal vein development of maize leaves into five stages. Feng et al. (2014) found that planting density significantly affected the structure of maize stem vascular bundles and that, with the increase in planting density, the number of large and small vascular bundles in the stem decreased. In addition, the number of large vascular bundles and that of total vascular bundles differed significantly between different density treatments. Xu et al. (2017) found that plant growth regulator EDAH significantly increased the number and area of vascular bundles. However, earlier studies mainly focused on the development and micro-structure of vascular bundles, and little is known on the functional genes regulating the vascular bundle system in maize, and the molecular mechanism on the vascular bundle-related traits in maize has remained largely blank. Huang et al. (2016) used a large maize-teosinte experimental population to perform a high-resolution QTL mapping for the number of vascular bundle in the uppermost internode of maize stem and validate the effect of one QTL qVb9-2 on chromosome 9.
In recent years, genome-wide association study (GWAS) has become an efficient tool to capture functional genes and favorable haplotypes for interested traits in maize (Tian et al., 2011; Li et al., 2013, 2019; Wang et al., 2016). For maize, due to the release of the B73 reference genome, GWAS application in agricultural traits of maize provides useful reference for revealing the phenotypic traits diversity and genetic architecture of vascular bundles in maize stalk. In this study, we performed GWAS for vascular bundle system using a natural population consisting of 248 maize inbred lines with abundant genetic diversity in 2017, 2018, and 2019. And the candidate genes adjacent to the significant SNPs were identified. This study lays the foundation for understanding the genetic architecture of vascular bundle-related traits in maize.
Materials and Methods
Plant Materials
A natural population panel containing 248 diverse maize inbred lines was used as research materials provided by the China Agricultural University and National Maize Improvement Center of China. The 248 maize inbred lines panel included not only some excellent back bone elite inbred lines in China but also some high-quality inbred lines introduced from abroad. The detailed information on this natural population can be found in Supplementary Table S1 (Yang, 2016). For all the 248 maize inbred lines, a randomized block design with two replications was used in this study. Each material inbred was planted in a plot of two 3.0-m-long rows with 0.60-m-inter-row space, using a population density of 75,000 plants per hectare at the Experimental Station of Hebei Agricultural University in Baoding (115.48° and 38.85°) in the summer of 2017, 2018, and 2019 and the Experimental Station of Shijiazhuang (115.12° and 37.54°) in the summer of 2017 and 2018, respectively. All the maize plants were in-followed standard local field management using local maize tillage methods throughout the whole growth periods.
Phenotypic Evaluation
One week after pollination, three individual plants with typical growth were selected for each inbred as three biological replicates. For each replicate, cross-section slices from the uppermost internode were manually made. The slices were stained with 5% (g/ml) m-trihydroxybenzene and concentrated hydrochloric acid (Huang et al., 2016). Then, the stained slices were imaged by the Zeiss Axioskop 40 microscope (Germany). Zen (blue edition) 2012 image processing software was used to collect data of vascular bundle-related traits, including the number of small vascular bundle (NSVB), the number of large vascular bundle (NLVB), average area of single small vascular bundle (ASVB), average area of single large vascular bundle (ALVB), and cross-sectional area (CSA). The vascular bundles of stalk are divided into two categories: small vascular bundles and large vascular bundles. The small vascular bundles are generally located in the 1~2 layer of the edge of the tissue, with relatively close arrangement, the area is small and some small vascular bundles are not fully developed; the large vascular bundles are located in the organization, loose arrangement, relatively large area, and complete structural development. The vascular bundle area and CSA were calculated in terms of near-ellipse ( and represent the major and minor axes of the ellipse, respectively; Yang, 2016).
Phenotypic Analysis
Phenotypic data were processed with the Microsoft Excel 2010, and descriptive statistical analysis and ANOVA were carried out with the IBM SPSS statistics v21.0 software (George and Mallery, 2013). The broad-sense heritability ( ) for each trait was estimated using the formula: ), as described by Knapp et al. (1985), where stands for the genetic variance, stands for genotype-by-environment interaction variance, for error variance, for the number of environments, and for the number of replications. Correlations analysis was performed using the “Performance Analytics” package in R. The boxplot was drawn using “ggplot2” package in R. The best linear unbiased prediction (BLUP; Henderson, 1975) values using the mixed linear model of the “lme4” package in R was calculated for each trait across five environments and adopted as the phenotypic values in the subsequent genome-wide association study.
Genotyping
The DNA of 248 maize inbred lines was extracted from fresh leaves with the CTAB method (Murray and Thompson, 1980). The sequencing libraries of 248 inbred lines were constructed and sequenced by genotyping-by-sequencing, and the qualified library was sequenced by the second-generation sequencer Illumina Hiseq2000 (Elshire et al., 2011). The derived short reads were compared to the reference genome of the second version of B73 by BWA software, and the SNPs were called by the SAMtools software to obtain the preliminary SNP markers. A total of 10,63,728 initial SNPs were obtained through strict sequencing data comparison and SNP-calling. Then, the SNPs with missing rate over 80% and minor allele frequency less than 5% were removed. Finally, a total of 83,057 SNPs were achieved and used in GWAS (Lai, 2017). The population structure of 248 inbred lines was detected by the Admixture 1.3 software (Chakraborty and Weiss, 1988), which was divided into five subpopulations: Lancaster, Lvda Red Cob, Tangsipingtou (TSPT), P group, Reid, and a mixed group. The Q model of population structure was obtained, and the K model of kinship was obtained by using the Analysis-Kindship in Tassel 5.0 software (Du et al., 2018).
Genome-Wide Association Analysis
GWAS was performed to identify the SNPs significantly associated with vascular bundle-related traits using 83,057 SNPs and the BLUP values of the 248 maize inbred lines. GWAS was performed with the FarmCPU model implemented in the GAPIT package in the R software, taken both K and Q matrix into account to decrease spurious association (Liu et al., 2016). Then, we used the GEC software (Li et al., 2012) to calculate effective marker number ( ), and the significance threshold was set as p ≤ 1/ .
Identification of Candidate Genes
Linkage disequilibrium (LD) decay of the same population was analyzed in the previous studies (Li et al., 2020; Liu et al., 2020). The studies showed that the LD decay distance for this natural population panel was 120 kb (r2 = 0.1). Then, we searched the flanking 120 kb upstream and downstream of each significant loci for candidate genes according to B73 reference genome version v2. The annotation of candidate genes was obtained from the MaizeGDB and NCBI.1 The candidate genes were uploaded to GENE ONTOLOGY Web site2 for GO analysis. The KOBAS 3.0 Web site3 was used to perform the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis.
Results
Diversity and Heritability of Vascular Bundle-Related Traits
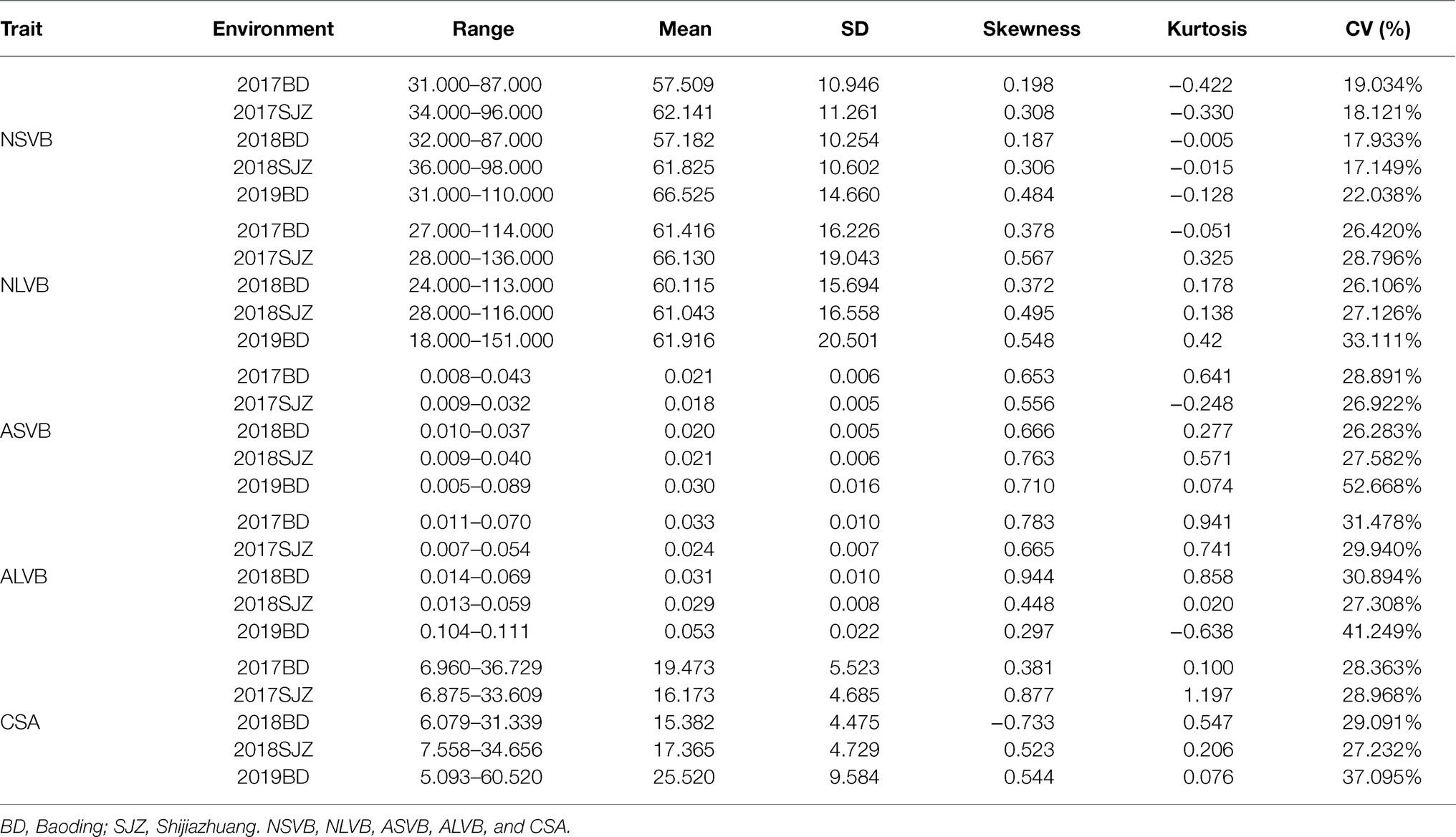
Substantial variation was observed for vascular bundle-related traits in different maize inbred lines (Figure 1). The statistical data for NSVB, NLVB, ASVB, ALVB, and CSA across the five environments showed that all five vascular bundle-related traits showed a normal distribution (Figure 2). The phenotype range of the above five traits was 17.933∼22.038, 26.106∼33.111, 26.283∼52.668, 27.308∼41.249, and 28.363∼37.095 (%), respectively (Table 1), exhibiting enriched genetic variation.
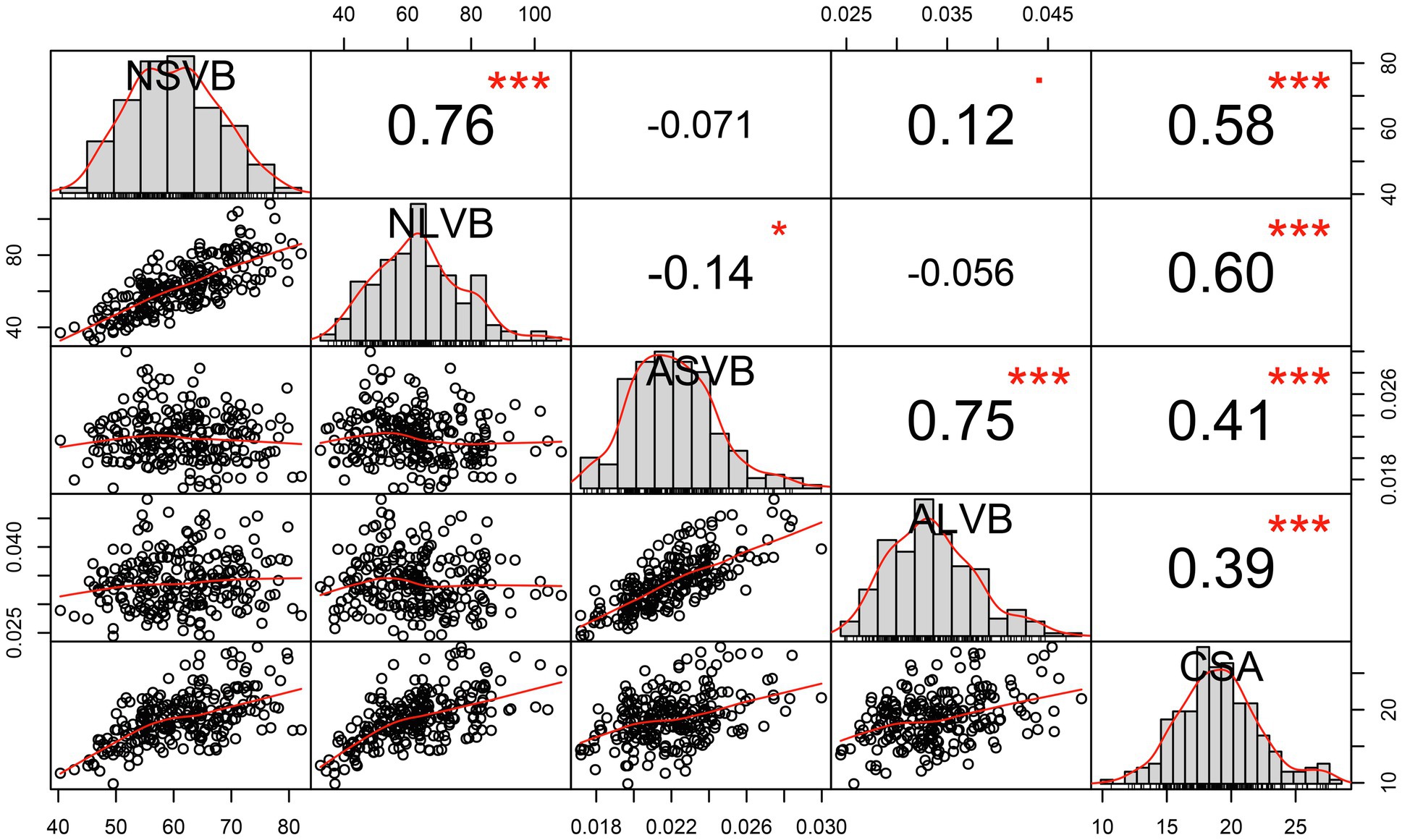
Figure 2. Correlation analysis of BLUP data for vascular bundle-related traits. NSVB, number of small vascular bundle; NLVB, number of large vascular bundle; ASVB, average area of single small vascular bundle; ALVB, average area of single large vascular bundle; and CSA, cross-sectional area. *Significant at 0.05 probability level. ***Significant at 0.001 probability level.
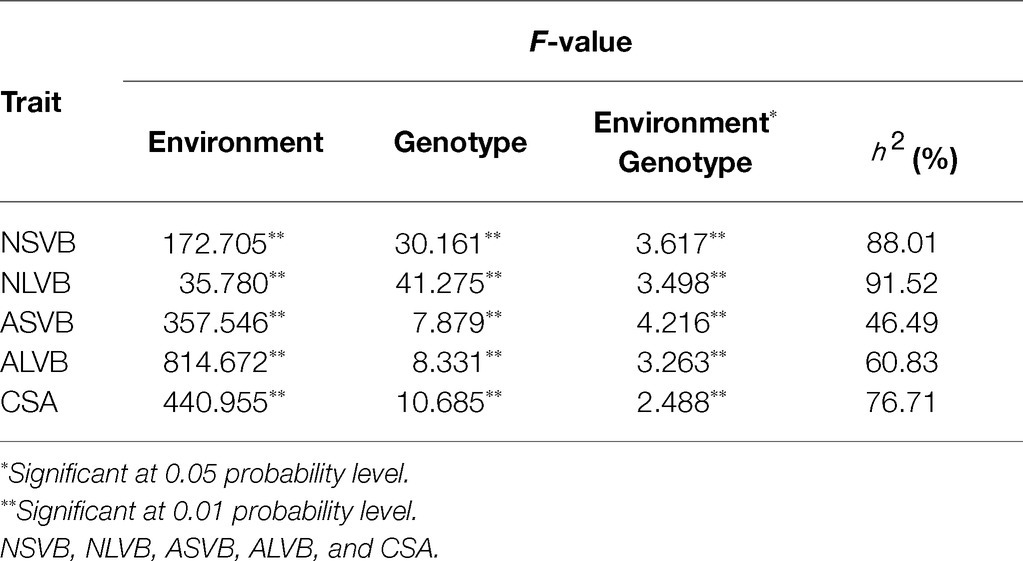
In the analysis of variance for the five traits, highly significant variations for genotypes (G), environments (E), and genotype-by-environment interaction were found (Table 2). This indicates the important roles of both genotypes, environment, and G × E interaction. The broad-sense heritability for NSVB, NLVB, ASVB, ALVB, and CSA across the five environments in the 248 inbred lines ranged from 46.49% (ASVB) to 91.52% (NLVB), indicating the predominant role of genetic factors for these traits (Table 2).
We calculated the BLUP values for each trait and observed significant correlations among them. CSA was positively correlated with NSVB (r = 0.58, p ≤ 0.001), NLVB (r = 0.60, p ≤ 0.001), ASVB (r = 0.41, p ≤ 0.001), and ALVB (r = 0.39, p ≤ 0.001). NSVB was positively correlated with NLVB (r = 0.76, p ≤ 0.001) and ASVB positively correlated with ALVB (r = 0.75, p ≤ 0.001). No significant correlations were detected between the vascular bundle area and the number of vascular bundles, except for the correlation between NLVB and ASVB (r = −0.14, p ≤ 0.05).
The 248 inbred lines used in this study can be divided into five subpopulations and one mixed group, which are designated as Reid, Lancaster, TSPT, Lvda Red Cob (LRC), P group, and Mixed group, respectively (Liu et al., 2012, 2014). To investigate the effect of population structure on vascular bundle, the phenotypic variations of vascular bundle-related traits were compared between different subpopulations. For NSVB and NLVB, the means in TSPT subpopulation were significantly higher than the other subpopulations (Figures 3A,B). For ASVB, ALVB, and CSA, no significant difference was observed among the subpopulations, indicating that population structure has little effect on these traits (Figures 3C–E). In summary, the vascular bundle-related traits show broad variations which are subject to population structure.
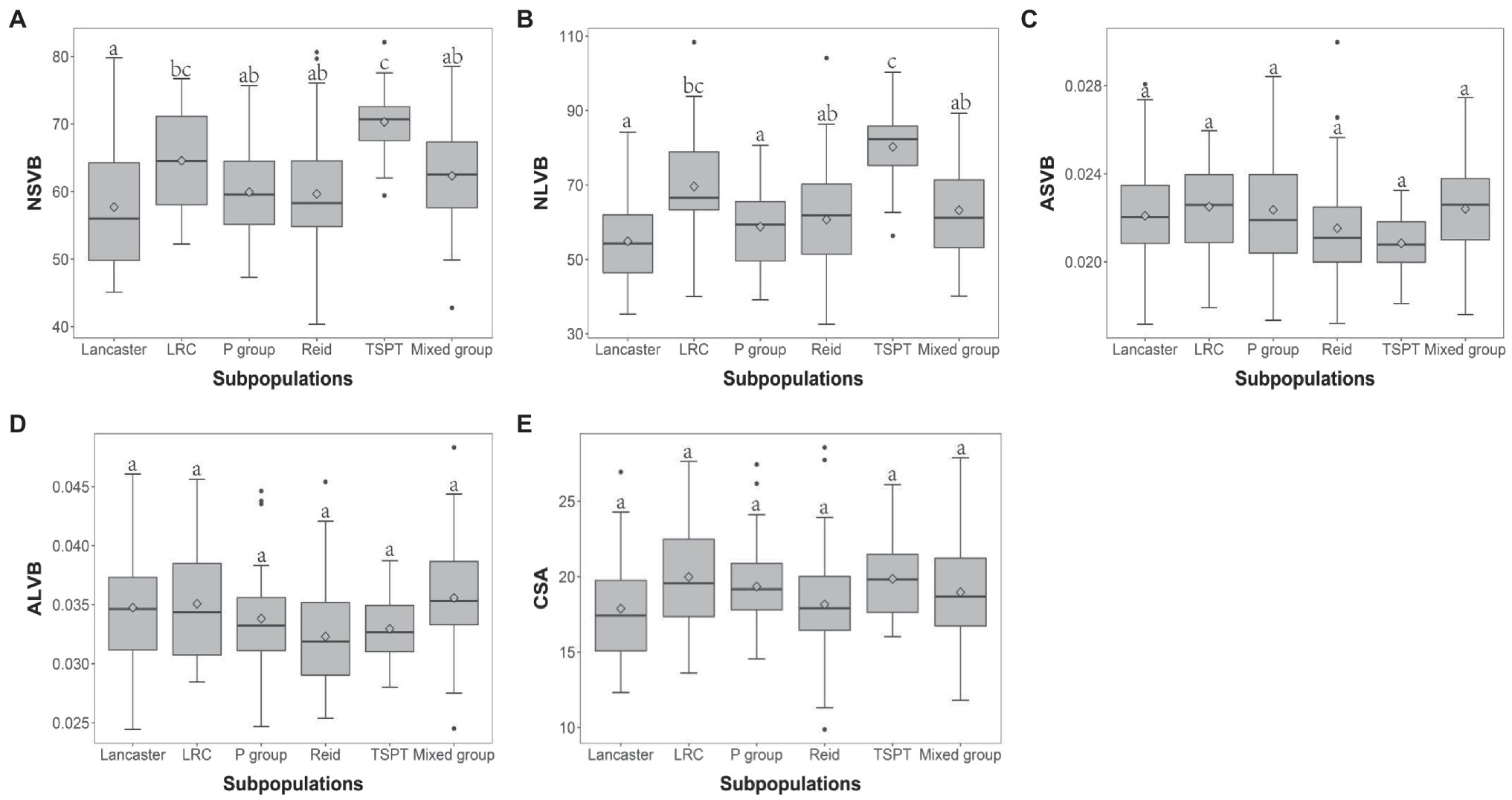
Figure 3. Boxplot of vascular bundle-related traits distribution in different subpopulations. ANOVA was applied to examine the difference of traits among subpopulations. Different numbers indicate significant difference at p ≤ 0.05. (A) NSVB; (B) NLVB; (C) ASVB; (D) ALVB; and (E) CSA.
Genome-Wide Association Analysis
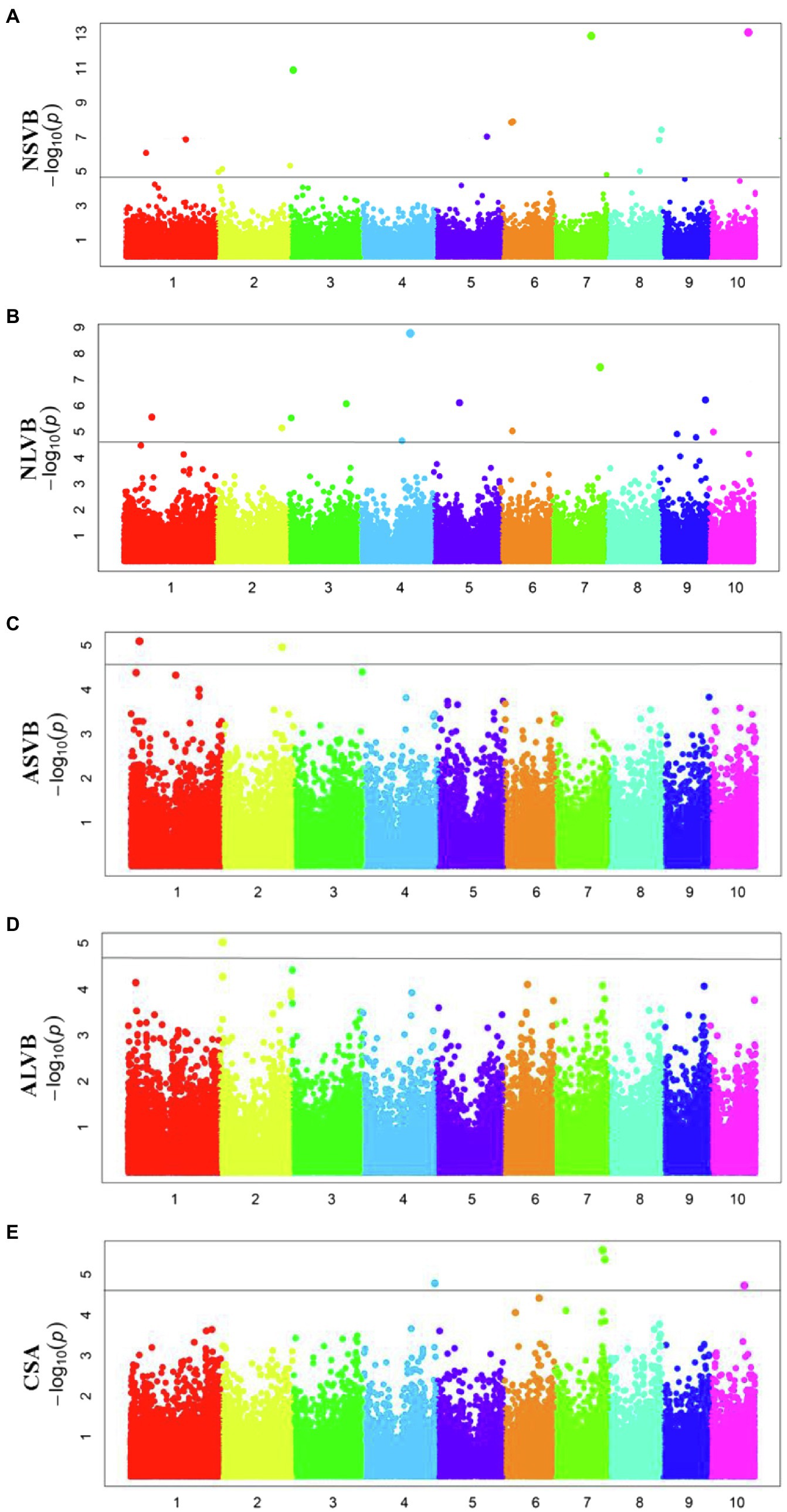
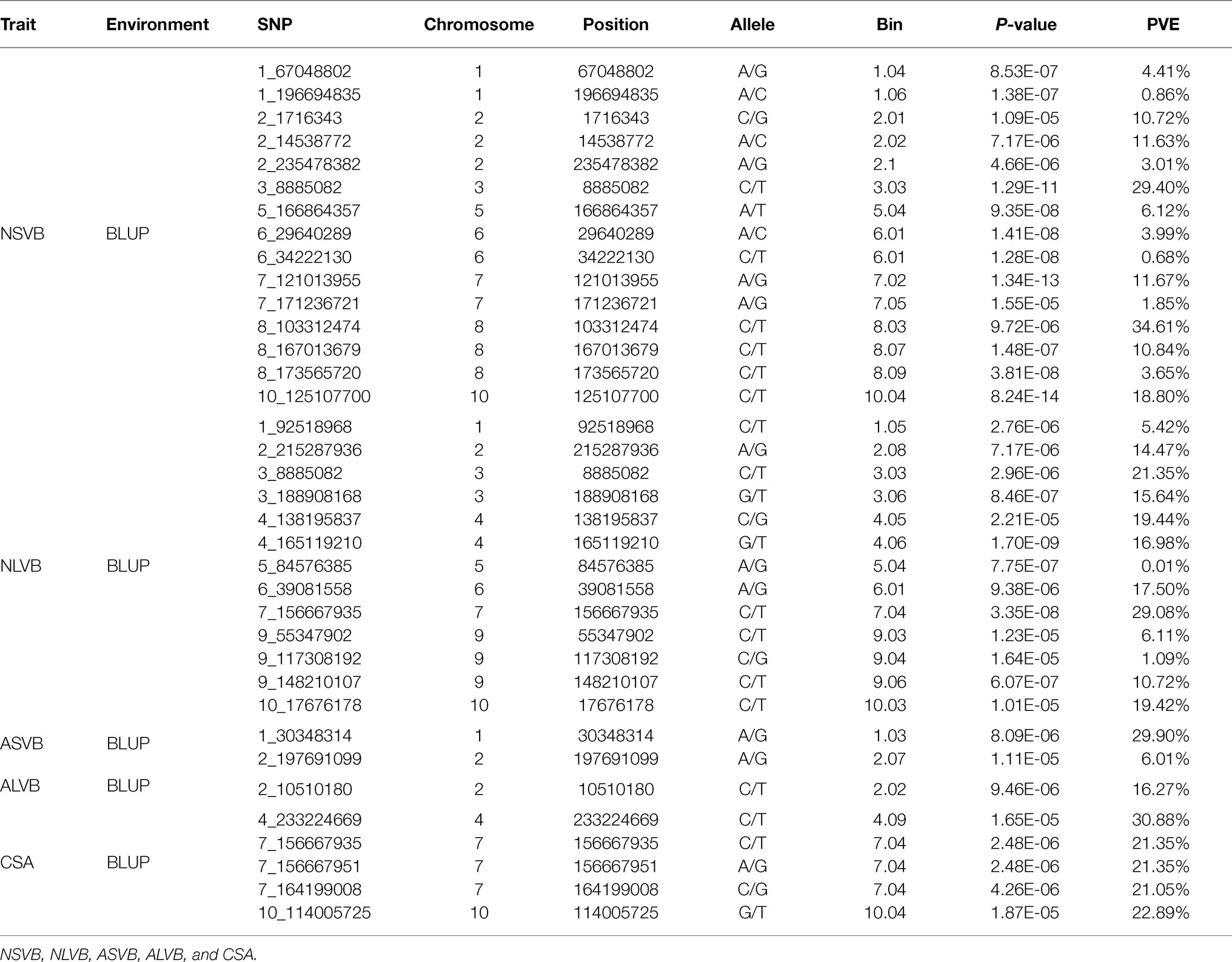
To minimize the effect of environmental variation, BLUP values across five environments were used for GWAS. The effective number which was calculated using the GEC software was 40,705 and p-value which was recommended using the GEC software was 2.46E-5. Thus, the threshold is –log10 (2.46E-5) = 4.61. In total, we identified 15, 13, 2, 1, and 5 SNPs significantly associated with NSVB, NLVB, ASVB, ALVB, and CSA, respectively, based on –log10 p = 4.61 (Figure 4; Table 3). The single phenotypic variation explained value of NSVB, NLVB, ASVB, ALVB, and CSA varied in ranges of 0.68%~29.40, 1.09%~29.08, 6.01%~29.90, 16.27, and 21.05%~30.88%.
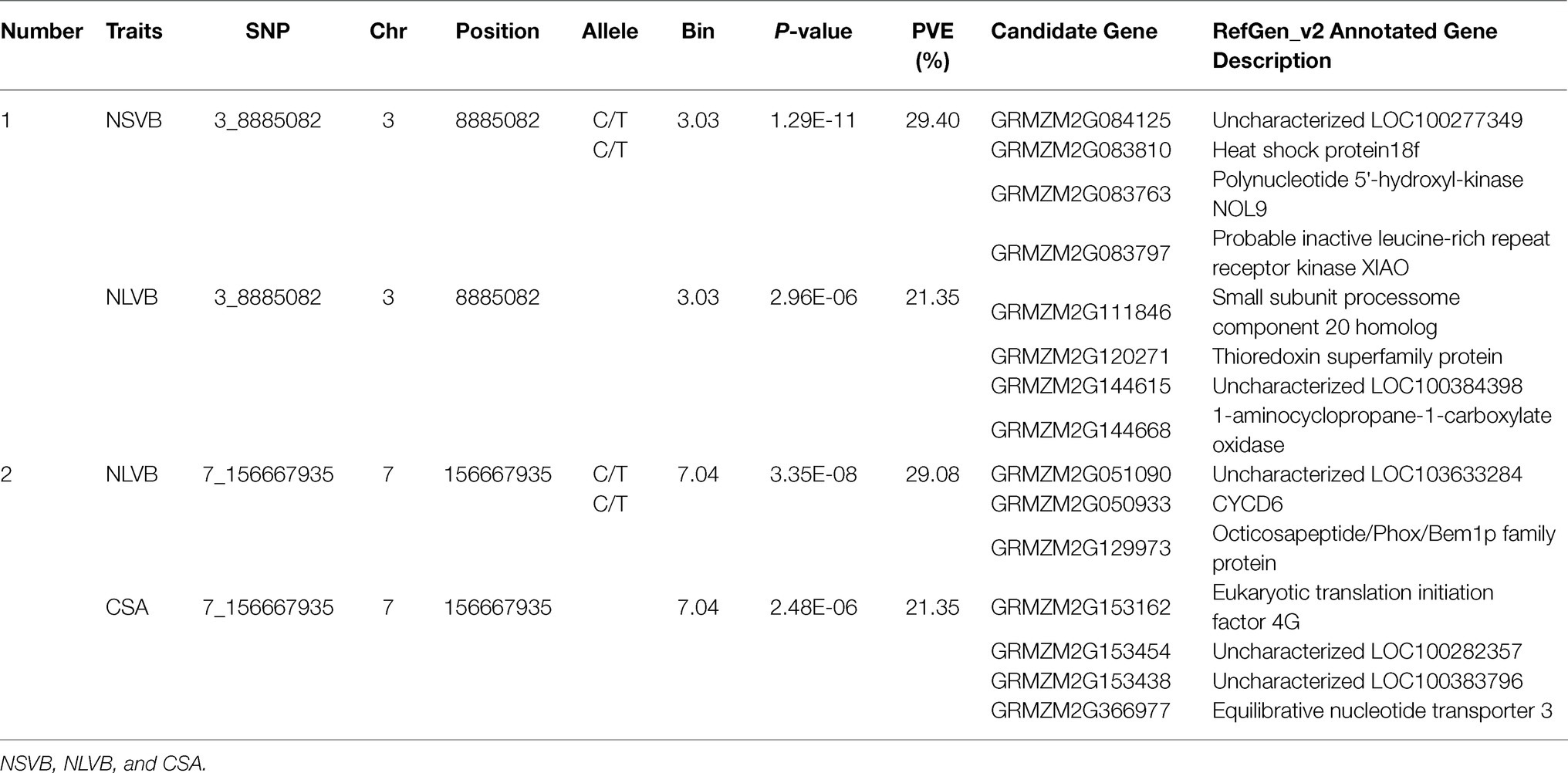
For five vascular bundle-related traits, we identified two pleiotropic loci that probably influenced more than one trait (hereinafter referred to as “co-loci”; Table 4). One located on Chromosome 3 (Chr3_8,885,082) was significantly associated with NSVB and NLVB, explaining from 21.35 to 29.40% of the phenotypic variation. The other one located at Chr7_156,667,935 was significantly associated with NLVB and CSA, explaining from 21.35 to 29.08% of the phenotypic variation.
Candidate Genes Associated With Significant SNPs
The physical locations of the significant SNPs were recorded according to the B73 RefGen_v24 based on the LD decay. A total of 210 candidate genes with gene descriptions were found (Supplementary Table S2). The number of candidate genes involved in the vascular bundle-related traits of NSVB, NLVB, ASVB, ALVB, and CSA was 75, 55, 11, 23, and 19. The candidate genes were uploaded to GENE ONTOLOGY Web site (see footnote 2) for GO secondary classification.
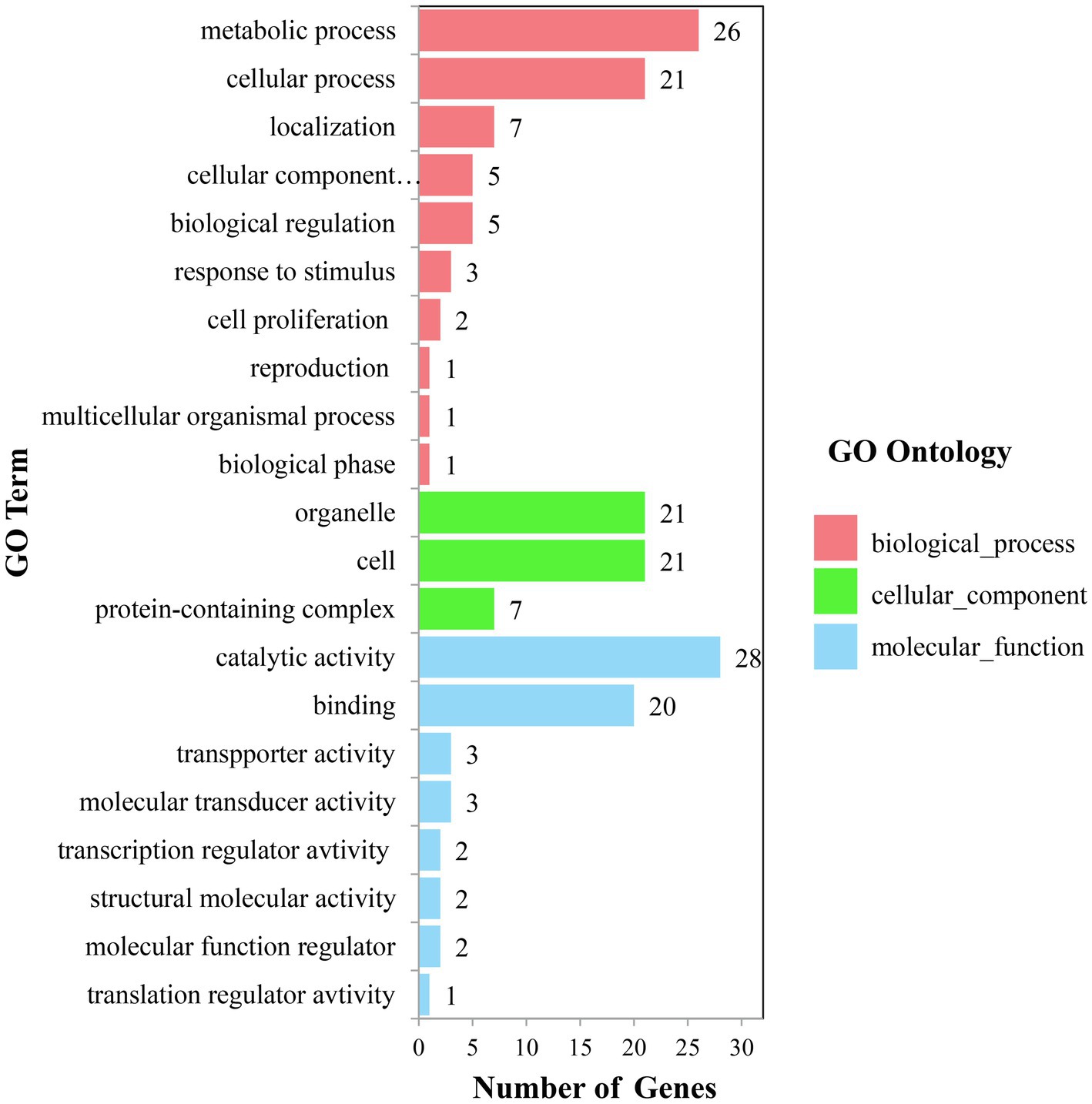
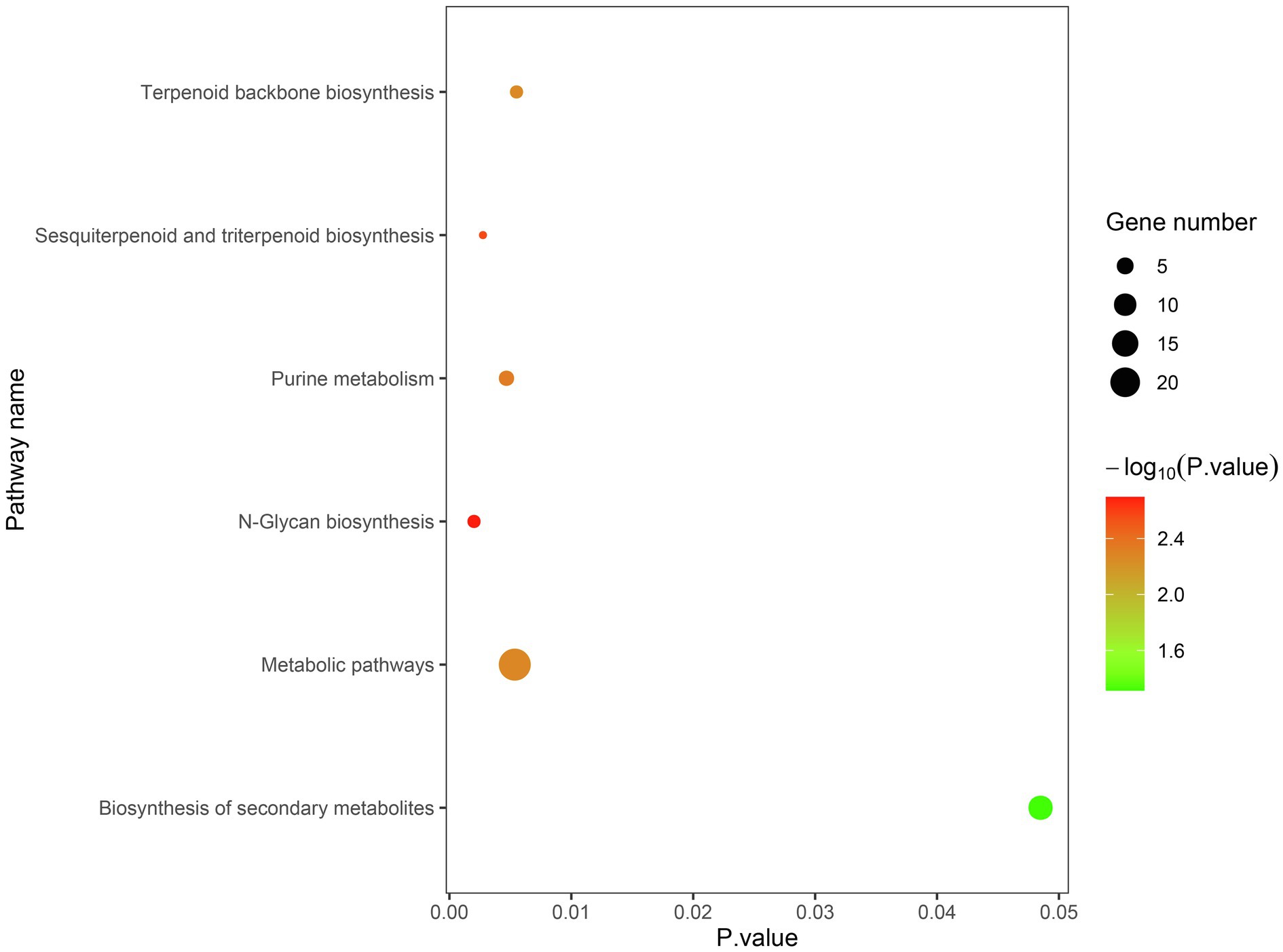
The candidate genes can be classified into ten biological processes, three cellular components, and eight molecular functions. Among them, the candidate genes in biological processes are mainly concentrated in the metabolic process and the cellular process, the candidate genes in cellular component are mainly concentrated in cell and organelle, and those in molecular function are mainly concentrated in catalytic activity and binding (Figure 5). As for the KEGG analysis of the candidate genes, a total of six pathways were identified. These pathways included the metabolic pathways, biosynthesis of secondary metabolites, purine metabolism, N-Glycan biosynthesis, terpenoid backbone biosynthesis, and sesquiterpenoid and triterpenoid biosynthesis, which could be related to vascular bundle (Figure 6). Combined with functional annotation of the candidate genes, we finally found five genes related to vascular development, three genes related to cell wall, and two genes related to the mechanical strength of the stem (Table 5).
Discussion
Genetic Basis of Vascular Bundle-Related Traits
Studies have shown that plant vascular cells continuously connected shoot organs with roots from top to bottom, and the vascular bundles were polar, continuous, and internally aligned (Sawchuk and Scarpella, 2013). Due to the strong consistency between the upper vascular bundles and the lower ones, as well as the relatively clearer structure in upper vascular bundles after staining, vascular-related traits were collected from the uppermost internode in this study.
The five vascular bundle-related traits exhibited wide phenotypic variation with normal distribution. ANOVA showed that the genotype effects, environment effects, and interactive effects between the genotype and environment were both significant for five traits. The heritability ( ) for these traits is very high, and ASVB was the least heritable of the traits included in this study. The correlations were observed among the area of vascular bundle and as were the number of vascular bundle. However, no significant differences were observed between the area of vascular bundle and the number of vascular bundles.
Subpopulations are often observed in natural maize populations. In this study, the maize panel was divided into five subpopulations, including TSPT, LRC, Lancaster, Reid, and P group (Lu et al., 2009; Yan et al., 2009; Yang et al., 2010, 2011). TSPT and LRC subpopulations were selected and improved from native resources in China (Ning et al., 2002; Feng et al., 2010; Xu, 2010), while Lancaster, Reid, and P groups were originated from the American germplasm resources (Guo et al., 2016). Therefore, the population structure may have imposed effects on the vascular bundles of maize. In this study, we observed no significant differences between subpopulations for CSA, ASVB, and ALVB, indicating that the area of vascular bundles is roughly analogous among each subpopulations. In contrast, the inbreds from TSPT and LRC possessed more vascular bundles, indicating that the vascular structure of native Chinese germplasm is more intense than that of American germplasm.
Analysis and Comparison of Genome-Wide Association Analysis of Vascular Bundle-Related Traits
Vascular bundle-related traits are important traits of crops, which are not only highly correlated with crop yield traits, but also related to crop resistance. At the same time, some genes related to vascular bundle development have been cloned in model plants Arabidopsis thaliana and rice (Emery et al., 2003; Zhong and Ye, 2004; Terao et al., 2010; Fei et al., 2019). However, little is known about the functional genes in maize. On this ground, it is necessary to study the genetic basis of vascular bundle-related traits in maize.
In this study, a total of 36 different loci that are significantly associated with vascular bundle were detected. These loci may be key regions for the regulation of maize vascular bundle development. Among them, two “co-loci” were detected, each explaining over 20.00% of the phenotypic variation. This indicates that the loci had a major genetic effect and were less affected by the environment. Therefore, the two “co-loci” were the focus of the vascular bundle molecular design breeding or vascular bundle QTL map cloning work.
The 36 loci were compared with those of the previous studies. In this study, there were two loci which were consistent with the results of Yang (2016), one was located at Chr7_156,667,935 associated with NLVB and CSA; and the other was located at Chr7_156,667,951 associated with CSA. The two loci were very close, indicating that they may share the same functional gene. Du (2018) also found the locus which located at Chr7_156,667,935, indicating that it was stable and important for the development of maize vascular bundles. In addition, another two loci were consistent with the results of Du (2018), one was located at Chr10_125,107,700 associated with NSVB; and the other was located at Chr7_164,199,008 associated with CSA. Huang et al. (2016) performed a high-resolution QTL mapping for the number of vascular bundle in the uppermost internode of maize stem using a large maize-teosinte experimental population and validated the effect of one QTL qVb9-2 on chromosome 9 and further fine mapped the QTL to a 1.8-Mb physical region. In this study, one locus controlling the number of vascular bundles was localized at Chr9_117.308192, about 1 Mb away from Huang et al. (2016) results.
Candidate Genes Analysis
In this study, candidate genes were collected in the range of 120 kb upstream and downstream of the detected 36 significant association loci. A total of 210 candidate genes were listed, of which 64 genes could be used for GO analysis. These candidate genes involved a variety of biochemical metabolic pathways, such as metabolic process, cellular process, cell, organelle, catalytic activity, binding, and so on. Among them, GRMZM2G314396 encodes NAC domain-containing protein 35, NAC transcription factors are a specific class of transcription factors in plants, regulating the growth and development of plants, such as secondary wall and root growth, plant senescence, and so on. And they respond to a variety of abiotic and biotic stresses (Zhang et al., 2019). Kubo et al. (2005) used microarray analysis to find seven transcription factor proteins which containing the NAC domain associated with vascular development in Arabidopsis.
GRMZM2G314396, GRMZM2G136400, and GRMZM2G136410 are related to calcium. Calcium plays an essential role as the second messenger in cells in various signaling transduction pathways by developmental and environmental (Evans et al., 2001; Tuteja and Mahajan, 2007). GRMZM2G314396 encodes calcium-dependent protein kinase (CDPK) and the CDPKs are one of the well-known Ca2+−sensor protein kinases involved in environmental stress resistance (Harper and Harmon, 2005). AtCPK28, one of the CDPKs in Arabidopsis, has been reported to regulate plant stem elongation and vascular development by altering the expression of NAC transcription regulators and gibberellin homeostasis regulators (Matschi et al., 2013). GRMZM2G136400 and GRMZM2G136410 encode CBL-interacting protein kinase (CIPK). Calcineurin B-like proteins (CBLs) and their target proteins–the CIPKs have emerged in a key Ca2+-mediated signaling network in response to stresses in plants (Chen et al., 2011a). Lee et al. (2005) found that expression of AtCIPK14, one of the CIPKs in Arabidopsis, is restricted predominantly to the vascular tissues.
GRMZM2G046070 encodes cinnamyl alcohol dehydrogenase 1, which is the last enzyme in lignin monomer synthesis pathway. Previous studies have shown that lignin is an important component of almost all vascular plant intact cell walls (Battle et al., 2000; Zhong et al., 2000). Zhu et al. (2014) showed that BnCAD1 expressed abundantly in the vascular bundle tissues in ramie. In addition, cinnamyl alcohol dehydrogenase was also found to relate to lodging in Arabidopsis (Jourdes et al., 2007), wheat (Ma, 2010; Chen et al., 2011b), rape (Huang et al., 2013), and so on.
Interestingly, we found some candidate genes related to the cell wall. As is known, cell elongation and cell wall thickening are involved in regulating lodging resistance in plants (Fan et al., 2018). Many studies have shown that vascular bundle-related traits affect the lodging resistance of crops, and vascular bundle-related traits are one of the breeding indexes for lodging resistance of crops (Bian et al., 2017; Hu et al., 2017). GRMZM2G045596 encodes polygalacturonase QRT3. Polygalacturonase, which can degrade pectin, is a plant cell wall structural protein (Abbott and Boraston, 2007). Xiao et al. (2017) overexpressed the PGX2 gene in Arabidopsis, resulting in an increase in lignin in the stems of transgenic plants. GRMZM5G831009 encodes purple acid phosphatase 22. Purple acid phosphatases are members of the metallo-phosphoesterase family identified from a wide range of plants and play vital roles in modulating plant carbon and phosphorus metabolism, cell wall synthesis, and so on. NtPAP12 of tobacco participates in the biosynthesis of the cell wall by catalyzing dephosphorylation of α-xylidase and β-glucosidase in the cell wall (Kaida et al., 2009). GRMZM2G136971 encodes leucine-rich repeat extensin-like protein 3. Baumberger et al. (2001) found LRX1, a new Arabidopsis gene that encodes a chimeric leucine-rich repeat/extensin protein. At the same time, their results suggested that LRX1 was a potential regulator of cell wall development.
Two candidate genes were found to be related to the mechanical strength of stem. GRMZM2G167520 encoded brittle stalk-2-like protein 6, and GRMZM2G167497 encoded brittle stalk-2-like protein 7. Ching et al. (2006) showed that the expression of Brittle stalk 2 (BK2) genes in maize stem, root, and leaf tissue affected the mechanical strength of maize stem. However, the expression was the highest in the vascular systems. The role of the BK2 genes in secondary wall formation is consistent. All in all, further studies are needed for the functional validation of these candidate genes to discover the possible mechanism of vascular bundle regulation.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, and further inquiries can be directed to the corresponding authors.
Author Contributions
HW and JG conceived the ideas, designed the methodology, and revised the manuscript. YZ conducted the field trials and collected the data. PH, LZ, WS, HL, and YH took part in experiment and analyzed the data. YZ and PH wrote the manuscript. All authors have read and approved the final manuscript.
Funding
This work was financially supported by the Science and Technology Innovation Team of Maize Modern Seed Industry in Hebei Province (21326319D), the National Key Research and Development Program of China (2016YFD0101204), and the Hebei Project Area Development Fund of National High-yield Grain Science and Technology Project (JY2019004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fpls.2021.699486/full#supplementary-material
Footnotes
1. ^https://www.ncbi.nlm.nih.gov/
2. ^http://www.geneontology.org/
References
Abbott, D. W., and Boraston, A. B. (2007). The structural basis for exopolygalacturonase activity in a family 28 glycoside hydrolase. J. Mol. Biol. 368, 1215–1222. doi: 10.1016/j.jmb.2007.02.083
Bai, X. F., Wu, B., and Xing, Y. Z. (2012). Yield-related QTLs and their applications in rice genetic improvement. J. Integr. Plant Biol. 54, 300–311. doi: 10.1111/j.1744-7909.2012.01117.x
Battle, M., Bender, M. L., Tans, P. P., White, W. C., Ellis, J. T., Conway, T., et al. (2000). Global carbon sinks and their variability inferred from atmospheric O2 and δ13C. Science 287, 2467–2470. doi: 10.1126/science.287.5462.2467
Baumberger, N., Ringli, C., and Keller, B. (2001). The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev. 15, 1128–1139. doi: 10.1101/Gad.200201
Bian, D. H., Liu, M. X., Niu, H. F., Wei, Z. B., Du, X., and Cui, Y. H. (2017). Effects of nitrogen application times on stem traits and lodging of summer maize (Zea mays L.) in the Huang-Huai-Hai plain. Sci. Agric. Sin. 50, 2294–2304. doi: 10.3864/j.issn.0578-1752.2017.12.010
Chakraborty, R., and Weiss, K. M. (1988). Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. Proc. Natl. Acad. Sci. U. S. A. 85, 9119–9123. doi: 10.1073/pnas.85.23.9119
Chen, X. F., Gu, Z. M., Xin, D. D., Hao, L. A., Liu, C. J., Huang, J., et al. (2011a). Identification and characterization of putative CIPK genes in maize. J. Genet. Genomics 38, 77–87. doi: 10.1016/j.jcg.2011.01.005
Chen, X. G., Shi, C. Y., Yin, Y. P., Wang, Z. L., Shi, Y. H., Peng, T. L., et al. (2011b). Relationship between lignin metabolism and lodging resistance in wheat. Acta Agron. Sin. 37, 1616–1622. doi: 10.3724/SP.J.1006.2011.01616
Ching, A., Dhugga, K. S., Appenzeller, L., Meeley, R., Bourett, T. M., Howard, R. J., et al. (2006). Brittle stalk 2 encodes a putative glycosylphosphatidylinositol-anchored protein that affects mechanical strength of maize tissues by altering the composition and structure of secondary cell walls. Planta 224, 1174–1184. doi: 10.1007/s00425-006-0299-8
Coaker, G. L., Meulia, T., Kabelka, E. A., Jones, A. K., and Francis, D. M. (2002). A QTL controlling stem morphology and vascular development in Lycopersicon esculentum Lycopersicon hirsutum (Solanaceae) crosses is located on chromosome 2. Am. J. Bot. 89, 1859–1866. doi: 10.3732/ajb.89.12.1859
Du, Y. Q. (2018). QTL Mapping of Vascular Bundle Related Traits in Maize Stem. China: Hebei Agricultural University.
Du, Q., and Wang, H. Z. (2015). The role of HD-ZIP III transcription factors and miR165/166 in vascular development and secondary cell wall formation. Plant Signal. Behav. 10:e1078955. doi: 10.1080/15592324.2015.1078955
Du, Y. Q., Yang, S., Zhu, L. Y., Zhao, Y. F., Huang, Y. Q., Chen, J. T., et al. (2018). Genome-wide association analysis of tassel branch number under high and low plant densities in maize (Zea mays L.). Mol. Plant Breed. 16, 5970–5977. doi: 10.13271/j.mpb.016.005970
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379. doi: 10.1371/journal.pone.0019379
Emery, J. F., Floyd, S. K., Alvarez, J., Eshed, Y. A., Hawker, N. P., Izhaki, A., et al. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. doi: 10.1371/journal.pone.0019379
Evans, N. H., McAinsh, M. R., and Hetherington, A. M. (2001). Calcium oscillations in higher plants. Curr. Opin. Plant Biol. 4, 415–420. doi: 10.1016/S1369-5266(00)00194-1
Fan, C., Li, Y., Hu, Z., Hu, H., Wang, G., Li, A., et al. (2018). Ectopic expression of a novel OsExtensin-like gene consistently enhances plant lodging resistance by regulating cell elongation and cell wall thickening in rice. Plant Biotechnol. J. 16, 254–263. doi: 10.1111/pbi.12766
Fei, C., Geng, X., Xu, Z. J., and Xu, Q. (2019). Multiple areas investigation reveals the genes related to vascular bundles in rice. Rice 12:17. doi: 10.1186/s12284-019-0278-x
Feng, G., Li, Y. Y., Jing, X. Q., Wang, L., and Lu, B. S. (2010). Key inbred lines cluster analysis and heterosis model analysis of different period maize hybrids in China. Rain Fed Crops 30, 63–67. doi: 10.3969/j.issn.2095-0896.2010.02.001
Feng, H. J., Zhang, S. P., Ma, C. J., Liu, P., Dong, S. T., Zhao, B., et al. (2014). Effect of plant density on microstructure of stalk vascular bundle of summer maize (Zea mays L.) and its characteristics of sap flow. Acta Agron. Sin. 40, 1435–1442. doi: 10.3724/SP.J.1006.2014.01435
Fujita, D., Trijatmiko, K. R., Tagle, A. G., Sapasap, M. V., Koide, Y., Sasaki, K., et al. (2013). NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. Proc. Natl. Acad. Sci. U. S. A. 110, 20431–20436. doi: 10.1073/pnas.1310790110
George, D., and Mallery, P. (2013). IBM SPSS Statistics 21 Step by Step: A Simple Guide and Reference. United Kingdom: Routledge.
Guo, Q. C., Zhang, Y. R., Kang, H. R., Liu, Z. K., Liu, H. Q., and Dou, B. D. (2016). Discussion on the introduction, selection and combination model of American maize germplasm. Mol. Plant Breed. 14, 3262–3272. doi: 10.13271/j.mpb.014.003262
Hardtke, C. S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411. doi: 10.1093/emboj/17.5.1405
Harper, J. F., and Harmon, A. (2005). Plants, symbiosis and parasites: a calcium signalling connection. Nat. Rev. Mol. Cell Biol. 6, 555–566. doi: 10.1038/nrm1679
Henderson, C. R. (1975). Best linear unbiased estimation and prediction under a selection model. Biometrics 31, 423–447. doi: 10.2307/2529430
Housley, T. L., and Peterson, D. M. (1982). Oat stem vascular size in relation to kernel number and weigh. I. Controlled environment. Crop Sci. 22, 259–263. doi: 10.2135/cropsci1982.0011183X002200020014x
Hu, H., Li, S. S., Hua, H., Sun, M. M., Kang, J., Xia, G. J., et al. (2017). Research on stalk morphological structure characteristics and its relationship between with the lodging of different wheat varieties. J. Triticeae Crops 37, 1343–1348. doi: 10.7606/j.issn.1009-1041.2017.10.10
Huang, C., Chen, Q. Y., Xu, G. H., Xu, D. Y., Tian, J. G., and Tian, F. (2016). Identification and fine mapping of quantitative trait loci for the number of vascular bundle in maize stem. J. Integr. Plant Biol. 58, 81–90. doi: 10.1111/jipb.12358
Huang, J. H., Li, W., Qu, C. M., Liu, L. Z., Xu, X. F., Wang, R., et al. (2013). Expression characteristics of key genes in lignin pathway among different lodging resistance lines of Brassica napus L. Acta Agron. Sin. 39:1339. doi: 10.3724/SP.J.1006.2013.01339
Jourdes, M., Cardenas, C. L., Laskar, D. D., Moinuddin, S. G. A., Davin, L. B., and Lewis, N. G. (2007). Plant cell walls are enfeebled when attempting to preserve native lignin configuration with poly-p-hydroxycinnamaldehydes: evolutionary implications. Phytochemistry 68, 1932–1956. doi: 10.1016/j.phytochem.2007.03.044
Kaida, R., Satoh, Y., Bulone, V., Yamada, Y., Kaku, T., Hayashi, T., et al. (2009). Activation of β-glucan synthases by wall-bound purple acid phosphatase in tobacco cells. Plant Physiol. 150, 1822–1830. doi: 10.1104/pp.109.139287
Knapp, S. J., Stroup, W. W., and Ross, W. M. (1985). Exact confidence intervals for heritability on a progeny mean basis. Crop Sci. 25, 192–194. doi: 10.2135/cropsci1985.0011183X002500010046x
Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., et al. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860. doi: 10.1101/gad.1331305
Lai, G. R. (2017). Genotype Analysis Using GBS Technology and High Density Genetic Map Construction in Maize. China: Hebei Agricultural University.
Lee, E. J., Iai, H., Sano, H., and Koizumi, N. (2005). Sugar responsible and tissue specific expression of a gene encoding AtCIPK14, an Arabidopsis CBL-interacting protein kinase. Biosci. Biotechnol. Biochem. 69, 242–245. doi: 10.1271/Bbb.69.242
Li, N., Lin, B., Wang, H., Li, X. M., and Chu, Z. H. (2019). Natural variation in ZmFBL41 confers banded leaf and sheath blight resistance in maize. Nat. Genet. 51, 1540–1548. doi: 10.1038/s41588-019-0503-y
Li, Z., Liu, W. T., Yang, S., Guo, J. J., Zhao, Y. F., Huang, Y. Q., et al. (2020). Genome-wide association study of flowering time related traits in maize (Zea mays L.). Mol. Plant Breed. 18, 37–45. doi: 10.13271/j.mpb.018.000037
Li, H., Peng, Z. Y., Yang, X. H., Wang, W. D., Fu, J. J., Wang, J. H., et al. (2013). Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 45, U43–U72. doi: 10.1038/ng.2484
Li, M. X., Yeung, J. M. Y., Cherny, S. S., and Sham, P. C. (2012). Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 131, 747–756. doi: 10.1007/s00439-011-1118-2
Liu, Y., Guo, J. J., Zhang, D. M., Zhao, Y. F., Zhu, L. Y., Huang, Y. Q., et al. (2014). Genetic diversity and linkage disequilibrium estimation among the maize breeding germplasm forassociation mapping. Int. J. Agric. Biol. 16, 851–861. doi: 10.1071/CP14071
Liu, X. L., Huang, M., Fan, B., Buckler, E. S., and Zhang, Z. W. (2016). Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 12:e1005767. doi: 10.1371/journal.pgen.1005767
Liu, W. T., Jian, L. Q., Guo, J. J., Zhao, Y. F., Huang, Y. Q., Chen, J. T., et al. (2020). Association mapping of ear-related traits and their general combining ability in maize. J. Plant Genet Res. 21, 706–715. doi: 10.13430/j.cnki.jpgr.20190801001
Liu, Z. Z., Wu, X., Liu, H. L., Li, Y. X., Li, Q. C., Wang, F. G., et al. (2012). Genetic diversity and.. population structure of important Chinese maize inbred lines revealed by 40 core simple sequence repeats (SSRs). Sci. Agric. Sin. 45, 2107–2138. doi: 10.3864/j.issn.0578-1752.2012.11.001
Lu, Y. L., Yan, J. B., Guimaraes, C. T., Taba, S., Hao, Z. F., Gao, S. B., et al. (2009). Molecular characterization of global maize breeding germplasm based on genome-wide single nucleotide polymorphisms. Theor. Appl. Genet. 120, 93–115. doi: 10.1007/s00122-009-1162-7
Ma, Q. H. (2010). Functional analysis of a cinnamyl alcohol dehydrogenase involved in lignin biosynthesis in wheat. J. Exp. Bot. 61, 2735–2744. doi: 10.1093/jxb/erq107
Matschi, S., Werner, S., Schulze, W. X., Legen, J., Hilger, H. H., and Romeis, T. (2013). Function of calcium-dependent protein kinase CPK28 of Arabidopsis thaliana in plant stem elongation and vascular development. Plant J. 73, 883–896. doi: 10.1111/tpj.12090
McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. doi: 10.1038/35079635
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Nátrová, Z. (1991). Anatomical characteristics of the uppermost internode of winter wheat genoypes differing in stem length. Biol. Plant. 33, 491–494. doi: 10.1007/BF02897726
Ning, J. L., Gao, H. M., Qu, G., Yu, B., and He, J. (2002). Utilization of inbred lines of luda red cob group in corn breeding and produciton in China. Rain Fed Crops 22, 63–65. doi: 10.3969/j.issn.2095-0896.2002.02.001
Qi, J., Qian, Q., Bu, Q. Y., Li, S. Y., Chen, Q., Sun, J. Q., et al. (2008). Mutation of the rice narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 147, 1947–1959. doi: 10.1104/pp.108.118778
Ray, D. K., Mueller, N. D., West, P. C., and Foley, J. A. (2013). Yield trends are insufficient to double global crop production by 2050. PLoS One 8:e66428. doi: 10.1371/journal.pone.0066428
Sakaguchi, J., and Fukuda, H. (2008). Cell differentiation in the longitudinal veins and formation of commissural veins in rice (Oryza sativa) and maize (Zea mays). J. Plant Res. 121, 593–602. doi: 10.1007/s10265-008-0189-1
Sang, Y., Deng, Z. Y., Zhao, L., Zhang, K. P., Tian, J. C., and Ye, B. X. (2010). QTLs for the vascular bundle system of the uppermost internode using a doubled haploid population of two elite Chinese wheat cultivars. Plant Breed. 129, 605–610. doi: 10.1111/j.1439-0523.2009.01727.x
Sawchuk, M. G., and Scarpella, E. (2013). Polarity, continuity and alignment in plant vascular strands. J. Integr. Plant Biol. 9, 824–834. doi: 10.1111/jipb.12086
Terao, T., Nagata, K., Morino, K., and Hirose, T. (2010). A gene controlling the number of primary rachis branches also controls the vascular bundle formation and hence is responsible to increase the harvest index and grain yield in rice. Theor. Appl. Genet. 120, 875–893. doi: 10.1007/s00122-009-1218-8
Tian, F., Bradbury, P. J., Brown, P. J., Hung, H., Sun, Q., Flint-Garcia, S., et al. (2011). Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 43, U159–U113. doi: 10.1038/ng.746
Tuteja, N., and Mahajan, S. (2007). Calcium signaling network in plants: an overview. Plant Signal. Behav. 2, 79–85. doi: 10.4161/psb.2.2.4176
Wang, X. L., Wang, H. W., Liu, S. X., Ferjani, A., Li, J. S., Yan, J. B., et al. (2016). Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 48, 1233–1241. doi: 10.1038/ng.3636
Xiao, C. W., Barnes, W. J., Zamil, M. S., Yi, H., Puri, V. M., and Anderson, C. T. (2017). Activation tagging of Arabidopsis POLYGALACTURONASE INVOLVED IN EXPANSION2 promotes hypocotyl elongation, leaf EXPANSION, stem lignification, mechanical stiffening, and lodging. Plant J. 89, 1159–1173. doi: 10.1111/tpj.13453
Xu, W. W. (2010). Effects of inbred line Dan 340 and its derivative lines on maize production in China. Rain Fed Crops 30, 255–257. doi: 10.3969/j.issn.2095-0896.2010.04.001
Xu, C. L., Gao, Y. B., Tian, B. J., Ren, J. H., Meng, Q. F., and Wang, P. (2017). Effects of EDAH, a novel plant growth regulator, on mechanical strength, stalk vascular bundles and grain yield of summer maize at high densities. Field Crop Res. 200, 71–79. doi: 10.1016/j.fcr.2016.10.011
Yan, J. B., Shah, T., Warburton, M. L., Buckler, E. S., McMullen, M. D., and Crouch, J. (2009). Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS One 4:e8451. doi: 10.1371/journal.pone.0008451
Yang, S. (2016). Identification of Lodging-Resistance and Mapping of Quantitative Trait Loci (QTL) for Stalk Vascular Bundle in Maize. China: Hebei Agricultural University.
Yang, X. H., Gao, S. B., Xu, S. T., Zhang, Z. X., Prasanna, B. M., Li, L., et al. (2011). Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol. Breed. 28, 511–526. doi: 10.1007/s11032-010-9500-7
Yang, X. H., Yan, J. B., Shah, T., Warburton, M. L., Li, Q., Li, L., et al. (2010). Genetic analysis and characterization of a new maize association mapping panel for quantitative trait loci dissection. Theor. Appl. Genet. 121, 417–431. doi: 10.1007/s00122-010-1320-y
Zhai, L. Y., Zheng, T. Q., Wang, X., Wang, Y., Chen, K., Wang, S., et al. (2018). QTL mapping and candidate gene analysis of peduncle vascular bundle related traits in rice by genome-wide association study. Rice 11:13. doi: 10.1186/s12284-018-0204-7
Zhang, H. Z., Bai, X. Q., and Zeng, Y. L. (2019). Biological functions of plant NAC transcription factors. Plant Physiol. J. 55, 915–924. doi: 10.13592/j.cnki.ppj.2019.0107
Zhong, R. Q., Morrison, W. H., Himmelsbach, D. S., Poole, F. L., and Ye, Z. H. (2000). Essential role of caffeoyl coenzyme a O-methyltransferase in lignin biosynthesis in woody poplar plants. Plant Physiol. 124, 563–577. doi: 10.1104/Pp.124.2.563
Zhong, R. Q., and Ye, Z. H. (2004). Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol. 45, 369–385. doi: 10.1093/Pcp/Pch051
Keywords: maize, stalk, vascular bundle, genome-wide association study, candidate genes
Citation: Zheng Y, Hou P, Zhu L, Song W, Liu H, Huang Y, Wang H and Guo J (2021) Genome-Wide Association Study of Vascular Bundle-Related Traits in Maize Stalk. Front. Physiol. 12:699486. doi: 10.3389/fpls.2021.699486
Edited by:
Sanghyeob Lee, Sejong University, South KoreaReviewed by:
Melaku Gedil, International Institute of Tropical Agriculture (IITA), NigeriaOnew Lee, Sejong University, South Korea
Copyright © 2021 Zheng, Hou, Zhu, Song, Liu, Huang, Wang and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Wang, hongwang1203@126.com; Jinjie Guo, guojinjie512@163.com
†These authors have contributed equally to this work and share first authorship
 Yunxiao Zheng1†
Yunxiao Zheng1† Peng Hou
Peng Hou Hong Wang
Hong Wang Jinjie Guo
Jinjie Guo