- 1Graduate School of Biostudies, Kyoto University, Kyoto, Japan
- 2School of Engineering and Applied Sciences, National University of Mongolia, Ulaanbaatar, Mongolia
- 3Gene Discovery Research Group, RIKEN Center for Sustainable Resource Science, Tsukuba, Japan
- 4Bioproductivity Informatics Research Team, RIKEN Center for Sustainable Resource Science, Yokohama, Japan
- 5Research Institute of Animal Husbandry, Mongolian University of Life Science, Ulaanbaatar, Mongolia
- 6Department of Applied Biological Chemistry, The University of Tokyo, Tokyo, Japan
Plants in Mongolian grasslands are exposed to short, dry summers and long, cold winters. These plants should be prepared for fast germination and growth activity in response to the limited summer rainfall. The wild plant species adapted to the Mongolian grassland environment may allow us to explore useful genes, as a source of unique genetic codes for crop improvement. Here, we identified the Chloris virgata Dornogovi accession as the fastest germinating plant in major Mongolian grassland plants. It germinated just 5 h after treatment for germination initiation and showed rapid growth, especially in its early and young development stages. This indicates its high growth potential compared to grass crops such as rice and wheat. By assessing growth recovery after animal bite treatment (mimicked by cutting the leaves with scissors), we found that C. virgata could rapidly regenerate leaves after being damaged, suggesting high regeneration potential against grazing. To analyze the regulatory mechanism involved in the high growth potential of C. virgata, we performed RNA-seq-based transcriptome analysis and illustrated a comprehensive gene expression map of the species. Through de novo transcriptome assembly with the RNA-seq reads from whole organ samples of C. virgata at the germination stage (2 days after germination, DAG), early young development stage (8 DAG), young development stage (17 DAG), and adult development stage (28 DAG), we identified 21,589 unified transcripts (contigs) and found that 19,346 and 18,156 protein-coding transcripts were homologous to those in rice and Arabidopsis, respectively. The best-aligned sequences were annotated with gene ontology groups. When comparing the transcriptomes across developmental stages, we found an over-representation of genes involved in growth regulation in the early development stage in C. virgata. Plant development is tightly regulated by phytohormones such as brassinosteroids, gibberellic acid, abscisic acid, and strigolactones. Moreover, our transcriptome map demonstrated the expression profiles of orthologs involved in the biosynthesis of these phytohormones and their signaling networks. We discuss the possibility that C. virgata phytohormone signaling and biosynthesis genes regulate early germination and growth advantages. Comprehensive transcriptome information will provide a useful resource for gene discovery and facilitate a deeper understanding of the diversity of the regulatory systems that have evolved in C. virgata while adapting to severe environmental conditions.
Introduction
Mongolia is a highland country with an average height of 1580 m, including steppe grassland, mountains, and desert. The Mongolian climate is cold and dry with long winters, short summers, and a wide temperature range between winter and summer. The precipitation level is extremely low and occurs mainly in summer between June and September. The main grasslands, including the steppe, mountain steppe, and forest areas, receive 200–300 mm precipitation annually (Suttie, 2006). As summer in the Mongolian grassland is very short and dry, almost all plants have prepared to germinate and grow quickly, making use of the small amount of rain and the short warm period, which starts in July. Plant growth ceases after August owing to low temperatures and frosts. These severe climatic conditions have suppressed and screened Mongolian grassland plants. Unique and high-performance plants have evolved by surviving this natural selection.
About 80% of Mongolian territory is covered by natural plants and used as natural pasture by nomadic groups, with extensive cultivation of livestock such as goats, sheep, cattle, horses, and camels. Mongolia is mainly engaged in animal husbandry; as of 2019 the number of livestock reached about 71 million, an increase of 6.4% compared to the previous year. As approximately 90% of the livestock rely on natural pastureland for their feed, extreme weather events such as droughts and ‘dzud’ (a local term for severe winter conditions that affect livestock) that causes decreasing of natural pasture have severe effects on the livestock in the nomadic pasture system. The highest annual growth of pasture plants occurs in August. However, in recent years, annual growth has decreased to 75% of the highest growth owing to drought and the degradation of grazing lands by overgrazing and exceeding the carrying capacity (Karban and Baldwin, 1997). Every season, Mongolian nomads move to convenient new areas to search for the best pasture to meet the nutritional demands of their livestock. These nomadic cultures might also be important for research on the growth mechanisms of Mongolian natural plants.
In recent years, most Mongolian grassland plants have been analyzed in the hopes of uncovering information useful to phytochemical and pharmacological research. Natural wild plants from 3160 species, 564 genera, and 128 families live in Mongolia, over a wide temperature range of four seasons (Grubov, 2008). Approximately 600 species have been identified as significant edible plants and 1100 species as medicinal plants. Furthermore, 150 species are rich vitamin resources, 200 contain essential oils, 250 contain tannins, more than 200 are used for dyeing, 231 are rich in flavonoids, 200 can be used as ingredients in traditional medicine, more than 480 are ornamental plants, 280 contain alkaloids, and 65 are used to prevent desertification (Jamyandorj et al., 2011). The Western Pacific Region of the World Health Organization have listed the most useful medicinal plants found in Mongolia (Shin, 2013).
The main Mongolian rangeland plants include 26 families and 73 genera, among which the Compositae and Gramineae families are the most abundant, both easily distributed by wind (Lapin et al., 2017). Agropyron cristatum is one of well eatable plant for animal husbandry in Mongolia and is also widely distributed in European and Asian grasslands. Additionally, A. cristatum is a possible gene donor for wheat improvement, including resistance to wheat streak mosaic virus and leaf rust disease (Sharma et al., 1984; Ochoa et al., 2015). In recent years, a comprehensive transcriptome analysis of A. cristatum has been performed for the investigation of functional genes (Zhou et al., 2019), and many stress-resistance genes have been identified (Zhang et al., 2015).
In this study, we screened Mongolian grassland plants for high growth ability and identified the Chloris virgata Dornogovi (DG) accession. To obtain a comprehensive transcriptome of C. virgata, RNA-seq was performed to gain insight into genetic regulation during the germination, early development, young development, and adult development stages of C. virgata, and the transcriptome of C. virgata is presented here for the first time. Complete genome sequencing has only been used to reveal a few complete genomes as the method faces several challenges, including the complexity of complete genomes, the sequencing costs, and the computational resources required; therefore, it is not suitable for non-model plants. Instead, RNA sequencing can be used for non-model plants without a reference genome, and has become the most convenient and cost-effective tool for gaining insight into transcriptome profiling and for detecting differentially expressed genes (DEGs) (Wang et al., 2009). For instance, RNA sequences for crops such as eggplants (Ramesh et al., 2016), peppers (Gordo et al., 2012), tobacco (Lei et al., 2014), corn (Lu et al., 2017), grapes (Xu et al., 2014), jute (Yang et al., 2017), and cotton (Wei et al., 2017), as well as wild plants such as Craterostigma plantagineum (Rodriguez et al., 2010) and A. cristatum (Zhang et al., 2015) have been published. However, only the complete chloroplast genome sequence of C. virgata is available in the National Center for Biotechnology Information (NCBI) database (Hereward et al., 2016).
In this study we establish, for the first time, a de novo transcriptome assembly for C. virgata at four different developmental stages. We present a total of 43,752,426 raw RNA-seq reads and use them for the de novo assembly and annotation of genes from C. virgata against Arabidopsis and rice. DEGs were identified at each of the four developmental stages and these data were used for phytohormone homolog searching. Our findings may serve as a basis for further gene discovery research on C. virgata.
Materials and Methods
Plant Materials and Growth Conditions
Seeds of the C. virgata DG accession were harvested from the native grassland of DG Province in Mongolia and stored at 25 ± 2°C. Rice (Oryza sativa L. cv. Nipponbare), wheat (Triticum aestivum L. cv. Norin No. 61), and Arabidopsis thaliana ecotype Columbia (hereafter Arabidopsis) were used as control plants. The 9 de-husked seeds per each plant were sterilized in 5% sodium hypochlorite and 0.07% Tween-20 for 20 min and rinsed three times with sterile water for 10 min. After sterilization, the seeds were sown on 1/2 Murashige and Skoog basal (MS, Duchefa-Biochemie, Haarlem, Netherlands) plates containing 1.5% sucrose and 0.9% phytoagar (Duchefa-Biochemie, Haarlem, Netherlands). After 2 days of incubation at 4°C in the dark, the seeds were incubated at 22°C with a 16-h light/8-h dark photoperiod for germination analysis. For the observation of plants grown in soil, the seedlings were transplanted into soil 7 days after germination (DAG) and grown at 22°C with a 16-h light/8-h dark cycle. These experiments was performed with three biological replicates.
Regrowth Potential Analysis
The leaf regrowth potential after animal bite was tested by cutting the leaves with scissors to mimic cutting by animal teeth. After 2 weeks of growth in soil, the plants were cut back to a height of 4 cm from the soil surface using scissors. Cutting was performed every 2 weeks for 12 weeks (six cutting events).
RNA Extraction
Total RNA was extracted from plants grown at four different developmental stages: the germination, early young development, young development, and adult development stages. C. virgata were grown in 1/2 MS medium supplemented with 1.5% sucrose and 0.9% phytoagar at 22°C under white light (a 6-h light/8-h dark cycle was used for long-day conditions). The C. virgata grown on 1/2 MS were harvested 2 days (germination stage) and 8 days (early young development stage) after germination. C. virgata grown in soil were harvested 10 days (young development stage) and 21 days (adult development stage) after transplantation of 7-day-old plants from 1/2 MS to the soil. For the whole-plant samples, only the shoots were collected and the roots cut away. Total RNA was extracted from the plant samples using the RNeasy Plant Mini kit (Qiagen).
cDNA Synthesis and RNA-Seq Analysis Using an Illumina Sequencing System
To create a cDNA sequencing library, mRNAs with poly (A) tails were isolated from the total RNA using the NEBNext® Poly (A) mRNA Magnetic Isolation Module (NEB E7490) and NEBNext® Ultra RNA Library Prep Kit for Illumina® (E7530). The first strand of cDNA was synthesized using random hexamer primers. The cDNA fragment was amplified using the 1.8 × Agencourt AMPure XP Beads linker and NEBNext Adaptor for Illumina. The Illumina NovaSeqTM sequencing system was used for sequencing. The raw reads dataset was submitted to the DNA Data Bank of Japan Sequence Read Archive (DDBJ SRA) under accession number DRA011714.
De novo Transcriptome Assembly and Sequence Clustering
The RNA-seq raw reads were trimmed using Trimmomatic (v0.39) (Bolger et al., 2014) with the following parameter settings: TLEADING:30 TRAILING:30 SLIDINGWINDOW:4:15 MINLEN:60. The trimmed reads were assembled using Trinity (v2.8.5) (Grabherr et al., 2011) with the parameters ‘seqType fq –SS_lib_type RF.’ Trinity-based contigs were further assembled using the PCAP assembler (Huang et al., 2003). The PCAP-based contigs were clustered using CD-HIT-EST (v.4.8.1) (Fu et al., 2012) with the identity parameter set to ≥80. Using the longest sequences in each of the CD-HIT-EST clusters as unique transcripts, protein-coding sequence regions were predicted using TransDecoder (v.5.5.0)1 with the results of a sequence similarity search and domain search against the UniProt and Pfam-A databases, conducted using BLASTx search (NCBI BLAST v2.8.1) and the hmmscan program in the HMMER package (v.3.2.1) (Johnson et al., 2010), respectively. Unique transcripts containing protein-coding sequences were used for downstream analyses as mapping references.
Functional Annotation and Gene Ontology Enrichment Analysis
To predict the gene function of the deduced protein sequences from the reference transcripts of C. virgata, their closest homologs in rice and Arabidopsis were searched for in the TAIR10 and IRGSP1.0 (Ensemble plants release-44) using the BLASTp program with a cutoff E-value < 10–5, and swissprot (v5) database using the CloudBlast function of OmixBox software (v.1.1.164) with a cutoff E-value < 10–5. The reference C. virgata transcripts were annotated with gene ontology (GO) terms using OmicsBox, and gene-set enrichment analysis was conducted using the GO terms of DEGs, based on the Fisher’s exact test function (false discovery rate, FDR < 0.03) in OmicsBox.
cDNA Synthesis and RNA-seq Analysis by Ion Proton Sequencing
The Thermo Fisher Ion ProtonTM sequencing system was used for RNA-seq analysis to identify DEGs in C. virgata. A Dynabeads mRNA Purification Kit and Ion Total RNA-Seq Kit v2 (Thermo Fisher Scientific) were used for the purification of mRNA from the total RNA samples and the construction of a cDNA library, respectively. The quality of the total RNA, mRNA, and cDNA libraries were analyzed with an Agilent Tape Station (Agilent Technologies). The cDNA libraries were pooled for emulsion PCR using an Ion PI Hi-Q Chef Kit (Thermo Fisher Scientific). The enriched samples were loaded onto an Ion PI chip (v3) using Ion Chef and sequenced using an Ion Proton instrument. The raw reads dataset was submitted to the DDBJ SRAunder accession number DRA011714.
Expression Profiling and Differentially Expressed Genes
The trimmed RNA-seq reads from the ion proton sequencing were mapped onto the reference transcript dataset using TMAP (v. 3.0.1)2. The expression level of each transcript was quantified based on read count data computed by the featureCounts program (Liao et al., 2014) and normalized to reads per million mapped reads (RPM). Differentially expressed transcripts between pairwise combinations of samples were identified using the Pairwise Differential Expression Analysis function based on edgeR (Robinson et al., 2009) with the read count data output from the Create Count Table function based on HT-seq (Anders et al., 2015) performed with OmixBox.
Phytohormone Similarity Sequence Searching and Alignment
Phytohormone similarity was assessed between C. virgata and the A. thaliana (accession number: GCF_000001735.4) and O. sativa (accession number: GCF_000005425.2) protein sequences downloaded from the NCBI. Similarity searches were conducted using BLASTx 2.9.0 + (E-value < 1e-5). Highly similar sequences were clustered using CD-HIT 4.8.1, with 99% identity, and each cluster was used for read alignment. Multiple sequence alignment was performed using MAFFT 7.470 (Katoh and Standley, 2013) with the <–auto> commands. All columns with gaps in more than 50% of the sequences were removed using TrimAL 1.4 and the maximum likelihood phylogenetic tree was created using IQ-Tree 1.6.12 (Nguyen et al., 2015) with 1000 bootstrap replicates (Hoang et al., 2018) and Model Finder (Kalyaanamoorthy et al., 2017).
Phylogenetic Analysis
We downloaded the protein sets of the whole chloroplast genomes of 17 species, including C. virgata, from the NCBI RefSeq and GenBank databases. Single-copy orthologs of all genomes were extracted using OrthoFinder 2.4.0 (Emms and Kelly, 2019) and all single-copy orthologs were aligned using MAFFT 7 (Katoh and Standley, 2013) <–auto> and TrimAL <automated>. The maximum likelihood tree was inferred by IQ-Tree 1.6 software (Nguyen et al., 2015) with 1000 bootstrap replicates (Hoang et al., 2018) and Model Finder (Kalyaanamoorthy et al., 2017).
Results
Chloris virgata Showed the Fastest Growth Phenotype of the Major Mongolian Grassland Plants
To screen for plants harboring fast growth potential, 40 species of major Mongolian grasslands plants were collected from the northern forest zone, middle grass zone, and southern Gobi desert zone of the Mongolian territory. Additionally, the agricultural crops, rice and wheat, and the representative molecular biology experimental plant, Arabidopsis, were used as controls during seed screening. The seeds were sown on 1/2 MS medium containing 0.9% agarose and 1.5% sucrose and incubated at 4°C for 2 days for vernalization. The seed germination treatment was started by moving the seeds to 22°C under 16-h light/8-h dark conditions. The first appearance of shoot were defined as a starting of germination and growth phenotypes were observed using a stereoscopic microscope. The germination of wheat and rice was observed 1 and 2 days, respectively, after the start of germination treatment. Arabidopsis germinated after 2 days. Of the Mongolian grassland plants, Agropyron cristatum and Medicago spp. that were used as feed for livestock germinated in 1 or 2 days. In contrast with these well-known plants, the C. virgata DG accession was observed to germinate just 5 h after the start of germination treatment and was identified as the fastest germinating plant of the 43 screened seeds (Table 1). C. virgata is an annual monocot belonging to the Poaceae family. The English common name of C. virgata is “feather finger grass” and it is also called ‘bulgan suul’ in Mongolia, which means “tail of sable.”
The complete chloroplast genomes of 14 common species and two Chloris species provided an analysis of the maximum likelihood relationships between C. virgata and the other 16 species. All chloroplast genomes were downloaded from NCBI RefSeq and the GenBank database using the chloroplast genome of C. virgata. The dicots A. thaliana and Medicago sativa were used as outgroups. Phylogenetic analysis of the 14 common species and two Chloris species revealed that the Chloris species are most closely related to Zoysia grasses (Supplementary Figure 2).
Only 5 h after the start of germination treatment, a tiny shoot appeared at the bottom of the C. virgata seed, whereas no shoots or roots emerged from the wheat or rice seeds (Figure 1A). For a detailed analysis of germination potential, germination efficiency was analyzed to measure the rate of shoot or root formation in each period after the start of germination treatment (Figure 1B). In the 5 h stage, the germination rate of C. virgata seeds was approximately 60%, which is significantly faster than that of the other plants (Figure 1B). In contrast, wheat showed a 10% germination rate at 10 h, and rice showed about a 30% germination rate at 24 h (Figure 1B). Growth potential is analyzed not only using germination speed, but also the shoot formation of each plant. Consequently, the shoot phenotypes and lengths of the C. virgata, wheat, and rice shoots were analyzed. Shoots length per seed length was measured for 7 days after the start of germination (Figure 1C). The C. virgata shoots elongated to 200 and 380% of seed length within 24 and 48 h of starting the germination treatment, respectively. The wheat and rice seeds only started to form shoots 24 and 48 h after the start of germination treatment, respectively. C. virgata continued to show faster shoot growth than the wheat or rice for 7 DAG (Figure 1C).
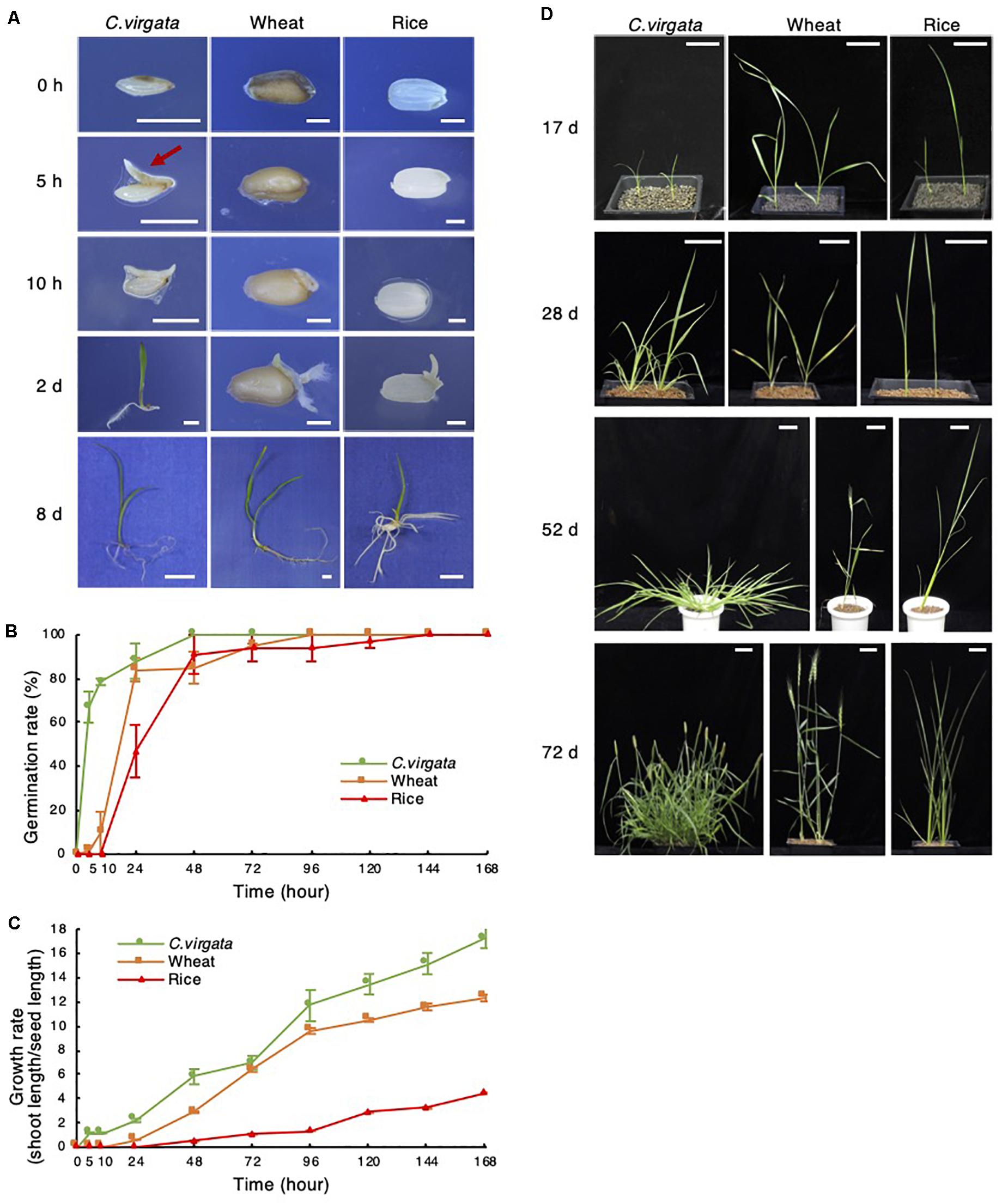
Figure 1. Chloris virgata possessed faster germination ability than wheat and rice. (A) Shoot and root formation in the germination stage of C. virgata, wheat, and rice. Each plant was germinated on 1/2 MS medium for 0, 5, 10 h, 2 days, or 10 days. Scale bars: 2 mm (0 and 10 h), 5 mm (2 days), and 1 cm (8 days). Seed germination rate (B) and growth rate (C) of C. virgata, wheat, and rice from 0 to 7 days after germination. Totally 27 plants were measured with three independent replicates. (D) Shoot and branch development from the early to young development stages. Each plant was germinated and grown on soil for 17, 28, 52, or 72 days. Scale bars: 5 cm.
To observe growth potential in later stages, the leaf and stem development of C. virgata, wheat, and rice were observed from the early young to adult development stages. Each plant seed was germinated on 1/2 MS medium for 7 days under 16-h light/8-h dark conditions, and the early young development plants were transplanted to soil and grown at 22°C under 16-h light/8-h dark conditions (Figure 1D). From 17 to 28 DAG, increased branch formation was observed in C. virgata. The active branch formation of C. virgata continued until day 52, and the branch number was higher than those of wheat or rice. C. virgata branch formation was observed until the seed formation stage at 72 days. The final branch number of C. virgata was found to be over 20 per plant, which is substantially more than the 3–5 branch formations in wheat and rice (Figure 1D).
Chloris virgata Showed a Strong Regrowth Phenotype
As nomadism is a traditional Mongolian way of life, many plant species growing on Mongolian grasslands are exposed to grazing. The nomadic culture in Mongolia dictates that C. virgata should preferably be eaten by sheep, but C. virgata showed regrowth after animal bite by Mongolian livestock. To investigate the regrowth potential of this species, the leaves and stems of C. virgata, wheat, and rice were cut to a height of 4 cm from the soil surface using scissors to mimick animal bite. The regrowth potential of the plants was observed for 2 weeks between cuttings. Initially, the 7-day-old C. virgata, wheat, and rice seedlings were transplanted to soil and grown for 2 weeks (Figure 2A). The first cutting treatments were then performed (Figure 2B, left). After the first cutting treatment, the leaves and stems of all the plants were regenerated and regrown over the next 2 weeks (Figure 2B, right). Then, the second cutting treatment was performed on the recovered plants (Figure 2C, left). After the second cutting, C. virgata and wheat showed better regrowth phenotypes, with higher branch numbers and shoot heights than the regrowth phenotypes after the first cutting treatment. The growth volume of rice after the first and second cutting treatments was less than those of C. virgata and wheat (Figure 2C, right). During the third to fifth regrowth period, C. virgata continued to regrow with the same shoot number and height as after the second cutting, but the regrowth shoot number and height of the wheat gradually decreased. Rice growth continued, but with low shoot numbers and heights (Supplementary Figures 1A–C, left). After the sixth cutting, the shoots of C. virgata still fully regrew, without any visible growth depression. In contrast, wheat regrowth declined after the sixth cutting, and the number and height of the shoots drastically decreased (Figure 2D, right). These results suggest that C. virgata has high regrowth ability against grazing, compared to the common commercial crops, wheat and rice.
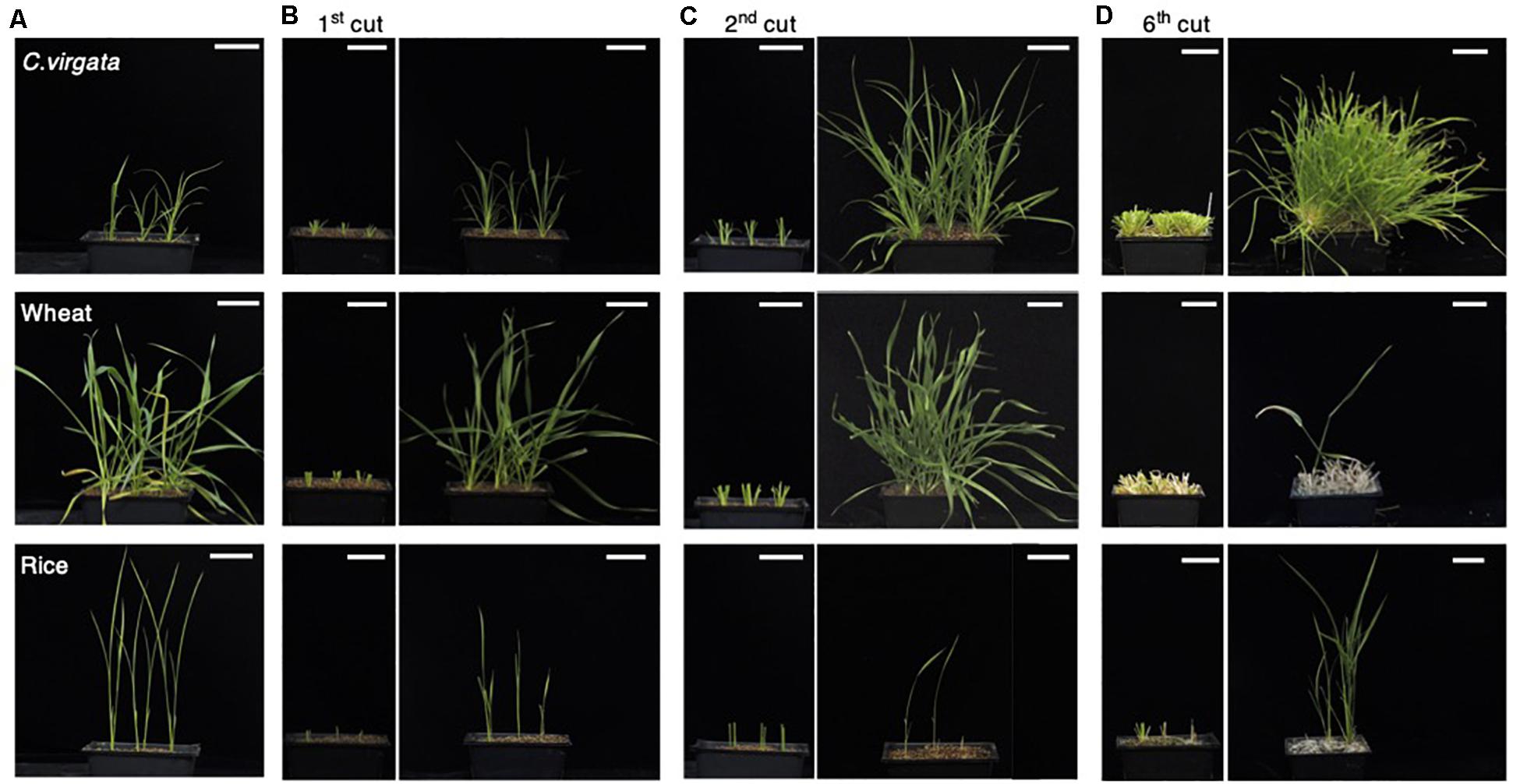
Figure 2. Chloris virgata possessed higher regeneration ability after shoot cutting than wheat and rice. (A) C. virgata, rice, and wheat were grown on soil for 2 weeks. (B) The shoots of the two-week-old plants were cut and removed (left). The shoots were regrown for 2 weeks after the first cutting treatment (right). (C) Regrown shoots after the first cutting treatment were recut a second time (left). The plants were regrown for 2 weeks after the second cutting treatment (right). (D) Remaining shoots after the sixth cutting treatment (left) and the regrown shoots after 2 weeks (right). Scale bars: 5 cm.
De novo Transcriptome Assembly and Functional Annotation of Chloris virgata
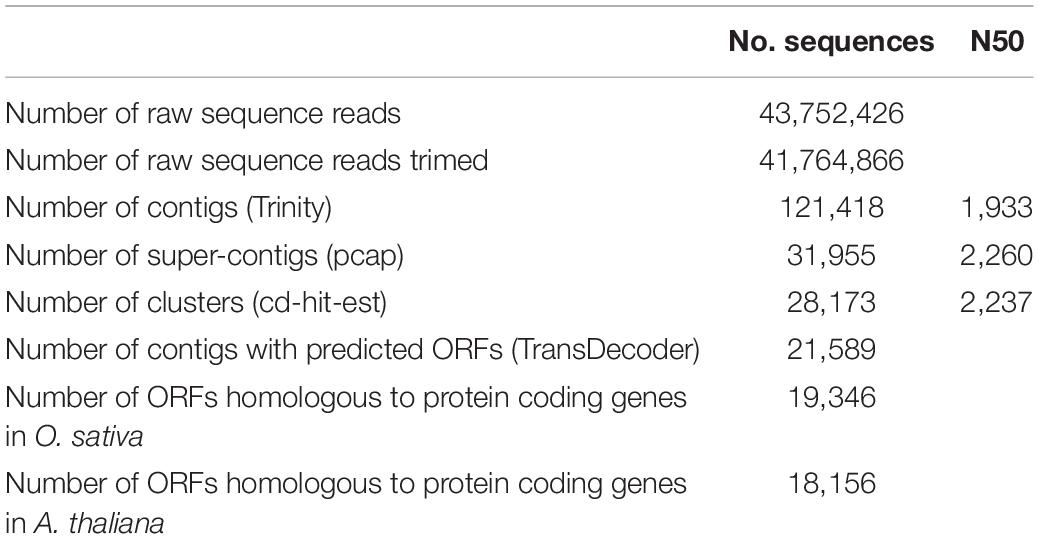
To analyze the molecular mechanisms of C. virgata growth activities, the total RNA extracted from samples of four independent growth stages were pooled and then subjected to Illumina RNA sequencing and de novo transcriptome assembly. A total of 43,752,426 raw sequence reads were generated from the C. virgata cDNA, and 41,764,866 filtered reads (Table 2). The quality-checked reads were assembled de novo using the Trinity program (Grabherr et al., 2011), resulting in 121,418 contigs with an N50 value of 1933 bp. These were assembled into 31,955 super-contigs with the PCAP program and clustered using the CD-HIT-EST program, resulting in 28,173 transcript clusters. The longest transcripts of the clusters were then subjected to open reading frames, resulting in 21,589 protein-coding transcripts, which were used in our downstream analyses as the reference transcript dataset. In the reference transcripts, 19,346 and 18,156 transcripts were homologous to protein-coding genes in O. sativa and Arabidopsis, respectively (Table 2).
Functional Annotation of Expressed Genes in Chloris virgata
The reference transcripts derived from the C. virgata transcriptome assembly were assessed using a homology search and GO annotation. The homology search was conducted against the NCBI non-redundant protein database for the reference transcripts, using the BLASTx program, and the highest hits were linked to the reference transcripts. This resulted in an aligned sequence homology of 75.5% with the NCBI-nr database. The data were further subjected to GO-based functional classification in OmicsBox software, to illustrate the annotated gene profiles in 60 GO categories related to biological process (20), molecular function (20), and cellular component (20) (Figure 3). Among the biological process categories, RNA metabolic process, cellular protein modification process, and regulation of cellular macromolecule biosynthesis were ranked the highest. Within the molecular function category, binding functions, including sequence-specific DNA binding, adenyl nucleotide binding, and double-stranded DNA binding, were ranked the highest. In addition, in the cellular component category, the chloroplast envelope, microtubule cytoskeleton, and nuclear chromosome were ranked the highest (Figure 3).
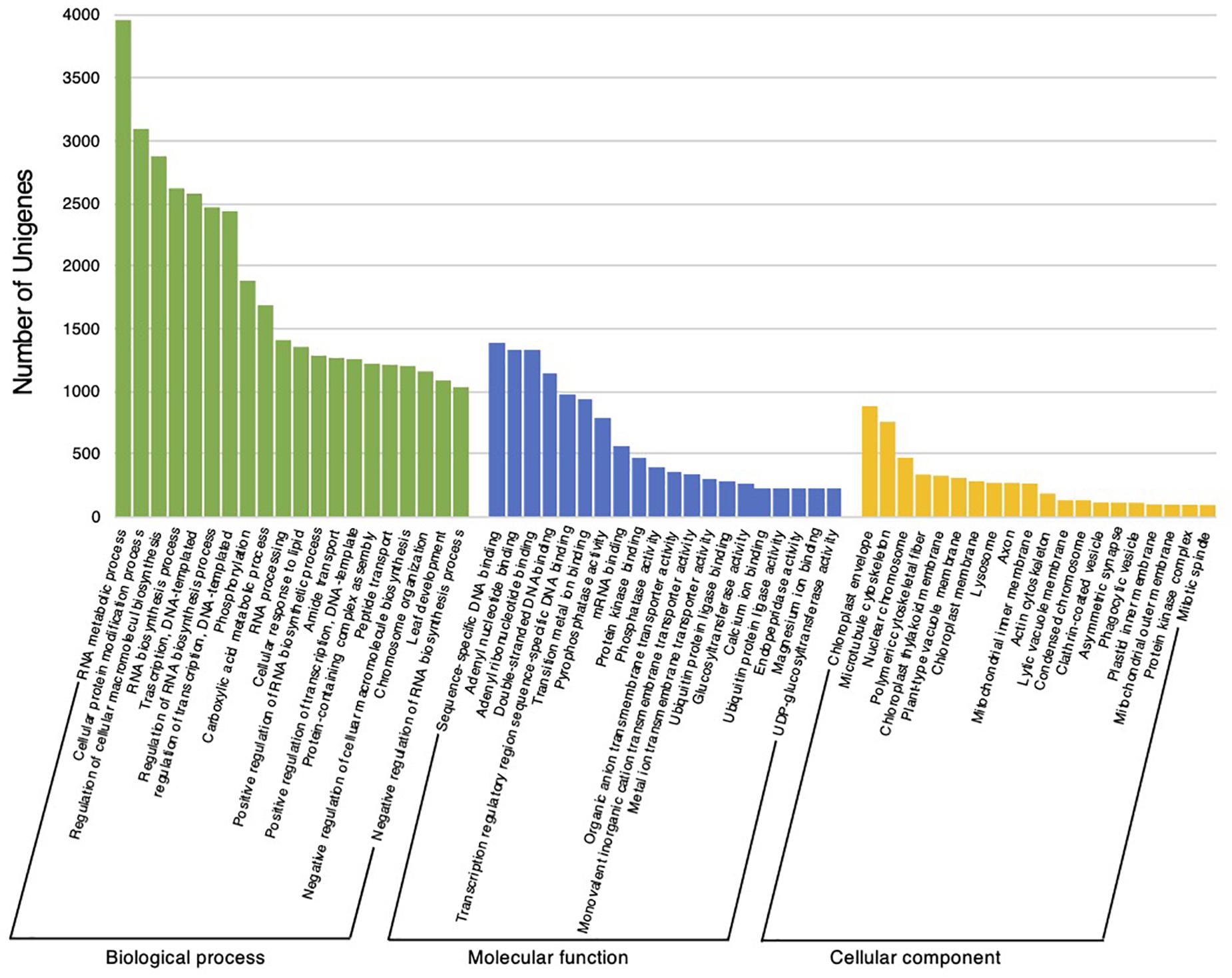
Figure 3. Functional classification of Chloris virgata assembled unigenes. Gene ontology terms for all unigenes sequences were assigned using a BLASTx search. The results were summarized into the following categories: biological process, molecular function, and cellular component.
Gene Expression Profiles in the Juvenile Development Stage of Chloris virgata
To investigate the molecular mechanism underlying the high growth performance of C. virgata (Figure 1), we compared the transcriptomes of samples from whole organs at the germination stage (2 DAG), early young development stage (8 DAG), young development stage (17 DAG), and adult development stage (28 DAG) (Figure 1D). Mapping the RNA-seq reads obtained by Ion Torrent sequencing to the reference transcripts, we illustrated a comprehensive gene expression map along with juvenile development in C. virgata. To further understand the expression changes in the four different developmental stages, we conducted GO enrichment analysis to assess overrepresented functions in the DEGs found in our pairwise comparisons of all possible sample combinations (Figure 3 and Table 2). We found that 85, 392, 77, and 31 GO terms in the biological process category were enriched (FDR < 0.03) in the germination stage, early young development stage, young development stage, and adult development stage, respectively (Figures 4–7 and Supplementary Data Sheets).
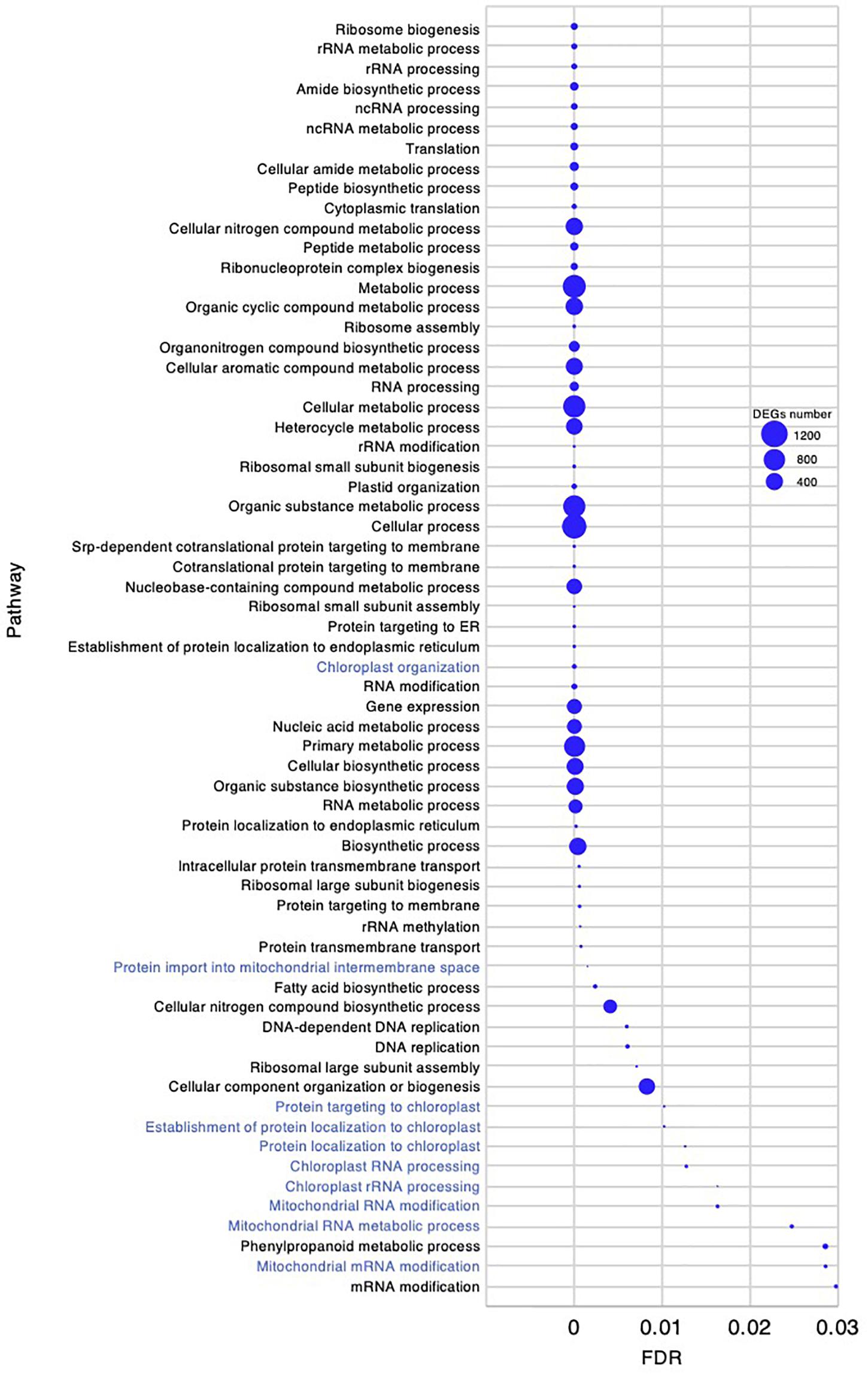
Figure 4. Gene ontology enrichment analysis in the germination stage of Chloris virgata. Gene ontology terms of differentially expressed genes (DEGs) enriched in the germination stage (2 days after germination), compared with the early young development stage (8 days after germination) of C. virgata grown on 1/2 MS medium. Gene ontology terms were selected from the biological process category. A false discovery rate (FDR) of 0.03 was used as the threshold. Blue circles show the number of DEGs in each Gene ontology term.
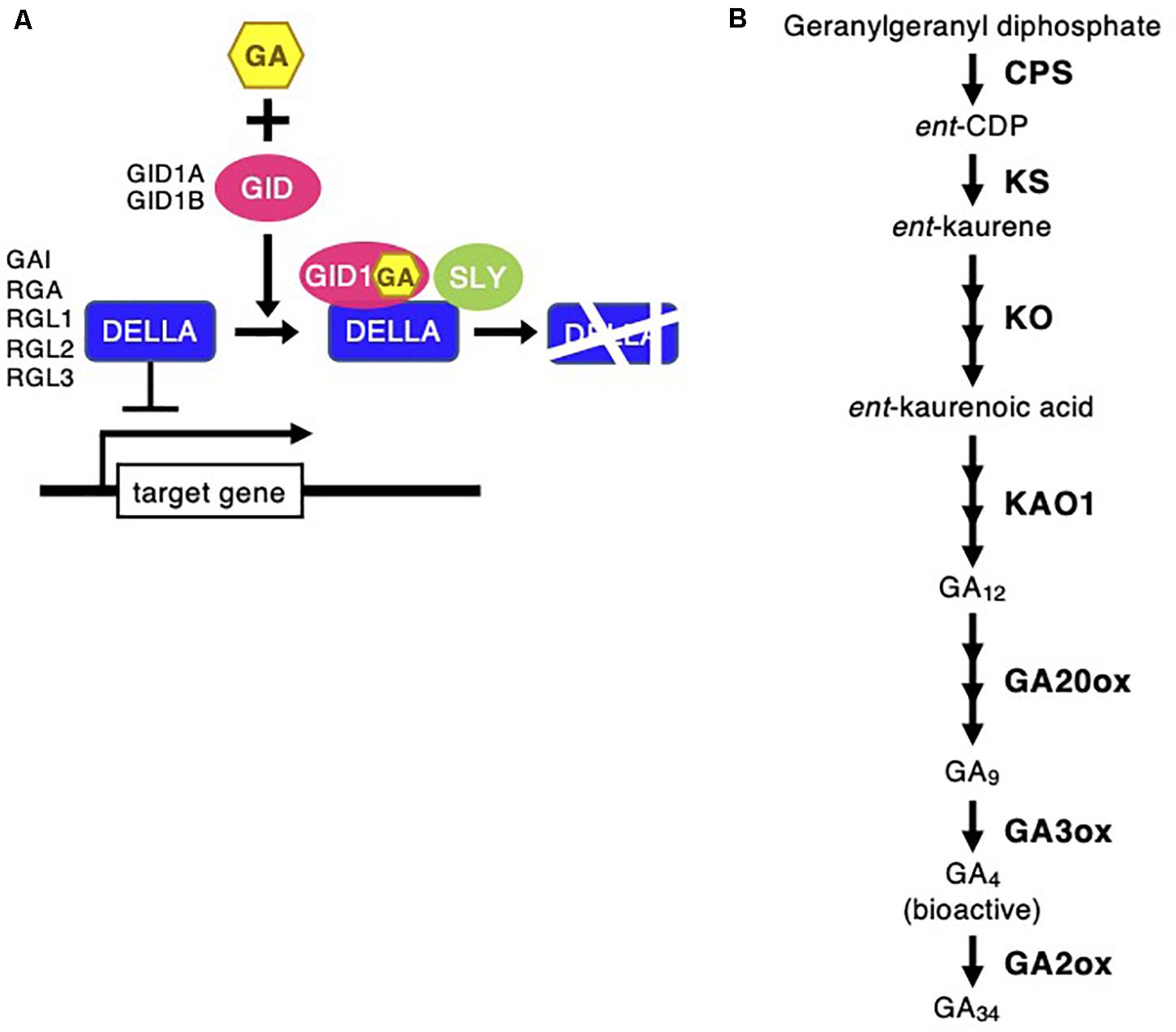
Figure 5. Gibberellin signaling and biosynthesis pathway following Chloris virgata. (A) The possible GA signaling pathway in C. virgata. GID, gibberellin-insensitive dwarf; GAI, gibberellic acid insensitive; RGA, repressor of GA; RGL, RGA-like; SLY, sleepy. (B) GA biosynthesis pathway in C. virgata. GAs are products of diterpenoid pathway and their formation from GGDP to bioactive GAs such as GA4. CPS, ent-copalyl diphosphate synthase; GGDP, geranylgeranyl diphosphate; ent-CDP, ent-copalyl diphosphate; KS, ent-kaurene synthase; KO, ent-kaurene oxidase; KAO, ent-kaurenoic acid oxidase. The arrows indicate each steps.
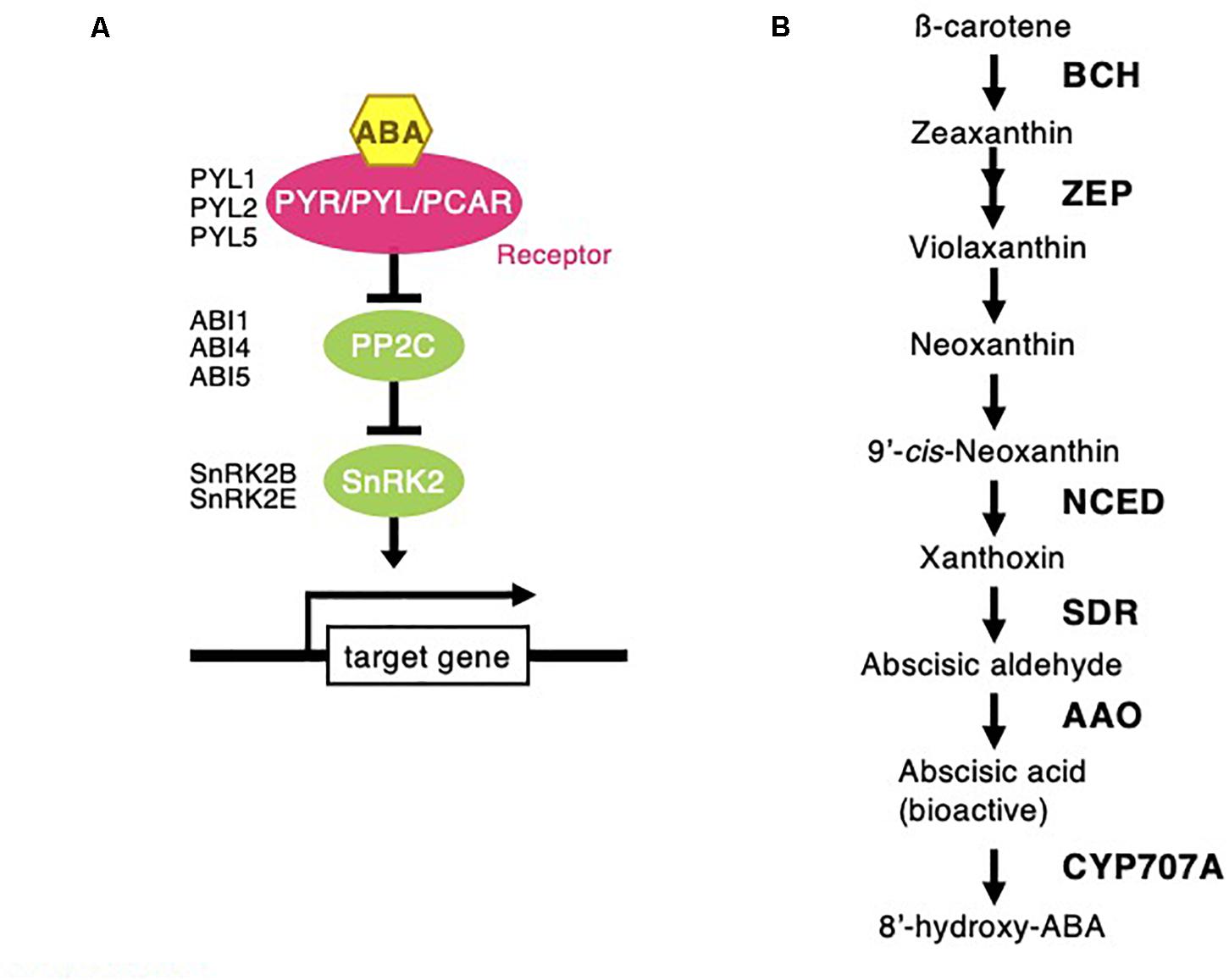
Figure 6. Abscisic acid signaling and biosynthesis pathway following Chloris virgata. (A) The possible ABA signaling pathway in C. virgata. PYL, pyrabactin resistance like; ABI, ABA insensitive; SnRK2, sucrose non-fermenting 1 related protein kinase. (B) ABA biosynthesis in C. virgata. ABA synthesis start form carotene by oxidative cleavage reaction. BCH, beta carotenoid hydroxylase; ZEP, zeaxanthin epoxidase; NCED, 9-cis-epoxycratenoid dioxygenase; ABA2, ABA deficient 2; AAO, abscisic aldehyde oxidase. The arrows indicate each steps.
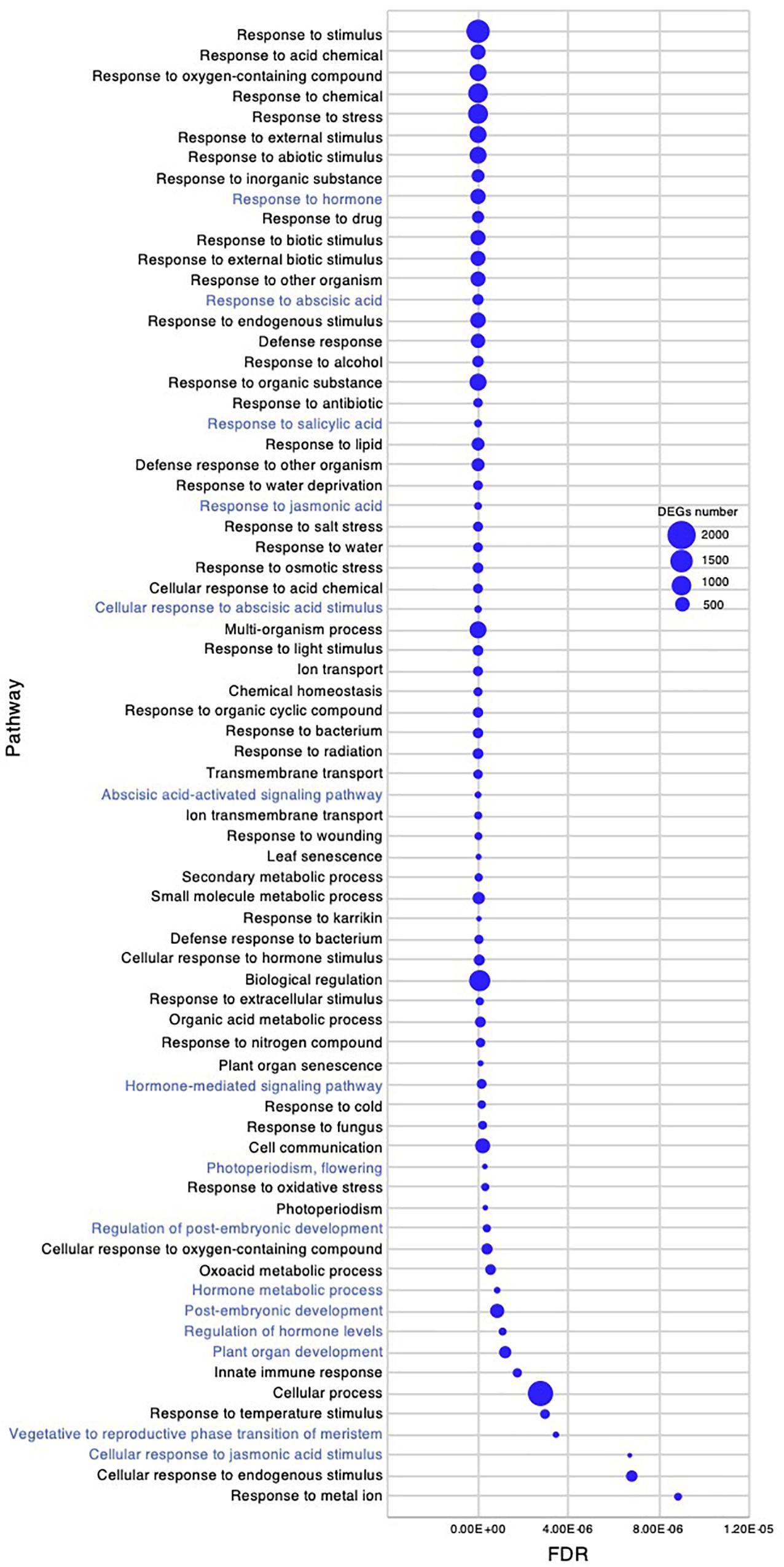
Figure 7. Gene ontology enrichment analysis in the early young development stage of Chloris virgata. Gene ontology terms of DEGs enriched in the young development stage, compared with the germination stage of C. virgata. False discovery rates were lower than 1.20E-05 and blue circles show gene number.
Identification and Quantification of Phytohormone Signaling and Biosynthesis Homologous Genes of Chloris virgata
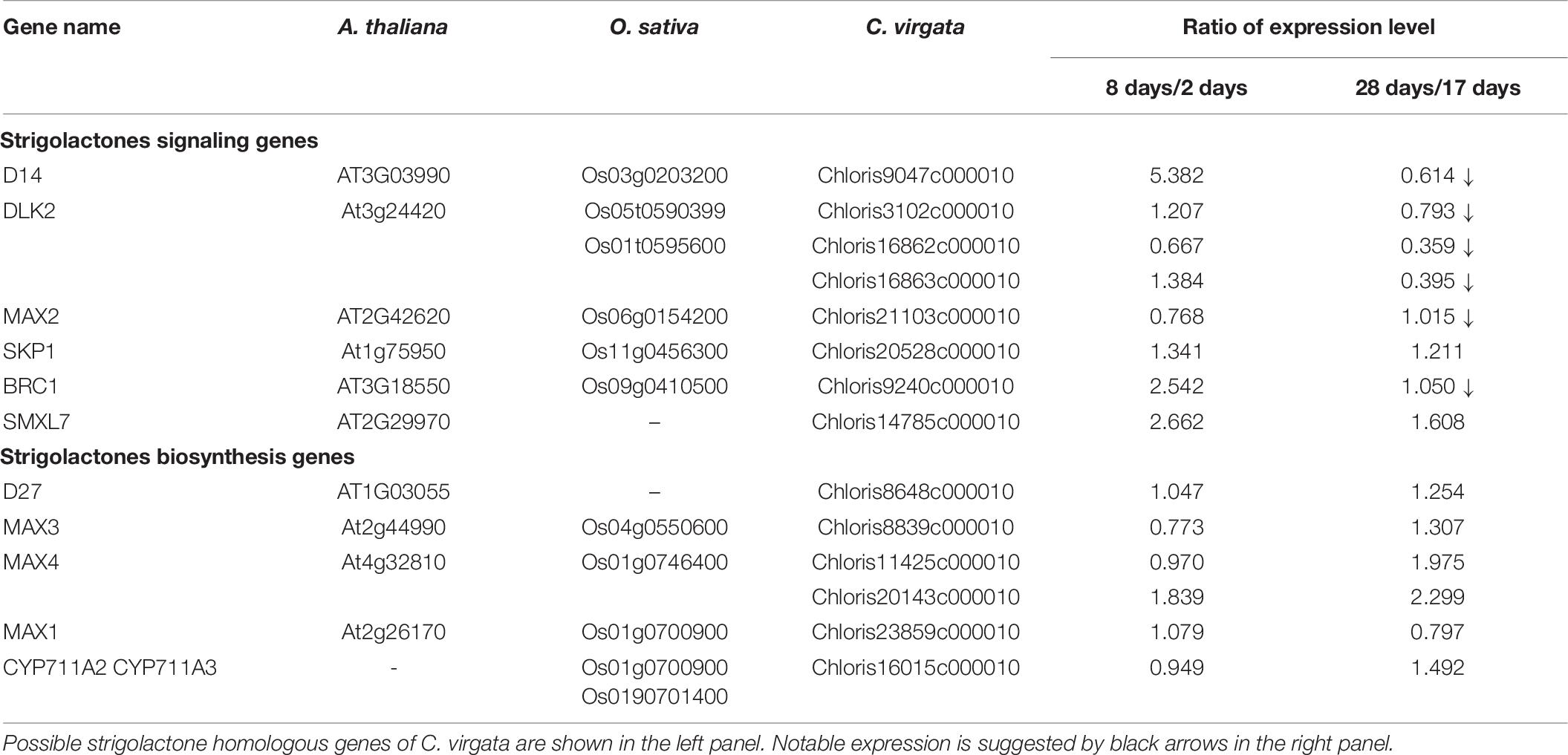
Phytohormones play an important role in plant development. Gibberellic acid (GA) activates and abscisic acid (ABA) suppresses plant germination. Brassinosteroids (BRs) promote shoot growth in the early young development stage and strigolactones (SLs) suppress branch formation in the adult development stage. To analyze the possible regulatory mechanisms of active germination, homologous genes of GA, ABA, BR, and SL signaling and biosynthesis were identified from the C. virgata unigenes database with BLASTx, using the phytohormone genes in Arabidopsis and rice. The expression levels of the identified GA, ABA, BR, and SL signaling and biosynthesis genes of C. virgata in the four independent developmental stages were analyzed using the Ion Proton Sequencer. Expression abundance was quantified based on the read count data computed using the featureCounts program and evaluated using the RPM values. The ratios of the expression levels of each gene in each development stage were calculated.
Functional Classification of Highly Expressed Transcripts and Expressed Genes Related to GA and ABA Functioning in the Germination Stage of Chloris virgata
Chloris virgata showed faster germination performance than the other Mongolian grassland plants (Table 1), wheat, or rice (Figures 1A,B). To investigate gene expression that may be related to the germination performance of C. virgata (Table 1), we examined the GO terms enriched in the germination stage. We found 85 enriched categories of biological processes with FDR < 0.03 (Supplementary Data 1). As there were too many enriched GO categories to present in one figure, the 64 categories with more than 6 DEGs are presented in Figure 4, while those with fewer DEGs were removed. Several GO terms related to chloroplasts were enriched, including “chloroplast organization,” “protein targeting to chloroplast,” “establishment of protein localization to chloroplast,” “protein localization to chloroplast,” “chloroplast RNA processing,” and “chloroplast rRNA processing.” The mitochondria-related GO categories “protein import into mitochondrial intermembrane space,” “mitochondrial RNA modification,” “mitochondrial RNA metabolic process,” and “mitochondrial mRNA modification” were also enriched in this stage.
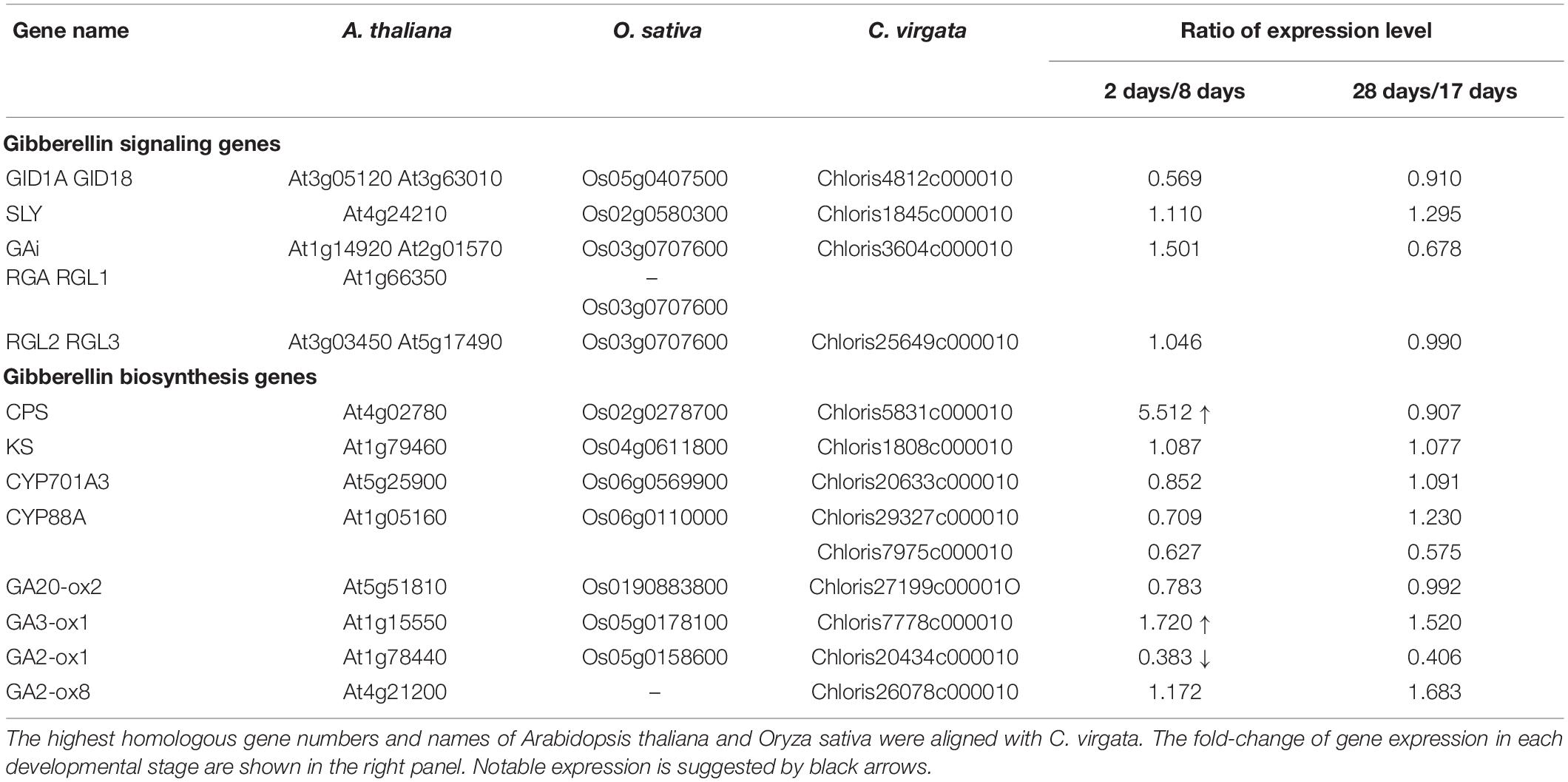
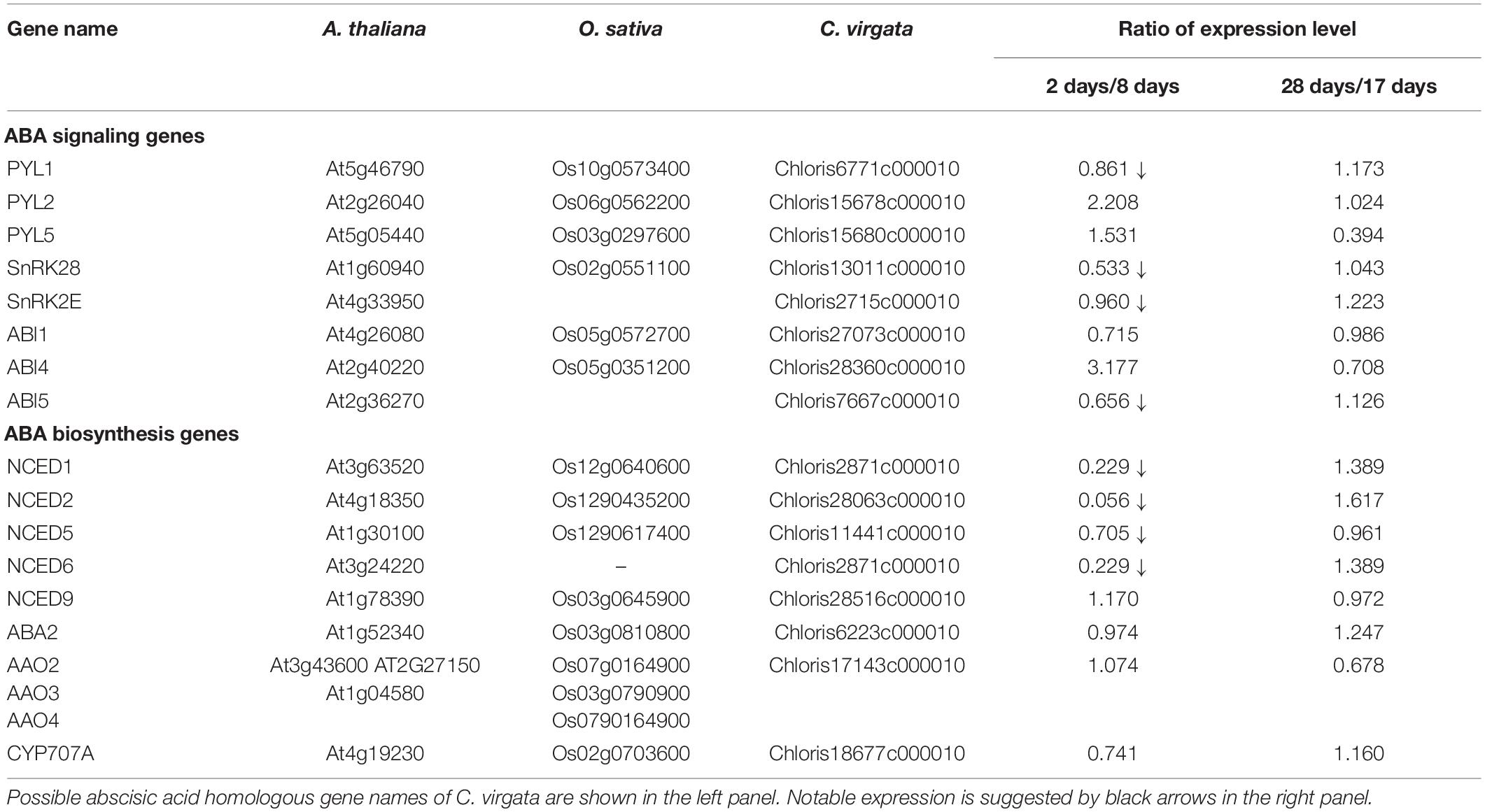
Phytohormones, GAs, and ABA antagonistically mediate several plant developmental processes, including seed maturation, seed dormancy and germination, primary root growth, and leaf development (Finkelstein et al., 2008). In particular, GA positively regulates seed germination, whereas ABA suppresses seed germination (Rajjou et al., 2012). To investigate the possible regulation of the germination stage of C. virgata by GA and ABA, the signaling and biosynthesis genes of GA and ABA in the Arabidopsis and rice genomes were used to identify homologous genes in the C. virgata unigenes database. Therefore, we examined the expression profiles of genes that are linked to GA and ABA signaling and biosynthesis in rice and Arabidopsis in the germination to early young development stage and in the young development to adult development stage of C. virgata (Tables 3, 4 and Supplementary Tables 1, 2). GA biosynthesis starts from the diterpenoid pathway and its formation is initiated by terpenoid cyclase (CPS) (Sun and Kamiya, 1994), and gibberellin 3-oxidase 1 (GA3-ox1). GA3-ox1 is involved in the final step of GA biosynthesis, when the active forms GA4 and GA1 are synthesized (Talon et al., 1990). Gibberrellin 2-beta-dioxygenase 1 (GA2-ox1) is metabolized from the active forms (GA4 and GA1) to inactivated GA34 and GA8 (Thomas et al., 1999; Figure 5). CPS (Chloris5831c000010) and GA3-ox1 (Chloris7778c000010) homologous genes in C. virgata were significantly upregulated and GA2-ox1 (Chloris20434c000010) homologs in C. virgata were downregulated during the germination stage (Table 3). In Arabidopsis, there are several protein families known as ABA receptor components, including pyrabactin resistance1 (PYR1) and pyrabactin resistance1-like proteins (PYL1) (Park et al., 2009). The plant-specific protein sucrose non-fermenting 1-related protein kinase 2 (SnRK2), mediates protein phosphorylation and the activation of the ABA signaling pathway. There are 10 members of SnRK2 in Arabidopsis and rice, including SnRK2B and SnRK2E (Boudsocq et al., 2004; Kobayashi et al., 2004). ABA insensitive 5 (ABI5) is a basic leucine zipper transcription factor that generally activates ABA signaling (Yu et al., 2015; Figure 6). The expression of the homologous genes of the ABA receptor component PYL1 (Chloris6771c000010), signal transduction genes SnRK2B (Chloris13011c000010) and SnRK2E (Chloris2715c000010), and transcription factor ABI5 (Chloris7667c000010) were downregulated in C. virgata during the germination stage (Table 4). In the first step in the ABA biosynthesis pathway, ABA is derived from carotene by an oxidative cleavage reaction involving 9-cis-epoxycarotenoid dioxygenase (NCED) in plastids (Schwartz et al., 1997; Iuchi et al., 2001; Figure 6). Expression of NCED1 (Chloris2871c000010), NCED2 (Chloris28063c000010), NCED5 (Chloris11441c000010), and NCED6 (Chloris2871c000010) was downregulated in the germination stage of C. virgata (Table 4).
Functional Classification of Highly Expressed Transcripts and Expressed Genes Related to Brassinosteroid Regulation in the Early Young Development Stage of Chloris virgata
Chloris virgata showed high growth performance with active elongation of shoots in the early young development stage, 7–8 DAG (Figures 1A,C). To investigate gene expression that may be related to the early young development stage of C. virgata, we examined enriched GO terms. The number of enriched GO categories in this stage was substantially higher than that in the other three development stages. A total of 392 enriched biological process categories with FDR < 0.03 were identified (Supplementary Data 2). As there were too many enriched GO categories to present in the figure, the 72 GO categories with FDR < 1.20E-05 and DEGs greater than 80 are presented in Figure 7. In the biological process category, several GO terms related to organ development were identified, including “photoperiodism, flowering,” “regulation of post-embryonic development,” “post-embryonic development,” “plant organ development,” and “vegetative to reproductive phase transition of meristem.” In the phytohormone-related GO categories, “response to hormone,” “response to ABA,” “response to ABA,” “response to salicylic acid,” “response to jasmonic acid,” “cellular response to ABA stimulus,” “ABA-activated signaling pathway,” “hormone-mediated signaling pathway,” “hormone metabolic process,” “regulation of hormone levels,” and “cellular response to jasmonic acid stimulus” were also enriched in this stage.
Brassinosteroids are phytohormones that are also categorized in a class of steroid hormones that are widely conserved from animals to plants. BRs regulate cell differentiation, cell division, and cell elongation, which together regulate the development and growth of plant leaves, stems, and roots in the young development stage (Nakano and Asami, 2014). To analyze the possible regulatory mechanisms that activate the development and growth of C. virgata by BR, the signaling and biosynthesis genes of BR in the Arabidopsis and rice genomes were used to identify homologous genes in the C. virgata unigenes database. Then, we examined the expression profiles of these genes in the four development stages (Table 5 and Supplementary Table 3).
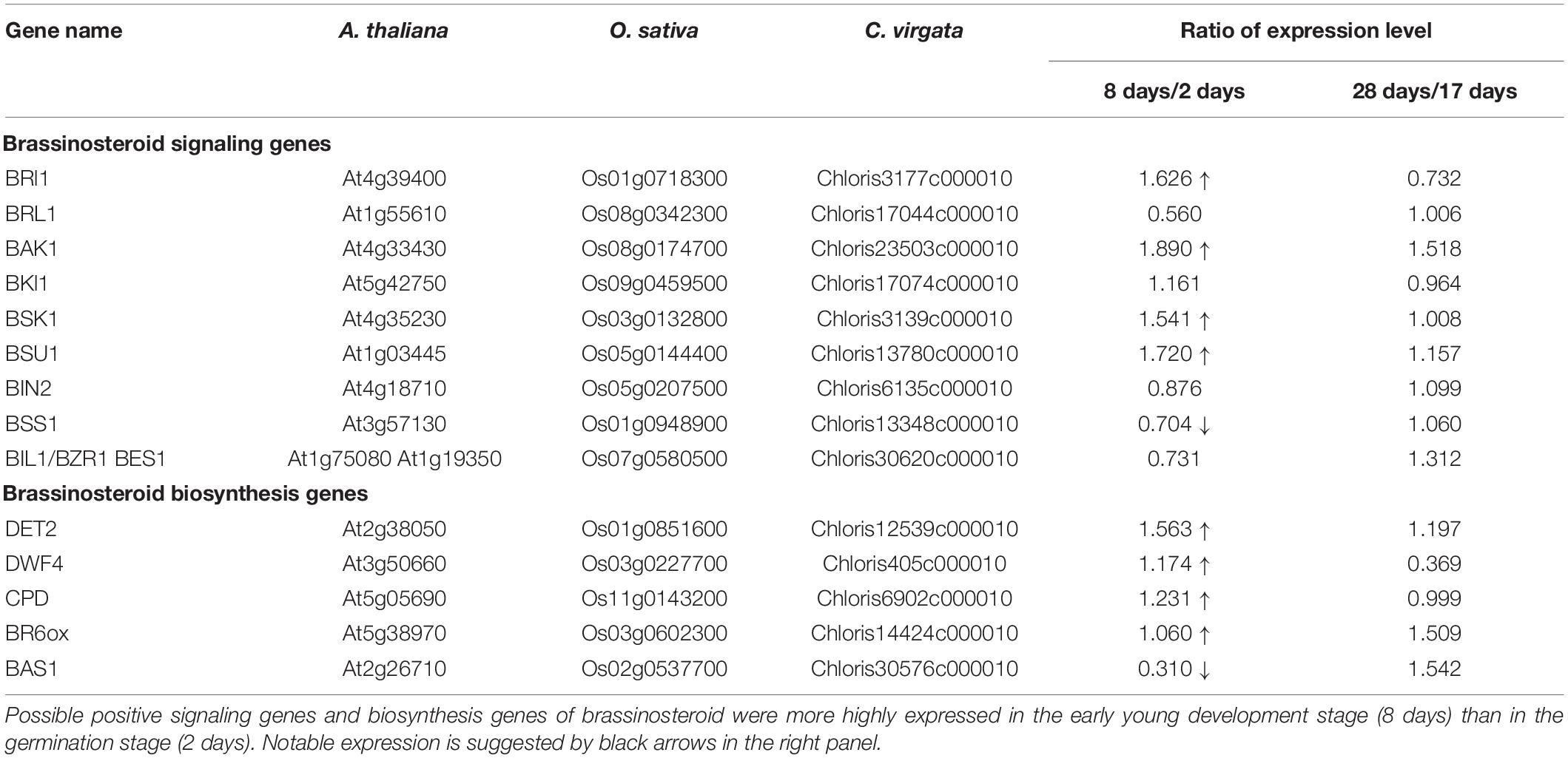
Brassinosteroids are accepted by plasma membrane protein complexes formed by the BR-type transmembrane receptor kinase BR insensitive 1 (BRI1) (Li and Chory, 1997), BRI1-associated receptor kinase (BAK1) (Li et al., 2002), and membrane-associated BR-signaling kinase 1 (BSK1) (Tang et al., 2008) as positive factors for BR signaling (Figure 8). The expression of these three positive BR receptor complex protein homologs in C. virgata (BRI: Chloris3177c000010, BAK1: Chloris23503c000010, and BSK1: Chloris3139c000010) was upregulated in the early young development stage of C. virgata (Table 5). In the middle stream of BR signaling, BRI1 suppressor 1 (BSU1) is a positive phosphatase (Mora-García et al., 2004) and brassinozole-sensitive-short hupocotyl 1 (BSS1) is a negative regulator of the BR master transcription factor brassinozole-insensitive-long hupocotyl 1 (BIL1) (Jun et al., 2010; Figure 8). Expression of the C. virgata BSU1 homolog (Chloris13780c000010) was upregulated and that of the BSS1 homolog (Chloris13348c000010) was downregulated at this stage. BR biosynthesis plays an important role in the activation of BR signaling (Table 5).
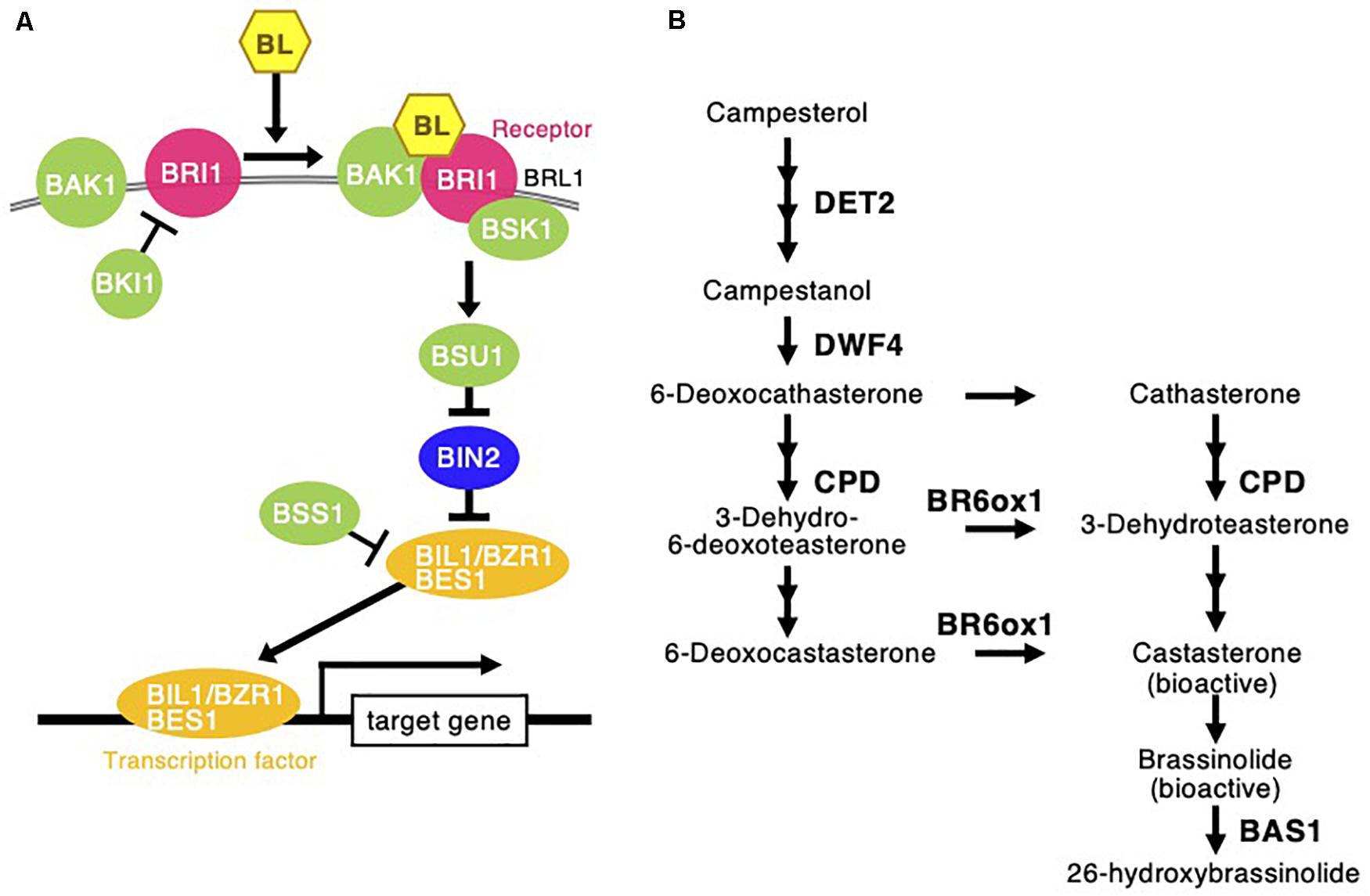
Figure 8. Brassinosteroid signaling and biosynthesis pathway following Chloris virgata. (A) The possible BR signaling transduction in C. virgata. BAK1, BRI1 associated receptor kinase; BRI1, BR insensitive 1; BRL1, BRI1 like; BSK1, BR signaling kinase 1; BKI1, BRI1 kinase inhibitor 1; BSU1, BRI1 suppressor 1; BIN2, BR insensitive 2; BSS1, blade on petiole 1; BES1, brassinazole resistant 2; BZR1, brassinazole resistant 1. (B) BR biosynthesis early C-22 oxidation and late C-6 oxidation pathway in C. virgata. DET2, de-etiolated 2; DWF4, dwarf 4; CPD, constitutive photomorphogenic dwarf; BR6ox, BR 6-oxidase; BL, brassinolide; BAS1, phyb activation tagged suppressor 1. The arrows indicate each steps.
De-etiolated 2 (DET2) is a homolog of steroid 5 α-reductase in humans. It works upstream of BR biosynthesis (Li et al., 1996). Dwarf 4 (DWF4) (Choe et al., 1998) and constitutive photomorphogenesis and dwarfism (CPD) (Szekeres et al., 1996) are major P450 oxidase in the intermediate step of BR biosynthesis. BR C-6 oxidase (CYP85A1) is also P450 oxidase in the last step of biosynthesis (Shimada et al., 2001). PhyB activation-tagged suppressor 1 (BAS1) metabolizes active brassinolide to non-active 26-hydroxybrassinolide (Neff et al., 1999; Figure 8). Expression of the C. virgata homologous genes of DET2 (Chloris12539c000010), DWF4 (Chloris405c000010), CPD (Chloris6902c000010), and BR6OX (Chloris14424c000010) were upregulated and that of BAS1 (Chloris30576c000010) was downregulated in the early young development stage of C. virgata (Table 5).
Functional Classification of Highly Expressed Transcripts and Expressed Genes Related to Strigolactone Functioning in the Adult Development Stage of Chloris virgata
The active branch formation of C. virgata started from the young development stage and continued throughout the adult development stage to the flowering stage. Increased branch formation during animal bite-mimicking cutting experiment was also observed (Figures 1D, 2). These observations suggest that C. virgata possesses a high branch formation potential. To investigate gene expression that may be related to high branch formation in C. virgata, we examined the GO terms in the young and adult developmental stages. A total of 77 biological process GO categories were enriched in the young development stage (FDR < 0.03) (Supplementary Data 3). As there were too many enriched GO categories in the young development stage to present in the figure, the 67 categories with FDR < 0.03 and DEGs > 10 are presented in Figure 9, including “cellular response to SL.”
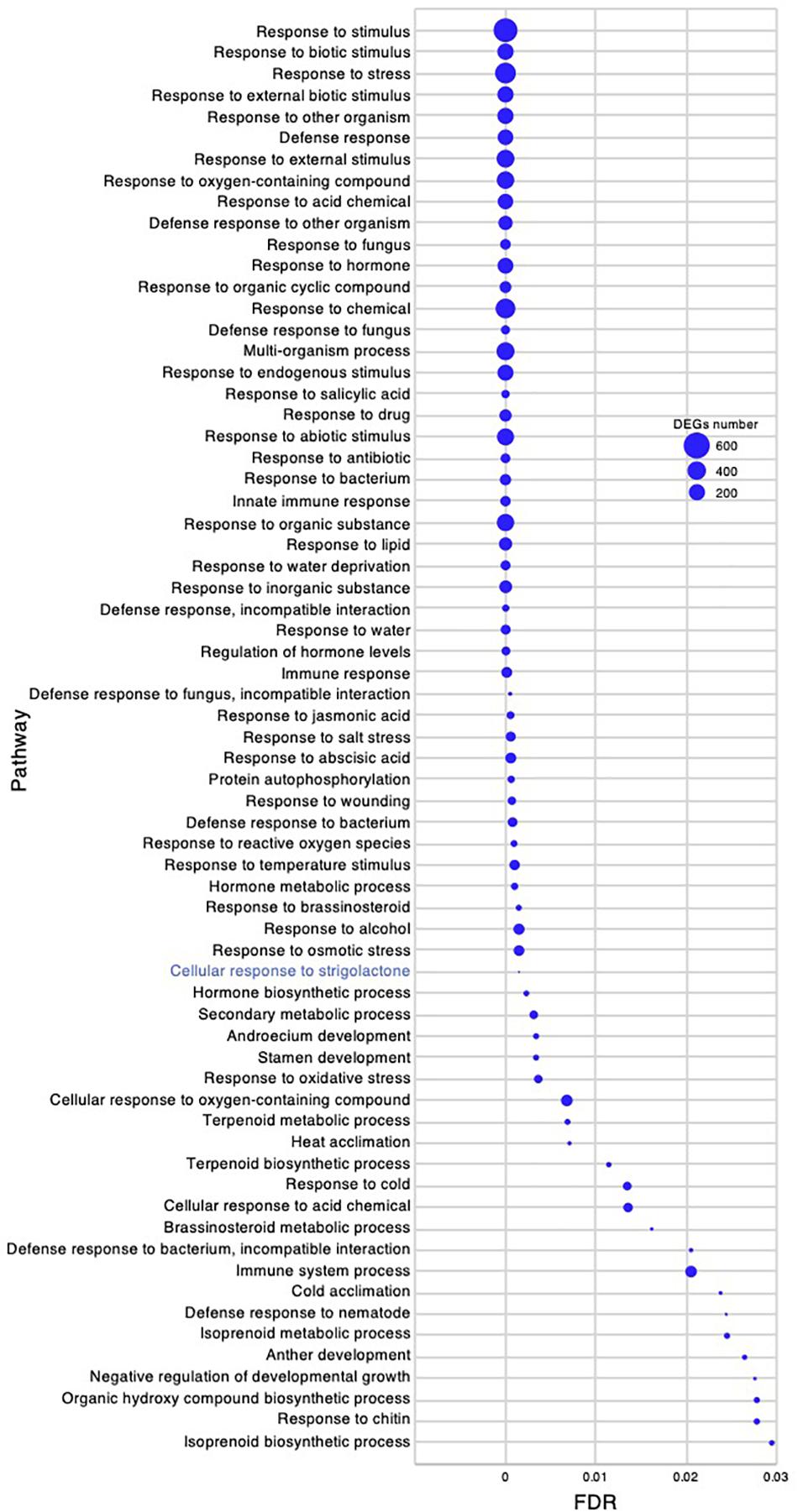
Figure 9. Gene ontology enrichment analysis in the young development stage of Chloris virgata grown on soil. Gene ontology terms of DEGs enriched in the young development stage (17 days after germination), compared with the adult development stage (28 days after germination) of C. virgata. False discovery rates were lower than 0.03 and blue circles show gene number.
In the adult development stage, 31 biological processes GO categories were enriched and had FDR < 0.03 (Supplementary Data 4). As there were fewer enriched GO categories than at the other stages, all 31 are presented in Figure 10. Of the enriched categories, the response to abiotic stimulus-related GO terms could be characteristically identified as “response to karrikin,” “response to red light,” “cellular response to far red light,” “cellular response to red light,” and “photomorphogenesis.”
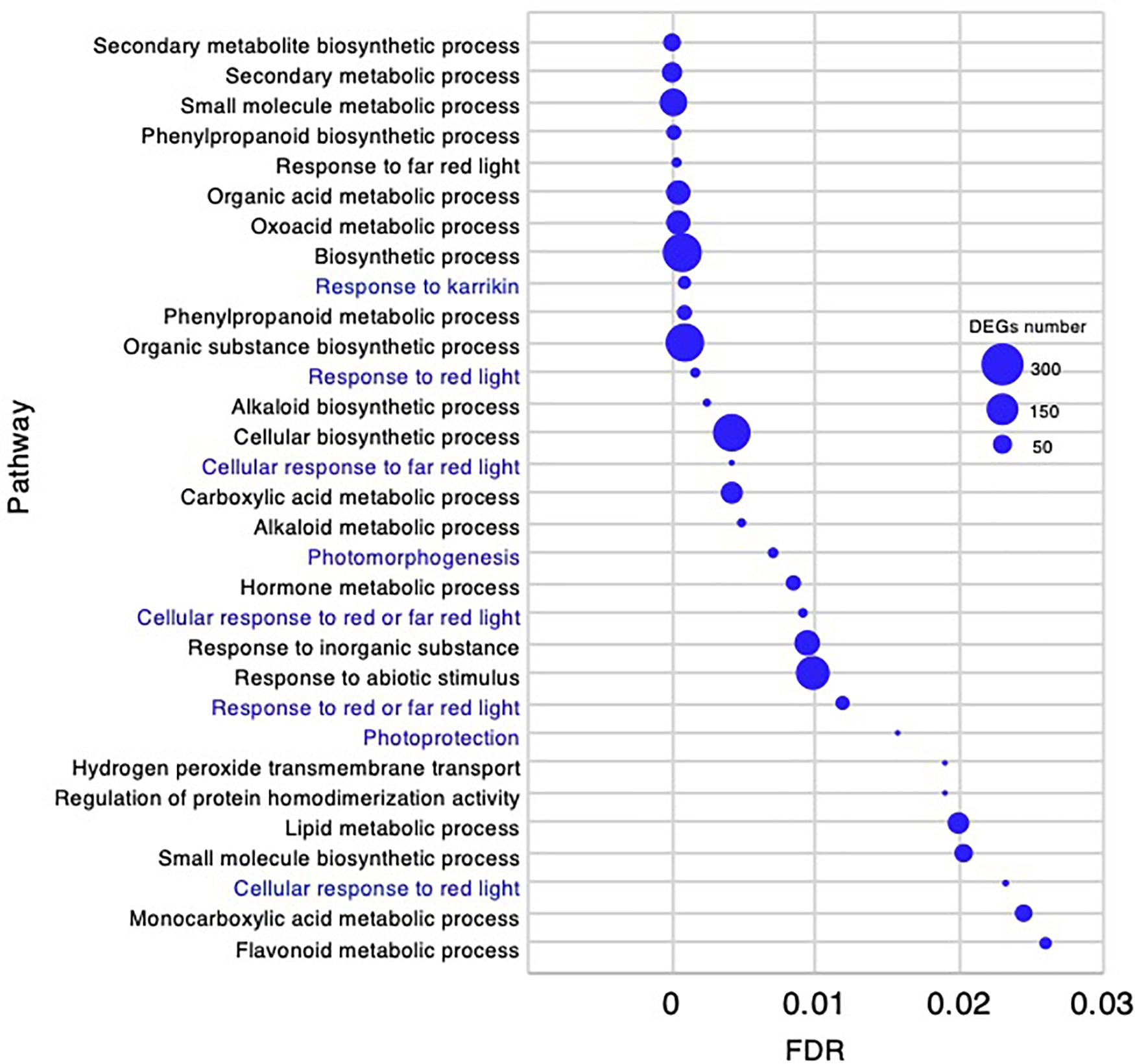
Figure 10. Gene ontology enrichment analysis in the adult development stage of Chloris virgata grown on soil. Gene ontology terms of DEGs enriched in the adult development stage compared with the young development stage of C. virgata. False discovery rates were lower than 0.03 and blue circles show gene number.
Strigolactones are phytohormones known to suppress plant branch formation (Umehara et al., 2008), and biosynthesis-deficient mutants have shown an increased number of tillers in rice (Ishikawa et al., 2005). To analyze the possible SL-related regulatory mechanism for the high branch formation of C. virgata, the signaling and biosynthesis genes of SLs in Arabidopsis and rice were used to identify homologous genes in the C. virgata unigenes database and the expression level of each gene in each development stages were calculated (Supplementary Table 4). DWARF14 (D14) has been identified as a receptor of SLs in Arabidopsis and rice (Arite et al., 2009), and DWARF14-LIKE2 (DLK2) also acts as an SL receptor. MORE AXILLARY GROWTH 2 (MAX2) forms a protein complex with D14 as a core signaling factor in the SL signaling pathway (Stirnberg et al., 2002; Végh et al., 2017). BRANCHED1 (BRC1) is a transcription factor that acts as a suppressor of branching (Aguilar-Martínez et al., 2007; Finlayson, 2007). Knock out of the SL receptors, MAX2 and BRC1, causes drastic promotion of branch formation in plants (Stirnberg et al., 2002; Aguilar-Martínez et al., 2007; Figure 11). The expression of the C. virgata homologous genes of D14 (Chloris9047c000010), DLK2 (Chloris3102c000010; Chloris16862c000010; Chloris16863c00010), MAX2 (Chloris21103c000010), and BRC1 (Chloris9240c000010) were downregulated in the adult developmental stage of C. virgata (Table 6).
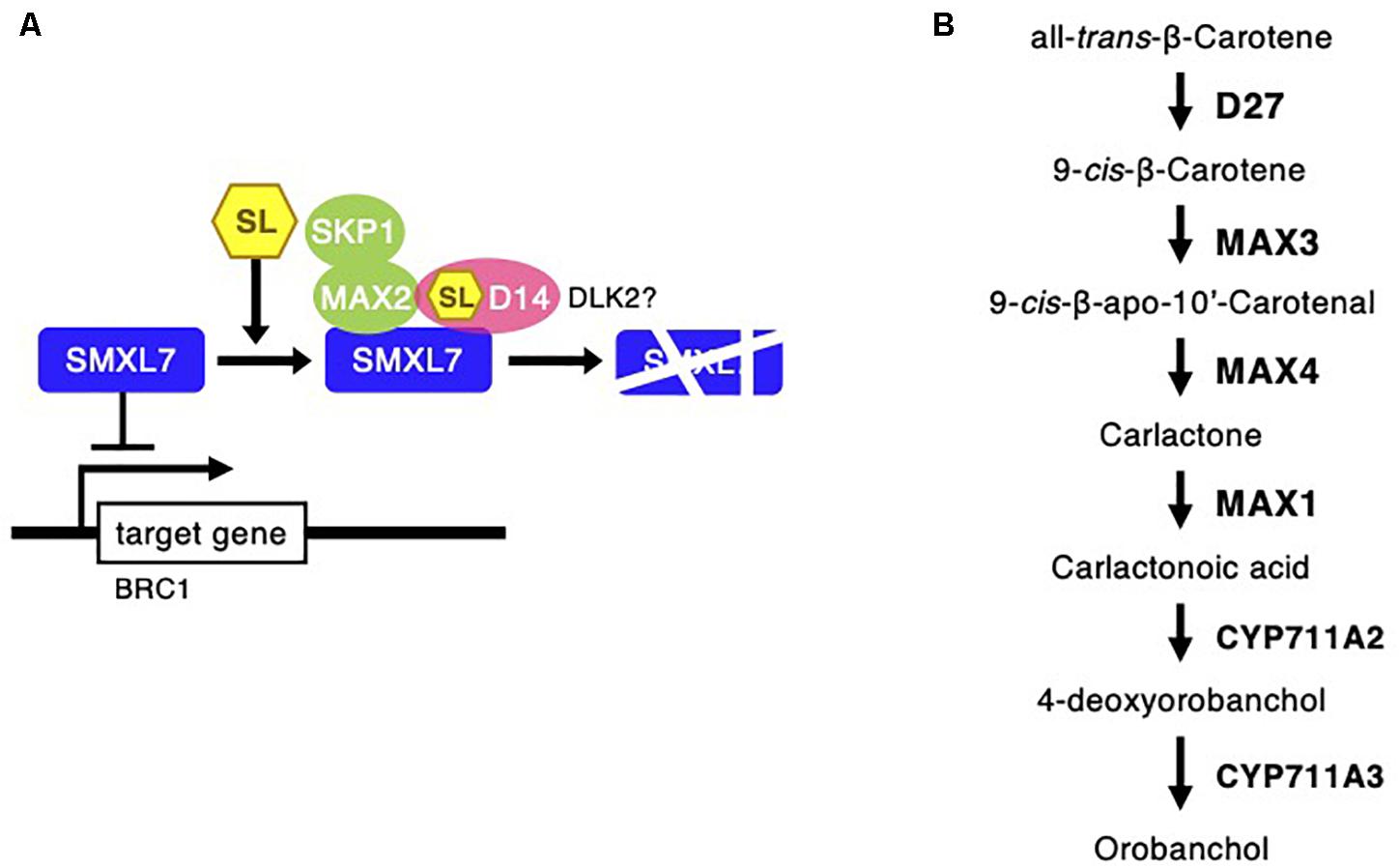
Figure 11. Strigolactone signaling and biosynthesis pathway following Chloris virgata. (A) The possible SLs signaling pathway in C. virgata. D14, dwarf 14; DLK2, D14 like 2; MAX2, more axillary branches 2; SKP1, S phase kinase-associated protein 1; SMXL7, suppressor of MAX2 1 like 7; BRC1, branched 1. (B) SLs biosynthesis pathway in C. virgata. D27, dwarf 27. The arrows indicate each steps.
Discussion
In this study, we identified the C. virgata DG accession as the fastest germination plant by seed screening 40 major Mongolian grassland plants. C. virgata also showed faster germination than the common agricultural crops, rice and wheat (Table 1). C. virgata is one of the major plants in the Mongolian grassland. There are approximately 50 species of Chloris that grow in most tropical areas worldwide, but only one species, C. virgata, has been found in Mongolia (Jigjidsuren and Johnson, 2003). Chloris gayana is a popular grass in pasture land, and its salt tolerance mechanism is to excrete salt via salt glands (Liphschitz et al., 1974; Kobayashi et al., 2007). C. virgata Sw., known as feather finger grass, is an annual grass of the Gramineae family, which is widespread across many habitats. The general phenotype is 5–50 cm tall, with semi-prostrate stems with 3–4 nodes; linear lanceolate leaflets, 5–15 cm long and approximately 2 mm wide; and arrowhead-type seeds, 2–3 mm long, with seed covers with long white hairs that can be carried by wind or water (Jigjidsuren and Johnson, 2003).
Previous research on C. virgata has been limited to a few fields based on specific ecological or physiological aspects. C. virgata is thought to be one of the major C4 summer grass weeds in Australia. It was found in 118,000 hectares of Australian crop farms, and a trial to decrease C. virgata growth using herbicide has been reported (Davidson et al., 2019; Mobli et al., 2020). C. virgata also tends to be resistant to alkaline soil conditions (Nishiuchi et al., 2010). Characteristics of C. virgata might suggest that it possesses tough viability, but direct research on its growth characteristics has been limited (Yang et al., 2008; Lin et al., 2016; Zhong et al., 2017).
Nevertheless, a previous report analyzed the phenotype and composition of fundamental components in the germination stage of six grassland species in Northeast China: C. virgata, Kochia scoparia, Lespedeza hedysaroides, Astragalus adsurgens, Leonurus artemisia, and Dracocephalum moldavica (Zhao et al., 2018). The report suggests that C. virgata started germination 10 h after imbibition, which was faster than the other four grassland plants. As we found that C. virgata germination started 5 h after germination treatment, these results were similar in both Mongolia and Northeast China. Furthermore, C. virgata quickly absorbed water during the first 2 h of imbibition, reached 73% water absorption. Dry C. virgata seeds contained approximately 62.2% starch, which was the highest ratio among the six grassland plants. Soluble sugars quickly increased after C. virgata germination. This starch composition might be one of the adaptations supporting the fast germination potential of C. virgata (Zhao et al., 2018).
When the apical meristem of the main stem in dicots and monocots is cut off, elongation of the lateral bud is generally observed, and these mechanisms are based on the phytohormones auxin and cytokinin as an apical dominance system (Domagalska and Leyser, 2011). In nomadic areas, all plants are exposed to the risk of animal bite. Several plants might die after a single animal bite, but other plants might be able to survive in this risky environment. In our survey of nomadic culture and previous research, C. virgata was shown to possess high regrowth ability after animal bite (Jigjidsuren and Johnson, 2003). We attempted to confirm the possible restoration power of C. virgata. As shown in Figure 2, C. virgata showed high regrowth potential after scissor cutting that mimicked animal bite, when compared with wheat and rice. The number of regrowth shoots of wheat and rice decreased with increased cutting, but C. virgata still maintained the ability to regrow after six cutting treatments. In a previous study, Medicago truncatula were cut off at 25 and 75% of the main and lateral shoots at the early (80 days old) and late (140 days old) development stages, and growth parameters such as shoot biomass and branch number were measured. The shoot biomass and branching number of M. truncatula showed recovery after 25% cutting during the early stage, but plants subjected to 75% cutting could not recover to the height they were before the treatment. Furthermore, the M. truncatula analysis was performed only once, and the phenotype after a second cutting has not been reported (Gruntman and Novoplansky, 2011). The analysis of plant growth after animal bite in grassland ecosystems has been performed in the field. Regrowth potential analysis of a Hordeum brevisubulatum population was conducted using three different cutting treatments (light cutting with a stubble height of 15 cm, medium cutting with a stubble height of 10 cm, and heavy cutting with a stubble height of 5 cm) and regrowth potential was calculated 1, 3, and 7 weeks after cutting. Heavily cut H. brevisubulatum showed regrowth to 25% of the height of an uncut plant 3 weeks after cutting and to 80% of that height 7 weeks after cutting (Yuan et al., 2020). In our study, C. virgata still showed regrowth to almost the same plant height after the sixth cutting. Our results indicating the regrowth advantages of C. virgata might contribute to the recovery of grasslands under severe stress from livestock husbandry in Mongolia.
In previous research, a few molecular biology studies have been performed and reported. Glyphosate is one of the strongest herbicides that binds to the enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) and inhibits amino acid metabolism in the shikimate pathway (Amrhein et al., 1980). In Australian grasslands, C. virgata showed resistance against glyphosate, which evolved and increased. Sequencing analysis of the EPSPS gene of the glyphosate-resistant C. virgata suggested several amino acid mutations at the herbicide target site (Ngo et al., 2018). In a previous study, 3168 expressed sequence tags (ESTs) were selected and sequenced from a cDNA library of NaHCO3-treated C. virgata, based on seeds originally collected in North China (Nishiuchi et al., 2010). Of these, 2590 ESTs indicated similarity to sequences in the NCBI database, and about 67% of the unigenes were annotated to genes in the rice genome. GO analysis revealed that 1,081 genes were annotated and distributed among 1245 terms in biological process, 1126 terms in molecular function, and 1415 terms in cellular component. In the GO functional categories for EST, 75 genes were annotated with the GO term “response to stress,” which might be owing to the experimental conditions, as these ESTs were identified from NaHCO3-treated C. virgata. A previous study also showed that the MT1 gene was upregulated in C. virgata under saline–alkaline conditions and heavy metal stress. Yeasts transformed with the MT1a gene of C. virgata showed tolerance to alkali stress. The ChvACT2 in C. virgata mRNA was upregulated more than twofold under alkali stress, such as that caused by NaCl, NaHCO3, CuSO4, ZnSO4, and CoCl2 (Nishiuchi et al., 2010). These reports are pioneering studies on the molecular biology of C. virgata. Based on the interesting phenotype of C. virgata as the fastest-growing plant, we began collecting more detailed genomic information.
In this study, transcriptome analyses of C. virgata at the germination (2 DAG), early development (8 DAG), young development (17 DAG), and adult development (28 DAG) stages were revealed. The cDNA library of these four groups of C. virgata yielded 21,589 contigs that were predicted to ORFs. Of these, 19,346 and 18,156 ORFs were matched to homologous genes in O. sativa and A. thaliana, respectively (Table 2).
In the germination stage of C. virgata, of the 85 enriched GO terms, six chloroplast- and four mitochondria-related GO terms were enriched (Figure 4 and Supplementary Data 1). A previous transcriptome analysis of the germination process of rice revealed that the GO terms involving chloroplasts and mitochondria were enriched 24 h after germination (Howell et al., 2009). Photosynthesis- and mitochondria-related proteins are also abundant in wheat and barley (Sreenivasulu et al., 2008; Yu et al., 2014). These results are similar to those of C. virgata. Furthermore, lipid metabolism-related GO terms were also enriched in the germination stages of C. virgata and rice. These systems are quickly induced and prepared to perform seed germination.
Plant germination is tightly regulated by phytohormone associations, GA, and ABA. In particular, GA and ABA play key roles in the germination stage and in the control of seed dormancy respectively. In this study, the ABA biosynthesis enzyme (NCED) family homologous genes (NCED1: Chloris2871c000010, NCED2: Chloris28063c000010, NCED5: Chloris11441c000010, and NCED6: Chloris2871c000010), regulatory component of the ABA receptor (RCAR) family homologous gene (PYL1: Chloris6771c000010), ABA signal transduction-related kinase SnRK2 homologous genes (SnRK2B: Chloris13011c000010 and SnRK2E: Chloris2715c000010), and basic leucine zipper (bZIP) transcription factor homologous gene (ABI5: Chloris7667c000010) were downregulated during the germination stage of C. virgata (Table 4). In previous studies, NCED5- and NCED6-deficient mutants showed higher germination efficiency than wild-type Arabidopsis (Frey et al., 2012). Our results show that the GA biosynthesis genes (CPS: Chloris5831c000010 and GA3-ox1: Chloris7778c000010) were significantly upregulated during the germination stage of C. virgata (Table 3). In Arabidopsis, GA3-ox1-deficient mutants were non-germinating until exogenous GA supplementation (Ogawa et al., 2003). Additionally, the mRNA expression of the GA deactivation enzyme AtGA2ox was extremely low during the early germination stage of Arabidopsis (Ogawa et al., 2003). Our results show that one of the GA deactivation enzymes, GA2-ox1 homologous gene (Chloris20434c000010), was expressed at extremely low levels during the germination stage of C. virgata (Table 3). The changes in the expression of ABA- and GA-related genes in the germination stage of C. virgata might be involved in the fast germination rate of C. virgata.
As shown in Figures 1A,C, C. virgata showed an active leaf elongation phenotype in the early developmental stage. At this stage, 392 biological process GO categories, including many organ development promotion-related categories, were overrepresented and enriched (Figure 7 and Supplementary Data 2). Based on these plant growth categories, the homologs of the biosynthesis enzyme of the plant growth and development promoting hormone BR (DET2: Chloris12539c000010, DWF4: Chloris405c000010, CPD: Chloris6902c000010, and BR6OX: Chloris144424c000010) were upregulated in the early young development stage of C. virgata (Table 5). In the early stages of BR research, the first BR biosynthesis gene (det2) mutants were isolated as dwarf phenotypes in Arabidopsis (Li et al., 1996; Szekeres et al., 1996). Furthermore, mutants deficient in the biosynthesis genes DWF4 and CPD showed dwarfism and were rescued by exogenously supplied BR (Szekeres et al., 1996; Fujiyama et al., 2019). Conversely, the BR catabolic enzyme BAS1 homologous gene (Chloris30576c000010) was downregulated in the early young development stage of C. virgata (Table 5). Overexpression of BAS1 caused dwarfism and decreased the interval content of brassinolide in Arabidopsis (Neff et al., 1999). Furthermore, the homologous genes of the transmembrane receptors of BR on the plasma membrane, BRI1 (Chloris3177c000010), BAK1 (Chloris23503c000010), and BR-signaling kinase, BSK1 (Chloris3139c000010), were upregulated in the early young development stage of C. virgata (Table 5). BRI1 is the major receptor of BR signaling, and mutations in BRI1 cause extreme dwarfism and cannot be rescued by exogenous BL (Wang et al., 2001; Caño-Delgado et al., 2004). BRI1 overexpression promotes the organ growth phenotype as an exogenous BR treatment (Wang et al., 2001). Overexpression of BAK1 promoted leaf growth, and BAK1-deficient mutants showed a semi-dwarfed phenotype that also exhibited reduced sensitivity to BR treatment (Li et al., 2002). BSK1 is a membrane-associated kinase, and overexpression of BSK1 rescued the dwarf phenotype of the BR-insensitive bri1 mutant (Tang et al., 2008). The BR-signaling positive phosphatase, BSU1, (Chloris13780c000010) homolog was upregulated in the early young development stage of C. virgata (Table 5). Overexpression of BSU1 rescued the bri1 dwarf phenotype (Mora-García et al., 2004). A negative regulator of the BR-signaling factor, BSS1, homologous gene (Chloris13348c000010) was downregulated in the early young development stage of C. virgata (Table 5). The bss1-1D mutant, which overexpressed BSS1, exhibited a BR biosynthesis inhibitor (Brz)-hypersensitive phenotype in hypocotyl elongation and exhibited no nuclear transfer of the BR master transcription factors, BIL1/BZR1 (Shimada et al., 2015). These results indicate that BR biosynthesis and signaling could promote the fast growth of C. virgata in the early young development stage.
In the adult development stage, C. virgata promoted branch formation (Figure 1D). Plant branch formation is negatively regulated by the phytohormone SL, which suppresses branch formation (Umehara et al., 2008). In the present study, the SL signaling homologous genes (D14: Chloris9047c000010; DLK2: Chloris3102c000010, Chloris16862c000010, and Chloris16863c000010; MAX2: Chloris21103c000010; and BRC1: Chloris9240c000010) were downregulated in the adult development stage (Table 6). DWARF14 and D14 have been identified as SL receptors, and d14-deficient mutants exhibit increased branching in rice (Arite et al., 2009). DLK2 is a divergent member of the D14 family that weakly binds or hydrolyzes SL ligands, and detailed regulations are still unclear in adult plants (Végh et al., 2017). The F-box leucine-rich repeat family protein MAX2 is a positive regulator of SL signaling and binds to the SL receptor D14. MAX2 is required to repress bud outgrowth at each node (Stirnberg et al., 2007). BRC1 is a positive transcription factor in SL signaling that is expressed in developing buds and leads to the suppression of bud development. The downregulation of BCR1 results in a high branching phenotype in Arabidopsis (Aguilar-Martínez et al., 2007). These results suggest that the downregulation of SL signaling gene expression promotes branch formation in the adult development stage of C. virgata.
Our report suggests that C. virgata possesses fast growth and high regrowth potential. Recently, Mongolian grasslands have been exposed to grazing pressure by the increasing number of livestock, such as sheep, goats, horses, cattle, camels, and other domestic animals. Climate change owing to global warming has also impacted and decreased the area of Mongolian grassland. We hope that the possible growth activity of C. virgata illustrated in this study will contribute to improving the robustness of the Mongolian grassland. Furthermore, we found that novel functional genes, including phytohormone signaling and biosynthesis genes, contribute to the rapid growth of C. virgata. A more detailed analysis of the novel genes of C. virgata could suggest possible functions in the improvement of growth and rapid germination in commercial crops, such as rice and wheat, and in Mongolian grassland plants.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ddbj.nig.ac.jp/, DRA011714.
Author Contributions
TN, AY, JB, B-OD, TA, and KS conceived and designed the experiments. BB, AY, and SK performed the experiments, phytohormone analog search, and phylogenetic analysis. GU, SJ, and TB performed the seed germination and seed collection. KM, FT, KoI, AK, MK, and KeI performed the sequencing and de novo transcriptome analyses. BB, AY, FT, KM, and TN contributed to the manuscript preparation. All the authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the Science and Technology Research Partnership for Sustainable Development (SATREPS), Japan Science and Technology Agency (JST)/Japan International Cooperation Agency (JICA) (grant number: JPMJSA1906), and JSPS KAKENHI [grant numbers: JP18H02140, JP19H03255 (to FT), and JP20K21437 (to FT)]. This work was also partially supported by the Core Research for Evolutionary Science and Technology (CREST) of the Japan Science and Technology Agency (JST) (to KM) and Cabinet Office, Government of Japan, Cross-ministerial Moonshot Agriculture, Forestry and Fisheries Research and Development Program, “Technologies for Smart Bio-industry and Agriculture” (funding agency: Bio-oriented Technology Research Advancement Institution, grant number: JPJ009237) (to KM).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.684987/full#supplementary-material
Footnotes
References
Aguilar-Martínez, J. A., Poza-Carrión, C., and Cubas, P. (2007). Arabidopsis Branched1 acts as an integrator of branching signals within axillary buds. Plant Cell 19, 458–472. doi: 10.1105/tpc.106.048934
Amrhein, N., Deus, B., Gehrke, P., and Steinrücken, H. C. (1980). The site of the inhibition of the shikimate pathway by glyphosate. Plant Physiol. 66, 830–834. doi: 10.1104/pp.66.5.830
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Arite, T., Umehara, M., Ishikawa, S., Hanada, A., Maekawa, M., Yamaguchi, S., et al. (2009). D14, a strigolactone-Insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 50, 1416–1424. doi: 10.1093/pcp/pcp091
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boudsocq, M., Barbier-Brygoo, H., and Laurière, C. (2004). Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 279, 41758–41766. doi: 10.1074/jbc.M405259200
Caño-Delgado, A., Yin, Y., Yu, C., Vefeados, D., Mora-García, S., Cheng, J. C., et al. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular defferentiation in Arabidopsis. Development 131, 5341–5354. doi: 10.1242/dev.01403
Choe, S., Dilkes, B. P., Fujioka, S., Takatsuto, S., Sakurai, A., and Feldmann, K. A. (1998). The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10, 231–243. doi: 10.1105/tpc.10.2.231
Davidson, B., Cook, T., and Chauhan, B. S. (2019). Alternative options to glyphosate for control of large echinochloa colona and Chloris virgata plants in cropping fallows. Plants 8, 245–256. doi: 10.3390/plants8080245
Domagalska, M. A., and Leyser, O. (2011). Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 12, 211–221. doi: 10.1038/nrm3088
Emms, D. M., and Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238–252. doi: 10.1186/s13059-019-1832-y
Finkelstein, R., Reeves, W., Ariizumi, T., and Steber, C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59, 387–415. doi: 10.1146/annurev.arplant.59.032607.092740
Finlayson, S. A. (2007). Arabidopsis TEOSINTE BRANCHED1-LIKE 1 regulates axillary bud outgrowth and is homologous to monocot TEOSINTE BRANCHED1. Plant Cell Physiol. 48, 667–677. doi: 10.1093/pcp/pcm044
Frey, A., Effroy, D., Lefebvre, V., Seo, M., Perreau, F., Berger, A., et al. (2012). Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 70, 501–512. doi: 10.1111/j.1365-313X.2011.04887.x
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
Fujiyama, K., Hino, T., Kanadani, M., Watanabe, B., Jae Lee, H., Mizutani, M., et al. (2019). Structural insights into a key step of brassinosteroid biosynthesis and its inhibition. Nat. Plants 5, 589–594. doi: 10.1038/s41477-019-0436-6
Gordo, S. M. C., Pinheiro, D. G., Moreira, E. C. O., Rodrigues, S. M., Poltronieri, M. C., de Lemos, O. F., et al. (2012). High-throughput sequencing of black pepper root transcriptome. BMC Plant Biol. 12:168. doi: 10.1186/1471-2229-12-168
Grabherr, M., Haas, B., Yassour, M., Levin, J., Thompson, D., Amit, I., et al. (2011). Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883.Trinity
Gruntman, M., and Novoplansky, A. (2011). Ontogenetic contingency of tolerance mechanisms in response to apical damage. Ann. Bot. 108, 965–973. doi: 10.1093/aob/mcr204
Hereward, J. P., Werth, J. A., Thornby, D. F., Keenan, M., Chauhan, B. S., and Walter, G. H. (2016). Complete chloroplast genome sequences of two species of Chloris grass, Chloris truncata Sw. and Chloris virgata R. Br. Mitochondr. DNA Part B Resour. 1, 960–961. doi: 10.1080/23802359.2016.1266705
Hoang, D. T., Chernomor, O., Von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Howell, K. A., Narsai, R., Carroll, A., Ivanova, A., Lohse, M., Usadel, B., et al. (2009). Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol. 149, 961–980. doi: 10.1104/pp.108.129874
Huang, X., Wang, J., Aluru, S., Yang, S., and Hillier, L. (2003). PCAP: a whole-genome assembly program. Genome Res. 13, 2164–2170. doi: 10.1101/gr.1390403.1
Ishikawa, S., Maekawa, M., Arite, T., Onishi, K., Takamure, I., and Kyozuka, J. (2005). Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 46, 79–86. doi: 10.1093/pcp/pci022
Iuchi, S., Kobayashi, M., Taji, T., Naramoto, M., Seki, M., Kato, T., et al. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27, 325–333. doi: 10.1046/j.1365-313X.2001.01096.x
Jamyandorj, J., Ligaa, U., Otgonbileg, K., and Saaral, N. (2011). Very Rare, Rare and Important Useful Plants, Cultivating in Khuduu-Aral of Kherlen. Ulaanbaatar: Ulaanbaatar Print.
Johnson, L. S., Eddy, S. R., and Portugaly, E. (2010). Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11:431. doi: 10.1186/1471-2105-11-431
Jun, J. H., Ha, C. M., and Fletchera, J. C. (2010). BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell 22, 62–76. doi: 10.1105/tpc.109.070763
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Karban, R., and Baldwin, I. T. (1997). Induced Responses to Herbivory. Chicago: University of Chicago Press.
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kobayashi, H., Masaoka, Y., Takahashi, Y., Ide, Y., and Sato, S. (2007). Ability of salt glands in Rhodes grass (Chloris gayana Kunth) to secrete Na+ and K+. Soil Sci. Plant Nutr. 53, 764–771. doi: 10.1111/j.1747-0765.2007.00192.x
Kobayashi, Y., Yamamoto, S., Minami, H., Kagaya, Y., and Hattori, T. (2004). Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16, 1163–1177. doi: 10.1105/tpc.019943
Lapin, K., Eipeldauer, A., Folly, G., Mank, D., and Bernhardt, K. (2017). The vegetation of North-Western mongolia: floristic checklist and conservation status of mongolian grassland flora. Mongolian J. Biol. Sci. 15, 13–22. doi: 10.22353/mjbs.2017.15.02
Lei, B., Lu, K., Ding, F., Zhang, K., Chen, Y., Zhao, H., et al. (2014). RNA sequencing analysis reveals transcriptomic variations in tobacco (Nicotiana tabacum) leaves affected by climate, soil, and tillage factors. Int. J. Mol. Sci. 15, 6137–6160. doi: 10.3390/ijms15046137
Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. doi: 10.1016/s0092-8674(00)80357-8
Li, J., Nagpal, P., Vitart, V., McMorris, T. C., and Chory, J. (1996). A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272, 398–401. doi: 10.1126/science.272.5260.398
Li, J., Wen, J., Lease, K. A., Doke, J. T., Tax, F. E., and Walker, J. C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. doi: 10.1016/S0092-8674(02)00812-7
Liao, Y., Smyth, G. K., and Shi, W. (2014). FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Lin, J., Shao, S., Wang, Y., Qi, M., Lin, L., Wang, Y., et al. (2016). Germination responses of the halophyte Chloris virgata to temperature and reduced water potential caused by salinity, alkalinity and drought stress. Grass Forage Sci. 71, 507–514. doi: 10.1111/gfs.12218
Liphschitz, N., Adiva-Shomer, I., Eshel, A., and Waisel, Y. (1974). Salt glands on leaves of rhodes grass (Chloris gayana Kth.). Ann. Bot. 38, 459–462. doi: 10.1093/oxfordjournals.aob.a084829
Lu, X., Zhou, X., Cao, Y., Zhou, M., McNeil, D., Liang, S., et al. (2017). RNA-seq analysis of cold and drought responsive transcriptomes of Zea mays ssp. Mexicana L. Front. Plant Sci. 8:136. doi: 10.3389/fpls.2017.00136
Mobli, A., Rinwa, A., and Sahil, and Chauhan, B. S. (2020). Effects of sorghum residue in presence of preemergence herbicides on emergence and biomass of Echinochloa colona and Chloris virgata. PLoS One 15:e0229817. doi: 10.1371/journal.pone.0229817
Mora-García, S., Vert, G., Yin, Y., Caño-Delgado, A., Cheong, H., and Chory, J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18, 448–460. doi: 10.1101/gad.1174204
Nakano, T., and Asami, T. (2014). Brassinosteroids signaling and biosynthesis. Plant Chem. Biol. 4, 128–144. doi: 10.1002/9781118742921
Neff, M. M., Nguyen, S. M., Malancharuvil, E. J., Fujioka, S., Noguchi, T., Seto, H., et al. (1999). Bas1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 96, 15316–15323. doi: 10.1073/pnas.96.26.15316
Ngo, T. D., Krishnan, M., Boutsalis, P., Gill, G., and Preston, C. (2018). Target-site mutations conferring resistance to glyphosate in feathertop Rhodes grass (Chloris virgata) populations in Australia. Pest. Manag. Sci. 47, 1094–1100. doi: 10.1002/ps.4512
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nishiuchi, S., Fujihara, K., Liu, S., and Takano, T. (2010). Analysis of expressed sequence tags from a NaHCO3-treated alkali-tolerant plant, Chloris virgata. Plant Physiol. Biochem. 48, 247–255. doi: 10.1016/j.plaphy.2010.01.024
Ochoa, V., Madrid, E., Said, M., Rubiales, D., and Cabrera, A. (2015). Molecular and cytogenetic characterization of a common wheat-Agropyron cristatum chromosome translocation conferring resistance to leaf rust. Euphytica 201, 89–95. doi: 10.1007/s10681-014-1190-5
Ogawa, M., Hanada, A., Yamauchi, Y., Kuwahara, A., Kamiya, Y., and Yamaguchi, S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15, 1591–1604. doi: 10.1105/tpc.011650
Park, S., Fung, P., Nishimura, N., Jensen, D. R., Zhao, Y., Lumba, S., et al. (2009). Abscisic acid inhibits PP2Cs via the PYR/PYL family of ABA- binding START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041.Abscisic
Rajjou, L., Duval, M., Gallardo, K., Catusse, J., Bally, J., Job, C., et al. (2012). Seed germination and vigor. Annu. Rev. Plant Biol. 63, 507–533. doi: 10.1146/annurev-arplant-042811-105550
Ramesh, K. R., Hemalatha, R., Vijayendra, C. A., Arshi, U. Z. S., Dushyant, S. B., and Dinesh, K. B. (2016). Transcriptome analysis of Solanum melongena L. (eggplant) fruit to identify putative allergens and their epitopes. Gene 576, 64–71. doi: 10.1016/j.gene.2015.09.064
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2009). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rodriguez, M. C. S., Edsgärd, D., Hussain, S. S., Alquezar, D., Rasmussen, M., Gilbert, T., et al. (2010). Transcriptomes of the desiccation-tolerant resurrection plant Craterostigma plantagineum. Plant J. 63, 212–228. doi: 10.1111/j.1365-313X.2010.04243.x
Schwartz, S. H., Tan, B. C., Gage, D. A., Zeevaart, J. A. D., and McCarty, D. R. (1997). Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276, 1872–1874. doi: 10.1126/science.276.5320.1872
Sharma, H. C., Gill, B. S., and Uyemoto, J. K. (1984). High levels of resistance in Agropyron species to barley yellow dwarf and wheat streak mosaic viruses. J. Phytopathol. 110, 143–147. doi: 10.1111/j.1439-0434.1984.tb03402.x
Shimada, S., Komatsu, T., Yamagami, A., Nakazawa, M., Matsui, M., Kawaide, H., et al. (2015). Formation and dissociation of the BSS1 protein complex regulates plant development via brassinosteroid signaling. Plant Cell 27, 375–390. doi: 10.1105/tpc.114.131508
Shimada, Y., Fujioka, S., Miyauchi, N., Kushiro, M., Takatsuto, S., Nomura, T., et al. (2001). Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis1. Plant Physiol. 126, 770–779. doi: 10.1104/pp.126.2.770
Sreenivasulu, N., Usadel, B., Winter, A., Radchuk, V., Scholz, U., Stein, N., et al. (2008). Barley grain maturation and germination: metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiol. 146, 1738–1758. doi: 10.1104/pp.107.111781
Stirnberg, P., Furner, I. J., and Ottoline Leyser, H. M. (2007). MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50, 80–94. doi: 10.1111/j.1365-313X.2007.03032.x
Stirnberg, P., Sande, K., and Van De Leyser, H. M. O. (2002). MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129, 1131–1141.
Sun, T. P., and Kamiya, Y. (1994). The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6, 1509–1518. doi: 10.1105/tpc.6.10.1509
Szekeres, M., Németh, K., Koncz-Kálmán, Z., Mathur, J., Kauschmann, A., Altmann, T., et al. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. doi: 10.1016/S0092-8674(00)81094-6
Talon, M., Koornneef, M., and Zeevaart, J. A. D. (1990). Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc. Natl. Acad. Sci. U.S.A. 87, 7983–7987. doi: 10.1073/pnas.87.20.7983
Tang, W., Kim, T.-W., Oses-Prieto, J. A., Sun, Y., Deng, Z., Zhu, S., et al. (2008). Brassinosteroid-Signaling Kinases (BSKs) mediate signal transduction from the receptor kinase BRI1 in Arabidopsis NIH Public Access. Science 321, 557–560. doi: 10.1126/science.1156973
Thomas, S. G., Phillips, A. L., and Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. U.S.A. 96, 4698–4703. doi: 10.1073/pnas.96.8.4698
Umehara, M., Hanada, A., Yoshida, S., Akiyama, K., Arite, T., Takeda-Kamiya, N., et al. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200. doi: 10.1038/nature07272
Végh, A., Incze, N., Fábián, A., Huo, H., Bradford, K. J., Balázs, E., et al. (2017). Comprehensive analysis of DWARF14-LIKE2 (DLK2) reveals its functional divergence from 0strigolactone-related paralogs. Front. Plant Sci. 8:1641. doi: 10.3389/fpls.2017.01641
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wang, Z. Y., Seto, H., Fujioka, S., Yoshida, S., and Chory, J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383. doi: 10.1038/35066597
Wei, Y., Xu, Y., Lu, P., Wang, X., Li, Z., Cai, X., et al. (2017). Salt stress responsiveness of a wild cotton species (Gossypium klotzschianum) based on transcriptomic analysis. PLoS One 12:e0178313. doi: 10.1371/journal.pone.0178313
Xu, W., Li, R., Zhang, N., Ma, F., Jiao, Y., and Wang, Z. (2014). Transcriptome profiling of Vitis amurensis, an extremely cold-tolerant Chinese wild Vitis species, reveals candidate genes and events that potentially connected to cold stress. Plant Mol. Biol. 86, 527–541. doi: 10.1007/s11103-014-0245-2
Yang, C. W., Jianaer, A., Li, C. Y., Shi, D. C., and Wang, D. L. (2008). Comparison of the effects of salt-stress and alkali-stress on photosynthesis and energy storage of an alkali-resistant halophyte Chloris virgata. Photosynthetica 46:273. doi: 10.1007/s11099-008-0047-3
Yang, Z., Dai, Z., Lu, R., Wu, B., Tang, Q., Xu, Y., et al. (2017). Transcriptome analysis of two species of jute in response to polyethylene glycol (PEG)- induced drought stress. Sci. Rep. 7, 16565–16576. doi: 10.1038/s41598-017-16812-5
Yu, F., Wu, Y., and Xie, Q. (2015). Precise protein post-translational modifications modulate ABI5 activity. Trends Plant Sci. 20, 569–575. doi: 10.1016/j.tplants.2015.05.004
Yu, Y., Guo, G., Lv, D., Hu, Y., Li, J., Li, X., et al. (2014). Transcriptome analysis during seed germination of elite Chinese bread wheat cultivar Jimai 20. BMC Plant Biol. 14:20. doi: 10.1186/1471-2229-14-20
Yuan, J., Li, H., and Yang, Y. (2020). The compensatory tillering in the forage grass hordeum brevisubulatum after simulated grazing of different severity. Front. Plant Sci. 11:792. doi: 10.3389/fpls.2020.00792
Zhang, J., Liu, W., Han, H., Song, L., Bai, L., Gao, Z., et al. (2015). De novo transcriptome sequencing of Agropyron cristatum to identify available gene resources for the enhancement of wheat. Genomics 106, 129–136. doi: 10.1016/j.ygeno.2015.04.003
Zhao, M., Zhang, H., Yan, H., Qiu, L., Baskin, C. C., and Job, D. (2018). Mobilization and role of starch, protein, and fat reserves during seed germination of six wild grassland species. Front. Plant Sci. 9:234. doi: 10.3389/fpls.2018.00234
Zhong, S., Chai, H., Xu, Y., Li, Y., Ma, J. Y., and Sun, W. (2017). Drought sensitivity of the carbon isotope composition of leaf dark-respired CO2 in C3 (Leymus Chinensis) and C4 (Chloris virgata and hemarthria altissima) grasses in northeast China. Front. Plant Sci. 8:1996. doi: 10.3389/fpls.2017.01996
Zhou, S., Zhang, J., Han, H., Zhang, J., Ma, H., Zhang, Z., et al. (2019). Full-length transcriptome sequences of Agropyron cristatum facilitate the prediction of putative genes for thousand-grain weight in a wheat-A. cristatum translocation line. BMC Genomics 20:1025. doi: 10.1186/s12864-019-6416-4
Keywords: plant growth, transcriptome, germination, plant hormone, regrowth, brassinosteroid, Chloris virgata
Citation: Bolortuya B, Kawabata S, Yamagami A, Davaapurev B-O, Takahashi F, Inoue K, Kanatani A, Mochida K, Kumazawa M, Ifuku K, Jigjidsuren S, Battogtokh T, Udval G, Shinozaki K, Asami T, Batkhuu J and Nakano T (2021) Transcriptome Analysis of Chloris virgata, Which Shows the Fastest Germination and Growth in the Major Mongolian Grassland Plant. Front. Plant Sci. 12:684987. doi: 10.3389/fpls.2021.684987
Received: 24 March 2021; Accepted: 26 April 2021;
Published: 28 June 2021.
Edited by:
Ken-ichiro Hayashi, Okayama University of Science, JapanReviewed by:
Hongning Tong, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, ChinaMasanori Okamoto, Utsunomiya University, Japan
Copyright © 2021 Bolortuya, Kawabata, Yamagami, Davaapurev, Takahashi, Inoue, Kanatani, Mochida, Kumazawa, Ifuku, Jigjidsuren, Battogtokh, Udval, Shinozaki, Asami, Batkhuu and Nakano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takeshi Nakano, nakano.takeshi.6x@kyoto-u.ac.jp
†Deceased November, 2018
 Byambajav Bolortuya
Byambajav Bolortuya Shintaro Kawabata
Shintaro Kawabata Ayumi Yamagami1
Ayumi Yamagami1 Fuminori Takahashi
Fuminori Takahashi Keiichi Mochida
Keiichi Mochida Minoru Kumazawa
Minoru Kumazawa Kentaro Ifuku
Kentaro Ifuku Gombosuren Udval
Gombosuren Udval Kazuo Shinozaki
Kazuo Shinozaki Tadao Asami
Tadao Asami Takeshi Nakano
Takeshi Nakano