- 1Research Center of Saline and Alkali Land of State Forestry and Grassland Administration, Chinese Academy of Forestry, Beijing, China
- 2State Key Laboratory of Tree Genetics and Breeding, Northeast Forestry University, Harbin, China
- 3College of Forest Resources and Environmental Science, Michigan Technological University, Houghton, MI, United States
In plants, GATA transcription factors (TFs) have been reported to play vital roles in to a wide range of biological processes. To date, there is still no report about the involvement and functions of woody plant GATA TFs in wood formation. In this study, we described the functional characterization of a Populus trichocarpa GATA TF, PtrGATA12, which encodes a nuclear-localized transcriptional activator predominantly expressing in developing xylem tissues. Overexpression of PtrGATA12 not only inhibited growths of most phenotypic traits and biomass accumulation, but also altered the expressions of some master TFs and pathway genes involved in secondary cell wall (SCW) and programmed cell death, leading to alternated SCW components and breaking forces of stems of transgenic lines. The significant changes occurred in the contents of hemicellulose and lignin and SCW thicknesses of fiber and vessel that increased by 13.5 and 10.8%, and 20.83 and 11.83%, respectively. Furthermore, PtrGATA12 bound directly to the promoters of a battery of TFs and pathway genes and activated them; the binding sites include two cis-acting elements that were specifically enriched in their promoter regions. Taken together, our results suggest PtrGATA12, as a higher hierarchical TF on the top of PtrWND6A, PtrWND6B, PtrMYB152, and PtrMYB21, exert a coordinated regulation of SCW components biosynthesis pathways through directly and indirectly controlling master TFs, middle-level TFs, and further downstream pathway genes of the currently known hierarchical transcription network that governs SCW formation.
Introduction
Wood accounts for the bulk of biomass produced by land plants, and has been considered to be an important renewable and environmentally friendly source of bioenergy (Ohlrogge et al., 2009). The major components of wood, secondary cell walls (SCWs), consist of a cross-linked matrix of cellulose, hemicellulose, and lignin biopolymers (Zhong et al., 2010a). To genetically improve the productivity and quality of woods for better utilization, it is imperative to identify the molecular regulatory mechanisms associated with differential regulation of SCW biosynthesis. Studies in herbaceous and woody plants have revealed that SCW formation is regulated by a hierarchical transcription regulation network (hGRN) that is comprised of more than four layers of genes (Mellerowicz and Sundberg, 2008; Zhong et al., 2008, 2010a, 2011; Zhong and Ye, 2010; Taylor-Teeples et al., 2015; Rao and Dixon, 2018). The currently known top-layered regulatory genes in this network are NAC SND1 (McCarthy et al., 2009), VND6/7 (Ohashi-Ito et al., 2010; Yamaguchi et al., 2010), and PtrWND2B/6B (Zhong et al., 2010a, 2011), which function as master regulators for xylem cell differentiation (Zhong et al., 2010a, 2011). Downstream of these NAC master TFs are the MYB46/83 and PtrMYB3/20, which function as intermediate level hub regulators (McCarthy et al., 2009, 2010; Ko et al., 2012), whose targets are SCW-associated genes, including other lower-level MYB TFs, such as MYB58, MYB63, PtrMYB125, PpDof5, and PdOLP1 (Zhou et al., 2009; Rueda-Lopez et al., 2017; Balmant et al., 2020; Li et al., 2020). At the terminal are a variety of bottom-layered pathway genes responsible of SCW component biosynthesis (Zhong et al., 2010a). Although many regulatory genes have been identified, it is critically important to identify more complements of this hGRN. In addition, we are especially interested in identifying the hub regulators and the high hierarchical regulators. Hub regulators can take multiple “commands” from the higher hierarchical levels, synthesize new components, and then pass them down to biosynthetic pathways, whereas the high hierarchical regulators are those that are capable of regulating multiple pathways. For this reason, we initiated a project to identify more middle-level “hub” regulators, and high hierarchical regulators that can differentially control multiple SCW component biosynthetic pathways, and believe that they are instrumental for genetical engineering wood composition and quality.
GATA TF family, which is characterized by a type-IV zinc-finger motif with a CX2CX17–20CX2C domain followed by a basic region facilitating DNA binding the WGATAR (W = T or A; R = G or A) in the promoter region, is widely distributed in fungi, metazoans, and plants (Lowry and Atchley, 2000). A GATA zinc-finger TF with 17–18 residues in the binding loop is a characteristic feature of animal and fungal GATA TFs, while a plant GATA zinc-finger TF possess 17–20 residues (Reyes et al., 2004; Behringer et al., 2014). Plant GATA TFs have been identified in multiple plant species, such as Arabidopsis thaliana (Zhang et al., 2018a), Oryza sativa (Reyes et al., 2004), Glycine max (Zhang et al., 2015), Gossypium raimondii (Zhang et al., 2019), Malus domestica (Chen et al., 2017), and Populus trichocarpa (An et al., 2020). GATA TFs have been reported to play important roles in a wide range of biological processes of plants, such as vegetative growth and development (An et al., 2020), seed dormancy and germination, flowering and shoot apical meristem development (Behringer and Schwechheimer, 2015), chloroplast development (An et al., 2014; Zhang et al., 2015), cell elongation (Shikata et al., 2004), response to light and stress (Zhang et al., 2015; Kobayashi and Masuda, 2016), hormone signaling (Zhang et al., 2015; Kobayashi et al., 2017), and N metabolism (An et al., 2014). Besides these, it has been reported that overexpression of AtGATA12 induces ectopic differentiation of xylem vessel elements and SCW deposition through activating VND7 expression in A. thaliana (Endo et al., 2015). Since AtVND7 is a relatively high hierarchical master regulator of lignin and cellulose pathway regulator (Zhong et al., 2008), AtGATA12 is more likely to be a hierarchical regulator. Although the major transcriptional programs regulating wood formation are conserved in vascular plants, some variations of ortholog gene functions might have evolved since wood anatomy and compositions vary greatly among different plant species (Zhong et al., 2010a). For instance, A. thaliana MYB103 preferentially induces the expression of genes for cellulose biosynthesis but not xylan and lignin (Zhong et al., 2008), whereas its poplar ortholog, PtrMYB128, is able to activate the genes related to entire SCW component biosynthesis (Zhong et al., 2011). In addition, P. trichocarpa PtrSND1-B1 mediates a four-layered hGRN while its counterpart in A. thaliana, AtSND1, mediates a three-layered hGRN (Chen et al., 2019), indicating that PtrGATA12 may mediates different target genes and a network especially when taking the more complicated wood components and the augmented wood formation in poplar into account, These facts taken together made us to choose GATA12 gene to study.
In this report, we demonstrated that PtrGATA12 (Potri.006G237700), a P. trichocarpa ortholog of the Arabidopsis GATA12, had vital roles in poplar woody formation. We showed that PtrGATA12 is prominently expressed in developing secondary xylem tissues, and is a transcriptional activator that functions in the nuclei. Overexpression of PtrGATA12 in poplar leads to arrests of growths and development of most phenotypic traits, alternations of SCW component and thickness, and increase of breaking forces of stem, accompanied by the changes of expression of TFs and pathway genes related to SCW biosynthesis. Furthermore, we revealed that PtrGATA12 was capable of directly activating a number of poplar wood-associated TFs and pathway genes through binding to the cis-elements in their promoter regions. Our findings suggested that the PtrGATA12, as a higher hierarchical transcription activator, plays significant roles in activating hemicellulose and lignin biosynthesis of SCW through directly or indirectly regulating a number of TFs and pathway genes that are elements of the hierarchical transcription regulatory network of wood formation, and can be used to genetically modify wood characteristics.
Materials and Methods
Plant Materials
The plantlets of P. trichocarpa clone Nisqually-1, whose genome was sequenced (Tuskan et al., 2006) early, were obtained from the Shanghai Institute for Biological Sciences, Chinese Academy of Sciences, and vegetatively propagated in our lab using tissue culture (Li et al., 2017).
One-year-old P. trichocarpa trees were propagated and planted in a mixture of turfy peat and sand (2:1 v/v) and grown under 16 h/8 h day/night photoperiod at 25°C in the greenhouse at Northeast Forestry University for 90 days. The primary shoot leaves, transition leaves, secondary leaves, primary xylem, transition xylem, secondary xylem, primary phloem, transition phloem, secondary phloem, and roots, which were used to analyze the tissue-specific expression patterns of PtrGATA12, were collected and immediately frozen in liquid Nitrogen and stored at −80°C. The RNA was isolated according to a previously published method (Liao et al., 2004) and later treated with DNase I (Qiagen) to remove genomic DNA (Kolosova et al., 2004).
Cloning PtrGATA12 From P. trichocarpa
Five microgram total RNA from secondary xylem of P. trichocarpa stems was used for synthesizing cDNAs using SuperScript II Reverse Transcriptase (Invitrogen). The full PtrGATA12 (Potri.006G237700) cDNA was amplified with gene-specific primers (Supplementary Table S1). The PCR products were cloned into pMD18-T vector (TaKaRa), and then transformed into Escherichia coli cells (DH5α) for validation by Sanger sequencing.
Subcellular Localization
The full-length coding region of PtrGATA12 without termination codon was amplified using specific primers (Supplementary Table S1) and then fused to the N-terminal of green fluorescent protein (GFP) driven by CaMV 35S promoter in pGWB5 vector. The two fusion constructs were delivered into onion epidermal cells via particle bombardment (GJ-1000). The GFP fluorescent images were photographed with confocal microscopy (Leica TCS SP5) at 24 h after bombardment.
Transcriptional Activation Assay
The transcriptional activation of PtrGATA12 on putative targets was corroborated using the yeast two-hybrid system. The complete CDS of PtrGATA12 was amplified using specifically designed primers (Supplementary Table S1). The amplified fragments were fused in-frame to the pGBKT7 vector to generate the pGBKT7-PtrGATA12 construct. The pGBKT7-PtrGATA12 and the pGBKT7 blank vector (as negative control) were transformed into AH109 yeast cells independently. The transformed AH109 yeast cells were plated onto SD/-Trp (growth control), SD/-Trp/-His/-Ade, and X-α-Gal media and incubated on at 30°C for 3–5 days to identify the transcriptional activation.
Transformation of P. trichocarpa for Generating PtrGATA12 Transgenic Lines
The PtrGATA12 was amplified with specific primers (Supplementary Table S1), and then inserted into the pROKII vector. The pROKII-PtrGATA12 was first transferred into Agrobacterium tumefaciens EHA105 using the freeze-thaw method. The transgenic method was described below based on previous study (Li et al., 2017). The genomic DNA of all kanamycin-resistant shoots amplified by regular PCR using the PROKII sequencing primers listed in Supplementary Table S2 to verify whether PtrGATA12 was integrated into poplar genome.
All tested PtrGATA12 transgene lines, and wild-type (WT) poplar were propagated and planted in a mixture of turfy peat and sand (2:1 v/v) and grown under 16 h/8 h day/night photoperiod at 25°C in the greenhouse. When the PtrGATA12 transgene lines grown for 90 days, they were subsequently used for further characterization.
Phenotypic Trait Measurement
The developmental stages of tissues were standardized by employing a plastichron index (PI; PI = 0 was defined as the first leaf greater than 5 cm in length; PI = 1 was the leaf immediately below PI = 0). Stems spanning PI = 5 to PI = 8 were cut and frozen in liquid nitrogen, and retained for PtrGATA12 transcript abundance and transcript abundance of the known TFs and pathway genes related to fiber formation, SCW biosynthesis, and PCD analysis. Stems spanning PI = 8 to PI = 10 were cut and used for SCW thickness, breaking force, and cell wall chemical composition analyses. The internode lengths were determined by distance between two adjacent leaf petioles along the stems. Leaves from PI = 4 to PI = 6 were measured for lengths and widths.
Before harvesting, we measured the height of each plant from the root to the tip of the tallest bud and the base diameters above ~3 cm of soil to calculate its biomass. The fresh weight is determined immediately after harvesting the whole plant. Then, the plant material was placed into the oven and incubated at 100°C for 10 min, and then at 75°C until the weight did not change. The final unchanged weight was recorded as the dry weight.
Determination of Break Forces
The breaking force, which has been reported to be correlated with the cellulose content in stem of maize (Dhugga, 2007), refers to the tensile or bending strength used to break stem. The breaking forces of stem segments were analyzed using YYD-1 plant stalk analyzer according to the manufacturer’s instructions (Zhejiang Top Instrument Co., Ltd.).
Scanning Electron Microscopy
Stem segments were prepared by freeze-drying for scanning electron microscopy (SEM; S-4800, HITACHI). Dry segments were mounted on aluminum stubs using carbon tape with conductive silver paint applied to the sides to reduce sample charging. The segments were then sputter-coated with gold in an E-100 ion sputter. Imaging was performed at beam accelerating voltages from 12.5 to 15 kV. The secondary wall thicknesses of fibers in the SEM micrographs were quantified in a randomly selected area of 45 cells using Image J software.1
Histological Analysis
The 90-day-old poplar basal stems were cut into 0.5 cm fragments and immersed into 4% paraformaldehyde at 4°C for 3 days, washed twice in 1x PBS for 15 min, and then dehydrated in a graded ethanol series (2 h each time). The stem fragment was then incubated in xylene: ethanol 1:3, 2:2, 3:1 in sequence, and then incubated in 100% xylene twice for 2 h each time. Following that, the stem fragments were incubated overnight in dimethylbenzene: paraplast 3:1 at 63°C, and then transferred into pure paraplast for an overnight treatment at 63°C.
The stem fragments (0.5 cm long) embedded in the paraplast were cut into stem sections (1 μm thick) with a Leica EM UC6 microtome, and then stained with 0.01% Calcofluor White, and the cellulose was observed with an inverted UV fluorescence microscope. Under this condition, only the secondary walls show bright fluorescence. At the same time, the stem sections, which were cut into 50 μm thick, were stained with phloroglucinol-HCl for observing the lignin, which takes on bright red under an optical microscope. To examine the xylan contents, stem sections (1 μm thick) were probed with LM10 monoclonal antibodies, which are capable of binding to 4-O-methylglucuronoxylan, and detected with fluorescein isothiocyanate-conjugated secondary antibodies. The fluorescence-labeled xylan signals were visualized and imaged with an Olympus DX51 light microscope.
Determination of Content of Cellulose, Hemicellulose, and Lignin
The determination of the contents of lignin, cellulose, and hemicellulose was conducted by following a previously published method (Liu et al., 2017) with a ANKOM 2000i Automatic fiber analyzer (Ankom).
Gene Expression Analysis of Poplar
Five microgram total RNA from xylem of stems spanning PI = 5 to PI = 8 of PtrGATA12 transgenic lines was used for synthesizing cDNA. Samples of cDNA were run in triplicate in an Applied Biosystems 7,500 Real-Time PCR System to determine the critical threshold (Ct) with the SYBR premix ExTaq kit (TaKaRa).
Primers used for quantitative RT-PCR (qRT-PCR) of the Potri.001G188500, Potri.006G237700, Potri.018G044900, and Potri.T158300 are listed in Supplementary Table S3. Analysis of expression levels of genes involved in cellulose biosynthesis pathways genes (PtrCESA4, PtrCESA7, and PtrCESA8; Suzuki et al., 2006), xylan biosynthesis pathways genes (PtrGT43A, PtrGT47C, and PtrGT8F; Zhou et al., 2006, 2007), lignin biosynthesis (PtrPAL4, PtrC4H1, PtrC3H3, Ptr4CL5, PtrCCoAOMT3, PtrCOMT2, PtrCCR2, PtrCAld5H2, and PtrCAD1), PCD (PtrXCP1, PtrXCP2, PtrRNS3, and PtrBFN1; Hussey et al., 2013; Cubria-Radio and Nowack, 2019), and well-known wood-associated TFs (PtrWND6A, PtrWND6B, PtrMYB2, PtrMYB21, PtrMYB157, PtrMYB221, PtrMYB28, PtrMYB152, PtrNAC105, PtrMYB128, PtrMYB52, and PtrMYB54) in poplars (Zhong et al., 2010a, 2011; Zhong and Ye, 2010; Hussey et al., 2013; Zhang et al., 2018b, 2020; Cubria-Radio and Nowack, 2019), were performed using qRT-PCR primers (Supplementary Table S3). PtrActin was employed as internal controls, and the delta-delta CT method was used to quantify gene expression levels relative to PtrActin (Taylor et al., 2019).
Transient Expression Assay
The full coding region of PtrGATA12 was cloned into pROKII under the control of the CaMV 35S promoter as the effector construct. The reporter construct contained the β-glucuronidase (GUS) reporter gene driven by a 2-kb promoter of genes, such as PtrWND6A, PtrWND6B, PtrMYB2, PtrMYB21, PtrMYB152, PtrMYB52, PtrCCoAOMT3, PtrCOMT2, PtrC3H3, PtrCAD1, PtrXCP1, and PtrGT47C, which were chosen based on the expression analysis results of genes related to wood formation in PtrGATA12 transgenic lines and the previous study of AtGATA12 in A. thaliana, the full coding region of PtrGATA12 was cloned into pROKII under the control of the CaMV 35S promoter as the effector construct. The reporter construct contained the GUS reporter gene driven by a 2-kb promoter of a TF for testing, which was amplified using the primer listed in Supplementary Table S4 from the P. trichocarpa genomic DNA. Both effector and reporter constructs were cotransfected into tobacco leaves by Agrobacterium tumefaciens-mediated transient transformation (Ji et al., 2014). After agroinfiltration, plants were covered with a transparent plastic cover and transferred into a growth chamber at 25°C with 16/8 h light/dark cycle for 3 days. The transfection leaves were soaked with 100 mM MG-132 (Wako Pure Chemical) for 6 h. Then, the total protein of leaves was extracted in the extraction buffer (Yoshizumi et al., 1999). The GUS activity was measured using the 4-MUG (4-methylumbelliferyl-β-D-glucuronide) assay, and was estimated as the mean of three independent assays.
Analysis of the Downstream cis-Elements of the PtrGATA12
Three tandem copies of the secondary NAC binding element (SNBE; Zhong et al., 2010b; McCarthy et al., 2011) and secondary MYB responding element (SMRE; Zhong et al., 2010b; Supplementary Table S5) were inserted into pHIS2 (Clontech) upstream of the reporter HIS3, respectively. The full CDS of PtrGATA12 amplified used primers in Supplementary Table S4 was inserted into pGADT7-Rec2 as the effector vector. Both constructs were co-transformed into Y187 yeast cells, which were plated onto TOD plus 60 mM 3-amonotrizole at 30°C for 3–5 days. The cotransformation of pGADT7-rec2-p53 and pHIS2-p53 were used as a positive control, and the pGADT7-rec2-PtrGATA12 and pHIS2-p53were used as a negative control. The interactions of these sequences with the PtrGATA12 were studied using yeast one-hybrid analysis as aforementioned. The identification of these motifs in the putative target genes promoter regions used ExactSearch tool (Gunasekara et al., 2016).
Statistical Analysis
The Dunnett’s test (SPSS 17.0) was used to test statistical significance of data. Difference between two groups of data for comparisons in this study were evaluated by statistical significance (*, 0.01 < p < 0.05) or very significance (**, p < 0.01).
Results
Isolation and Characterization of P. trichocarpa Ortholog of AtGATA12
To identify P. trichocarpa ortholog of AtGTAG12 (AT5G25830), a combined phylogenetic tree was first generated using 39 PtrGATA proteins and 30 AtGATA proteins. According to the results of phylogenetic analysis of GATAs in A. thaliana (Reyes et al., 2004), we also divided 39 PtrGATAs into four subgroups (I, II, III, and IV), which had 18, 10, 9, and 2 PtrGATA members, respectively (Figure 1A). Among the subgroup I, four poplar proteins, Potri.001G188500, Potri.006G237700, Potri.018G044900, and Potri.T158300, shared a clade with AtGTAG12, suggesting that there were four poplar orthologs of AtGTAG12 (Figure 1A).
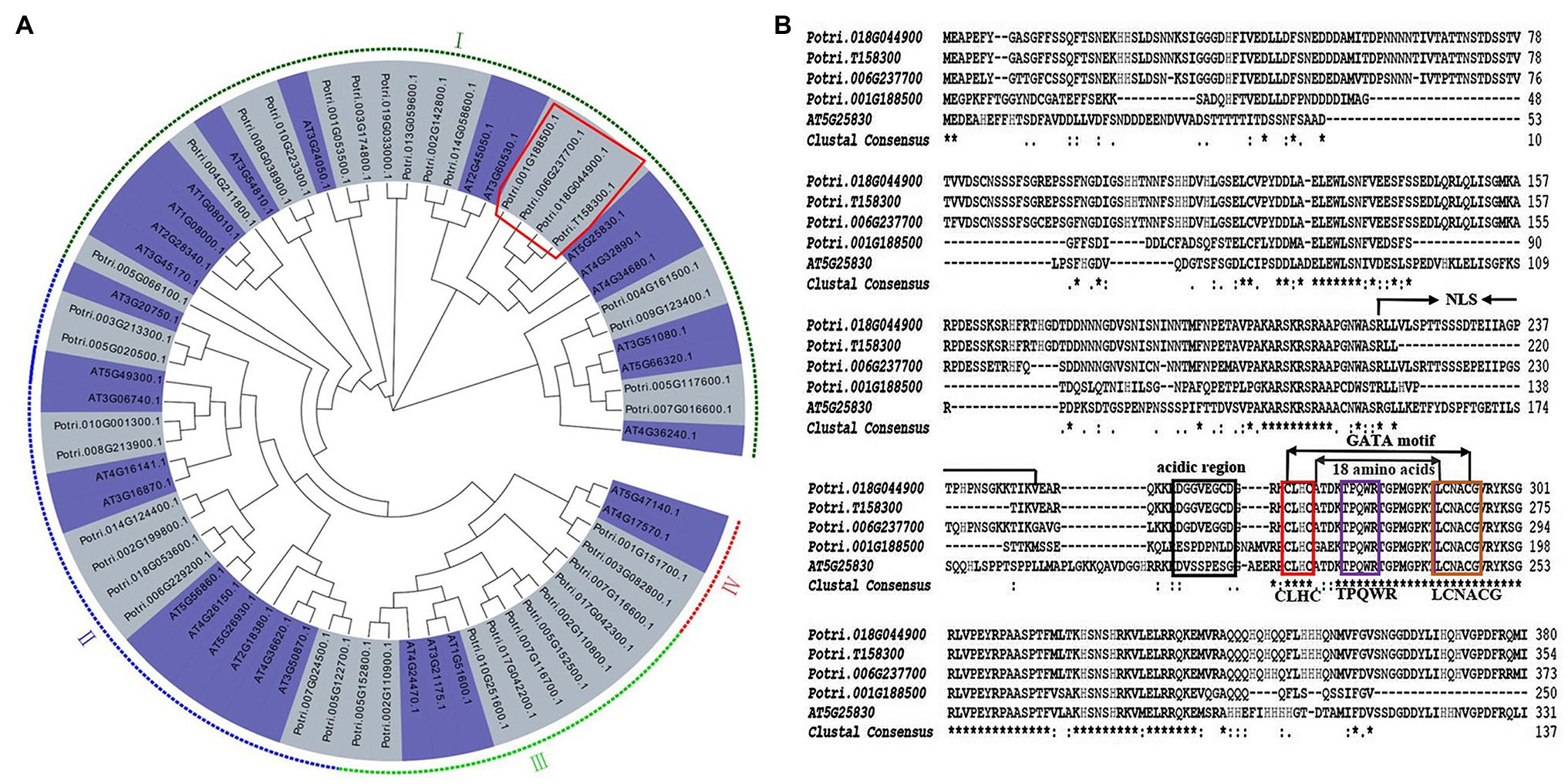
Figure 1. Phylogenetic analysis and protein sequence alignment of PtrGATA12. (A) Phylogenetic analysis of PtrGATA12 with other GATA proteins in Populus trichocarpa and Arabidopsis thaliana. PtrGATA12 proteins were shown in a red rectangle. The tree showed four major phylogenetic subfamilies (subfamilies I to IV) indicated with different colored backgrounds. (B) AtGATA12 protein (AT5G25830) of A. thaliana was aligned with PtrGATA12 proteins from P. trichocarpa (Potri.001G188500, Potri.006G237700, Potri.018G044900, and Potri.T158300). The conserved CLBC, TPQWR, and LCNACG in GATA motif are indicated by red box, blue box, and brown box, respectively; NLS, the putative domains for nuclear localization signals; the boxed region indicates the putative activation domain.
The multiple alignments revealed that these four PtrGATA proteins exhibited 86, 96, 96, and 96% similarity of amino acid sequence with AtGTAG12, respectively. Moreover, these PtrGATA proteins, which contain the integrated conserved GATA domain according to the zinc finger configuration C-x2-C-x18-C-x2-C, have the following features: (1) the presence of two pairs of Cys residues within the predicted zinc finger domain that are each separated by two amino acids (CLHC); (2) a loop of 18 amino acids between the secondary and third Cys residues; (3) conservation of the amino acid sequence LCNACG around the second Cys pair; and (4) the presence of conserved TPQWR motifs within the X18loop (Figure 1B). In addition, these PtrGATA proteins contained a region with 34 aa (aa211–245) that might serve as a nuclear location signal (NLS), and an acidic amino-terminal domain that might act as an activation domain (Figure 1B).
To determine which of these four PtrGATAs had the similar functions as ATGATA12 in the wood formation, we further analyzed their expression levels in various tissues of poplar by qRT-PCR. The result showed that these four genes were differently expressed at detectable levels in all examined tissues, suggesting that they would play different roles in the development of diverse tissues of poplars. Among them, the transcript levels of Potri.006G237700 in the transition and secondary xylem were higher than those in any other studied tissues or those of other three PtrGATA transcripts in the same tissues. This preferential expression in developing secondary xylem tissues suggests that Potri.006G237700 is most likely to be involved in SCW biosynthesis among these four genes as ATGATA12 is (Figures 2A–D). Thus, the Potri.006G237700 was given the nomenclature of PtrGATA12, in resemblance to ATGATA12, and chosen for functional characterization related to wood formation as described below.
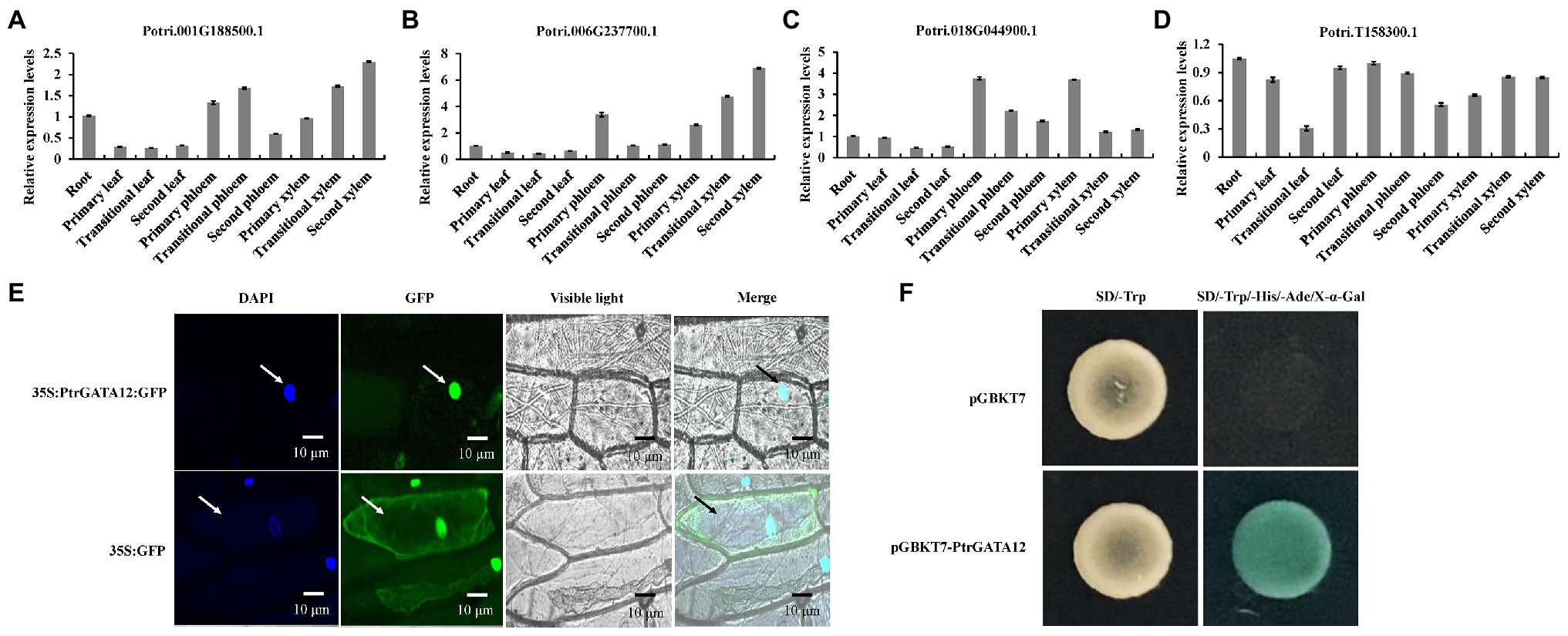
Figure 2. Tissue-specific expression patterns, subcellular localization and transcriptional activity of PtrGATA12. (A) The tissue-specific expression patterns of Potri.001G188500 gene as determined by quantitative RT-PCR (qRT-PCR) analysis. (B) The tissue-specific expression patterns of Potri.006G237700 gene as determined by qRT-PCR analysis. (C) The tissue-specific expression patterns of Potri.018G044900 gene as determined by qRT-PCR analysis. (D) The tissue-specific expression patterns of Potri.T158300 gene as determined by qRT-PCR analysis. PtrActin was used as a control. Error bars represent the SD of three biological replicates. (E) Subcellular localization of PtrGATA12. Confocal images manifested the localization of PtrGATA12-green fluorescent proteins (GFP) in the nuclei of onion epidermal cells. DAPI, a nuclear staining dye; Merged: the merged images of bright-field, GFP, and DAPI staining. Arrows indicate cells located in the epidermis of onions. (F) Transcriptional activation analysis of PtrGATA12 fused with the GAL4 DNA binding domain (GAL4DB) in yeast shows its potential to activate the expression of the His-3 and X-α-Gal reporter genes.
Subcellular Localization and Transcriptional Activation Activity of PtrGATA12
The presence of a nuclear localization signal in the PtrGATA12 may indicate that the protein is likely to localize in the nuclei (Figure 2E). To verify this, the PtrGATA12 coding region was fused to the N-terminus of the GFP gene under the control of the cauliflower mosaic virus (CaMV) 35S promoter and transferred into onion epidermal cells using the particle gun bombardment. Localization of the fusion protein was then visualized with a fluorescence confocal microscope. As seen in Figure 2E, the PtrGATA12-GFP fusion protein was exclusively colocalized to DAPI-stained nuclei, indicating that PtrGATA12 encodes a nuclear-localized protein. In addition, the presence of acidic amino-terminal domain next to GATA domain in the C-terminal region suggests that PtrGATA12 is likely a transcriptional activator. To verify this, we fused PtrGATA12 with the GAL4 DNA-binding domain and test its potential to activate the reporter gene expression in yeast. It was found that PtrGATA12 could activate the expression His-3 and X-α-Gal reporter genes (Figure 2F), indicating that it is indeed a transcriptional activator. These results demonstrated that PtrGATA12 is a transcriptional activator located in nuclei.
Alternation of Growth-Related Traits in PtrGATA12 Transgenic Poplar
To investigate the biological roles of PtrGATA12, we expressed PtrGATA12 under the control of the 35S promoter in wild-type (WT) P. trichocarpa. In total, seven transgenic poplar lines, which exhibited normal phenotype as WT, were generated and corroborated to harbor the transformed PtrGATA12 by genomic PCR (Figure 3A). The PtrGATA12 expression levels in these transgenic lines were then quantitatively analyzed using qRT-PCR. Three of these PtrGATA12 transgenic lines, OE-1, OE-3, and OE-6, which had higher expression levels of PtrGATA12 than other transgenic lines, were chosen for further characterization (Figure 3B). Although these three PtrGATA12 transgenic lines did not exhibit abnormal phenotypes, the growth potentials of them were slightly attenuated compared with WT (Figure 3C). For example, the leaf lengths and widths, internode lengths, and heights of PtrGATA12 transgenic lines were reduced by 11.4 and 10.3, 12.0, and 13.4% on average than those of the WT, respectively (Figures 4A–D). The fresh and dry weights decreased by 21.7 and 15.6% on average in PtrGATA12 transgenic lines than in WT, respectively (Figures 4E,F). In addition, it was worth noting that the diameters of stems above ground and breaking forces of stems increased by 21.0 and 14.9% on average in PtrGATA12 transgenic lines compared with WT, respectively (Figures 4G,H). These results indicated that PtrGATA12 overexpression may to some extent repress vegetative growth and decrease biomass for transgenic lines.
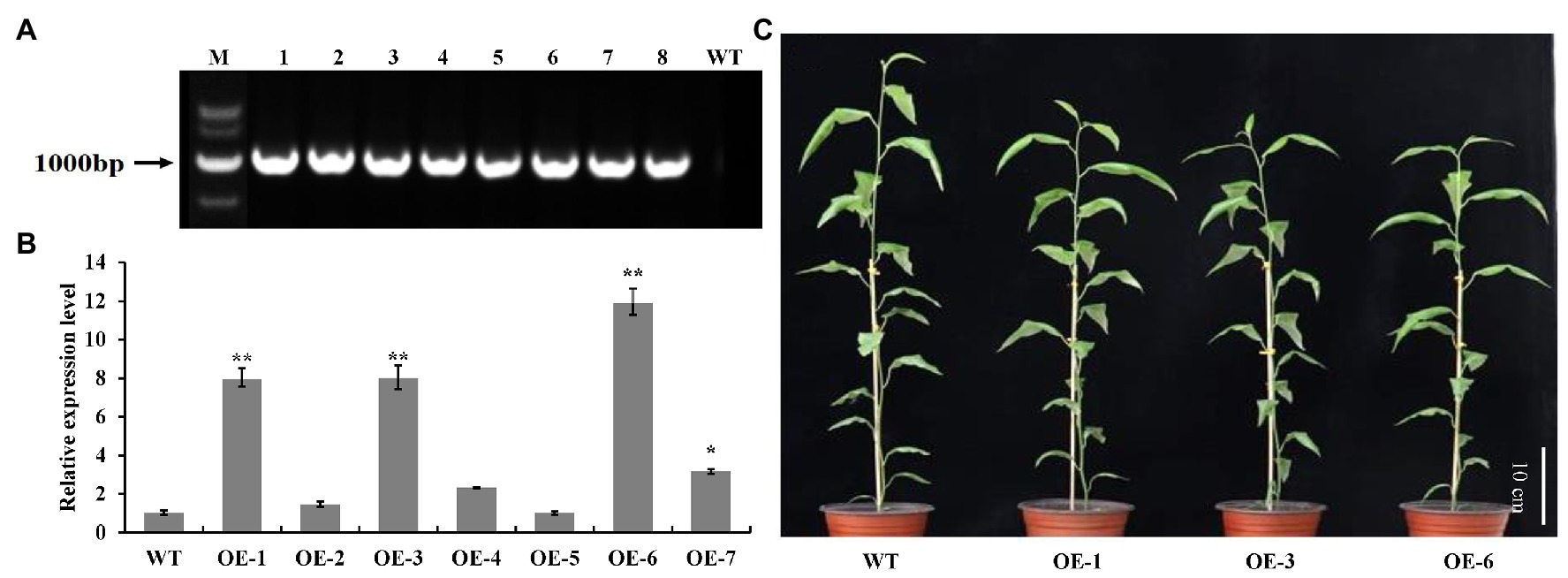
Figure 3. Identification and transgenic lines of PtrGATA12 in P. trichocarpa. (A) PCR detection of PtrGATA12 transgenic lines. M, DNA marker DL5000; 1, positive control; 2–8, PCR products of PtrGATA12 transgenic lines; WT, wild-type P. trichocarpa. (B) qRT-PCR detection of PtrGATA12 transgenic lines. PtrActin was used as a control. Each error bar represents the SD of three biological replicates. Asterisks indicate levels of significance (Dunnett’s test; *, 0.01 < p < 0.05, **, p < 0.01). (C) Three-month-old wild-type (WT) and PtrGATA12 transgenic lines (OE-1, OE-3, and OE-6). Scale bars = 10 cm.
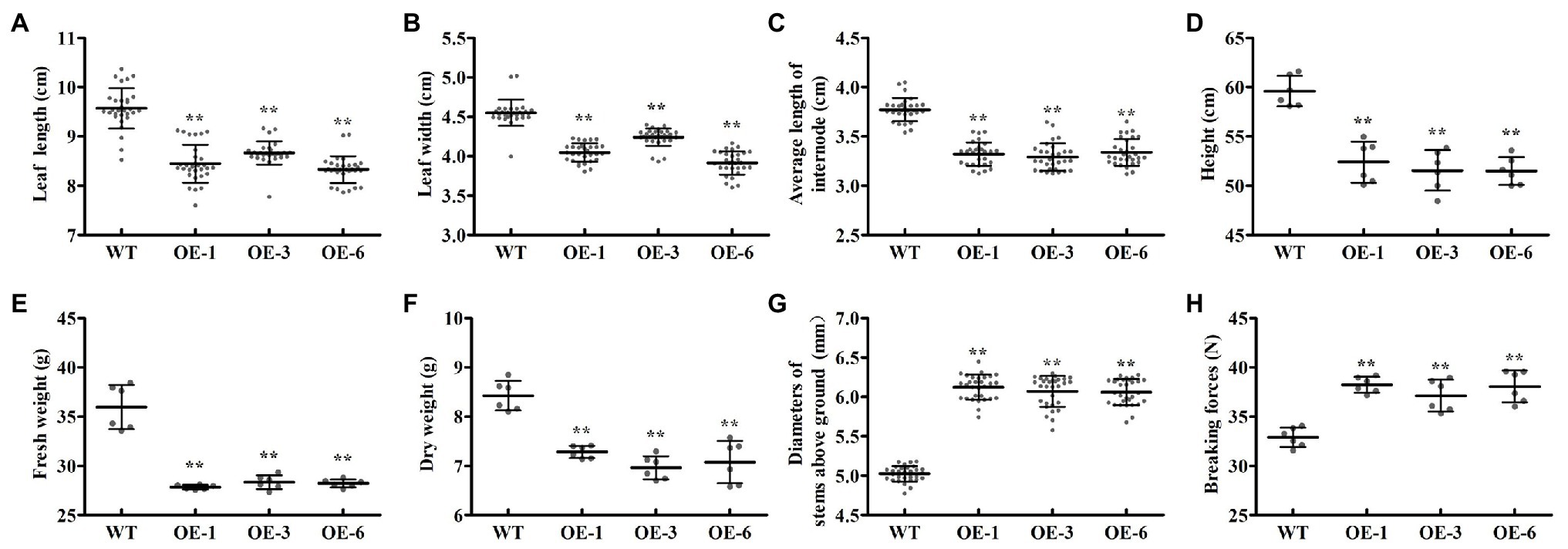
Figure 4. Effect of PtrGATA12 overexpression on growth-related traits in P. trichocarpa. (A–H) Represent the leaf lengths, leaf widths, average lengths, plant heights, fresh weights, dry weights internode base diameters, and breaking forces of 90 days old WT and PtrGATA12 transgenic lines (OE-1, OE-3, and OE-6), respectively. Each error bar represents the SD of three biological replicates. Asterisks indicate levels of significance (Dunnett’s test; **, p < 0.01).
Changes in the Characteristics of Wood Caused by PtrGATA12 Overexpression in P. trichocarpa
To investigate whether PtrGATA12 had similar function as AtGATA12 does in wood formation, we analyzed the wood characteristics in the bottom stems of the aforementioned three PtrGATA12 transgenic lines. The scanning electron microscope of stem cross-sections revealed that the SCW thicknesses of fiber and vessel increased by 20.83 and 11.83% on average in the PtrGATA12 transgenic lines than those of WT, respectively (Figures 5A–J).
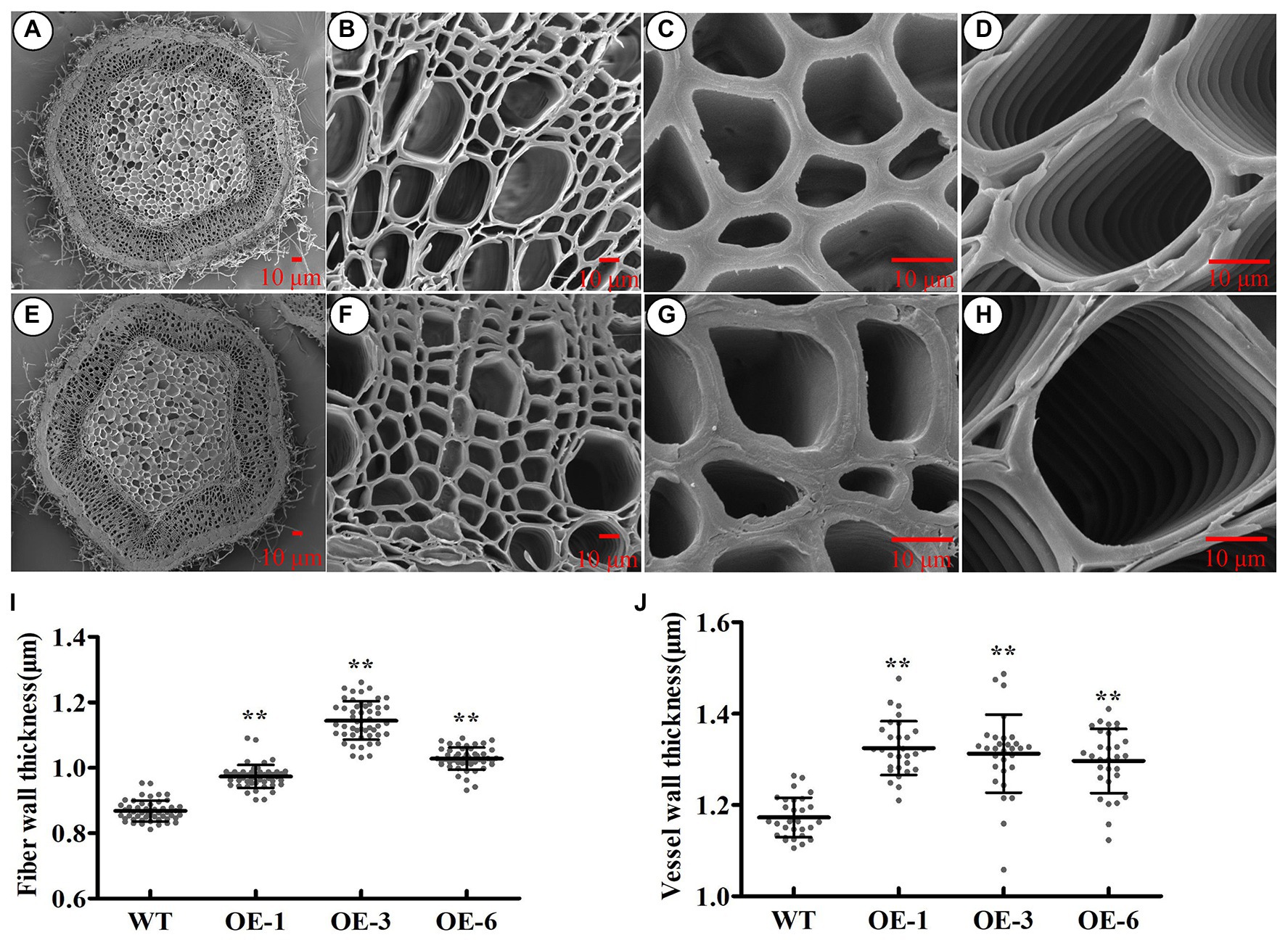
Figure 5. Effect of PtrGATA12 overexpression on the secondary wall thickness of stems in P. trichocarpa. (A–H) The scanning electron microscope of cross stem sections of 90 days old wild-type (A–D) and PtrGATA12 transgenic lines (E–H). Scale bars = 10 μm. (C,G) The scanning electron microscope of fiber walls in wild-type and PtrGATA12 transgenic lines. (D,H) The scanning electron microscope of vessel walls in wild-type and PtrGATA12 transgenic lines. (I) The fiber wall thickness of cross stem sections in WT and PtrGATA12 transgenic lines (OE-1, OE-3, and OE-6). (J) The vessel wall thickness of cross stem sections in WT and PtrGATA12 transgenic lines. Error bars represent SD of three biological replicates. Asterisks indicate levels of significance (Dunnett’s test; *, 0.01 < p < 0.05, **, p < 0.01).
To identify which component (e.g., cellulose, lignin, or hemicellulose) contributed to SCW thickening, the phloroglucinol-HCl and calcofluor white were used to stain lignin and cellulose, respectively, and the monoclonal antibody LM10 was used to label xylan immunologically. As a result, the intensities of phloroglucinol-HCl and antibody LM10 were more striking in PtrGATA12 transgenic lines than in WT, while the intensities of calcofluor white staining were only slightly stronger in PtrGATA12 transgenic lines than in WT (Figures 6A–F). These results demonstrated that the lignin and hemicellulose contents increased while the cellulose deposition had no obvious increase in the PtrGATA12 transgenic lines than in WT.
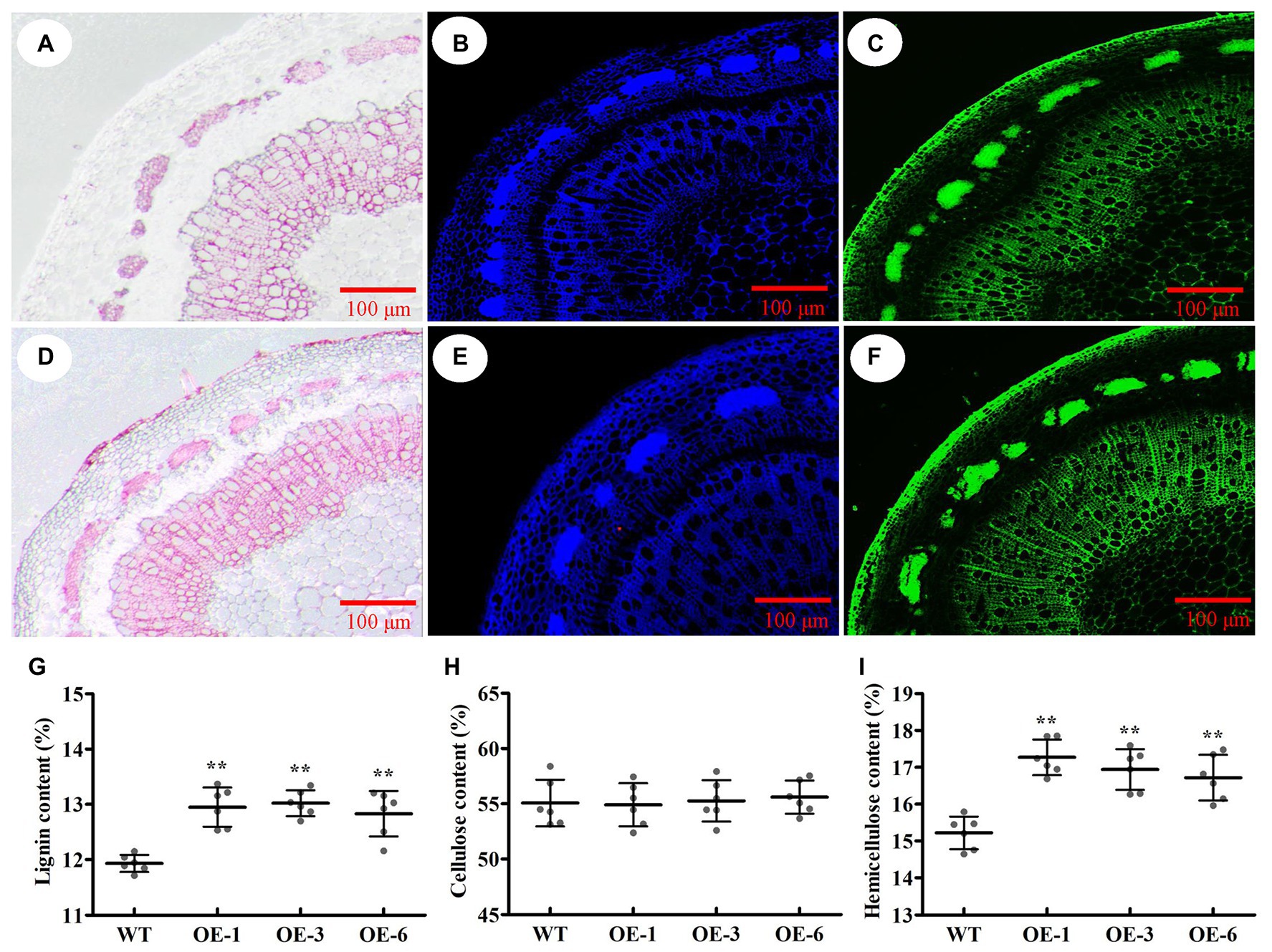
Figure 6. Effects of PtrGATA12 overexpression on components of secondary cell wall of stems in P. trichocarpa. (A,D) Phloroglucinol-HCl staining (red color) stem sections of 90 days old wild-type (A) and PtrGATA12 transgenic lines (D). (B,E) Calcofluor white staining (blue color) stem sections of wild-type (B) and PtrGATA12 transgenic lines (E). (C,F) Monoclonal antibody LM10 (green color) stem sections of wild-type (C) and PtrGATA12 transgenic lines (F). Scale bars = 100 μm. (G) The lignin content of stems in WT and PtrGATA12 transgenic lines (OE-1, OE-3, and OE-6). (H) The cellulose content of stems in WT and PtrGATA12 transgenic lines. (I) The hemicellulose content of stems in WT and PtrGATA12 transgenic lines. Error bars represent SD of three biological replicates. Asterisks indicate levels of significance (Dunnett’s test; **, p < 0.01).
To accurately assess these alternations of SCW compositions, the chemical analysis of SCW compositions was conducted using an automatic fiber analyzer and the results showed 11.5 and 8.4% increase in hemicellulose and lignin contents, respectively, and no-significant increases of cellulose content in PtrGATA12 transgenic lines as compared to the WT (Figures 6G–I). Taken together, these results implicated that PtrGATA12 is involved in regulating biosynthesis of some components especially lignin and hemicellulose of SCW.
Wood Formation Pathway Genes and TFs Affected by PtrGATA12 Overexpression in Poplar
To investigate the molecular basis of the alternations of wood characteristics described above, we further studied the expressions of genes related to SCW biosynthesis in the stems of PtrGATA12 transgenic lines. The results from qRT-PCR analysis demonstrated that the transcript levels of lignin biosynthesis genes, including PtrC3H3, PtrCCoAOMT3, PtrCOMT2, and PtrCAD1, and xylan biosynthesis genes, such as PtrGT47C, were significantly increased in the PtrGATA12 transgenic lines compared with the WT (Figure 7). On the contrary, the cellulose biosynthesis genes, such as PtrCESA4, PtrCESA7, and PtrCESA8, exhibited no obvious upregulation in PtrGATA12 transgenic lines compared with WT. In addition, it is worth mentioning that PtrXCP1, whose counterpart in A. thaliana is specifically expressed in xylem tissues at the onset of PCD, was also significantly upregulated in PtrGATA12 transgenic lines compared with WT (Figure 7).
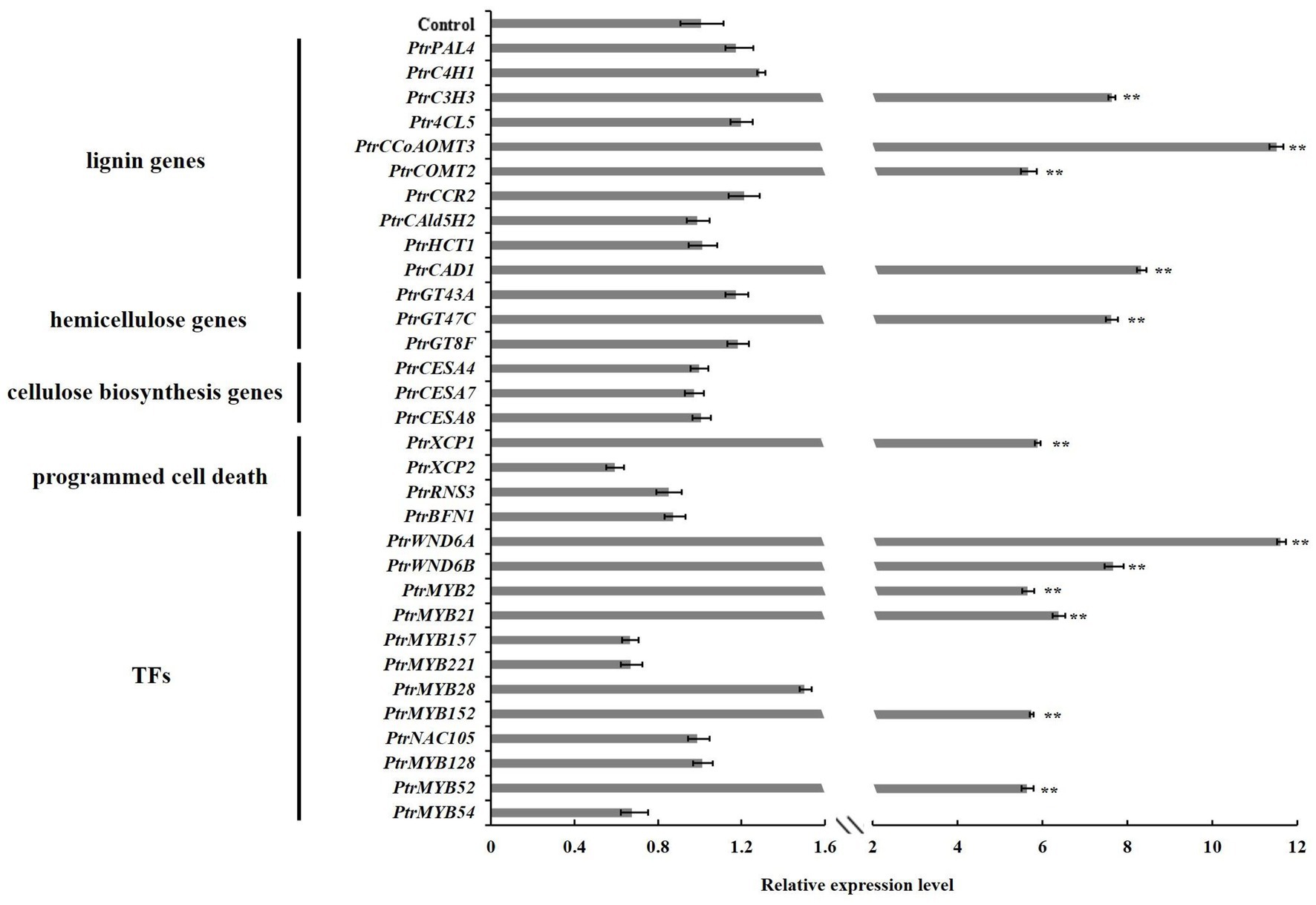
Figure 7. Expression analysis of wood formation pathway genes and their regulatory TFs in 90 days old wild-type and PtrGATA12 transgenic lines. Control represents the normalized expression level (namely 1 in this case) of each wood formation gene examined in wild-type plants. Error bars represent SD of three biological replicates. Asterisks indicate levels of significance (Dunnett’s test; **, p < 0.01).
Given the fact that AtGATA12 is an upstream TF of AtVND7 (Endo et al., 2015), we investigated whether PtrGATA12 could affect the expressions of poplar orthologs of AtVND7, PtrWND6A, and PtrWND6B as well as other well-known wood-associated TFs in transgenic poplar. The qRT-PCR analysis revealed that the transcript abundances of master switches, such as PtrWND6A, PtrWND6B, PtrMYB2, and PtrMYB21 (Zhang et al., 2018b, 2020), were significantly upregulated in PtrGATA12 transgenic lines compared with WT (Figure 7). In addition, the TFs specifically activating lignin biosynthesis, such as PtrMYB152, were significantly upregulated in PtrGATA12 transgenic lines compared with WT. The expression levels of PtrNAC105 and PtrMYB128 involved in activation of cellulose biosynthesis (Zhang et al., 2020) showed no obvious increases in PtrGATA12 transgenic lines compared with WT (Figure 7). Moreover, the TFs related to activation of PCD, such as PtrMYB52 (Zhang et al., 2018b), was significantly upregulated in PtrGATA12 transgenic lines compared with WT. These results are aligned well with the alternations of SCW characteristics observed in PtrGATA12 transgenic lines.
Regulation of Poplar TFs and Pathway Genes Involved in Wood Formation by PtrGATA12 in Tobacco
Based on fact that PtrGATA12 significantly altered the expression of a number of wood-associated genes in the poplar transgenic lines, we investigated whether PtrGATA12 can directly control these genes using a transient expression system in tobacco. We selected those TFs and pathway genes whose expression had a fold change of more than five times in PtrGATA12 poplar transgenic lines compared with WT for testing. The 2-kb proximal promoter regions of these genes amplified from P. trichocarpa genomic DNA were linked to the GUS reporter gene to create the reporter constructs, and the full-length cDNA of PtrGATA12 was ligated to 35S promoter to generate the effector construct (Figures 8A,B). The reporter and effector constructs were co-transfected into tobacco leaves by Agrobacterium-mediated method. The subsequent assay of the GUS activity in the transfected leaves demonstrated that PtrGATA12 significantly activated the activities of PtrWND6A, PtrWND6B, PtrMYB21, and PtrMYB152 promoters (Figure 8C), albeit to different levels. It was notable that PtrGATA12 significantly activated the activities of promoters of lignin biosynthesis genes, PtrC3H3 and PtrCAD1 (Figure 8C). However, PtrGATA12 had no regulatory effects on the activities of PtrMYB2, PtrMYB52, PtrCCoAOMT3, PtrCOMT2, PtrXCP1, and PtrGT47C promoters (Figure 8C). These results suggested that PtrGATA12 could directly control some TFs and pathway genes during SCW biosynthesis.
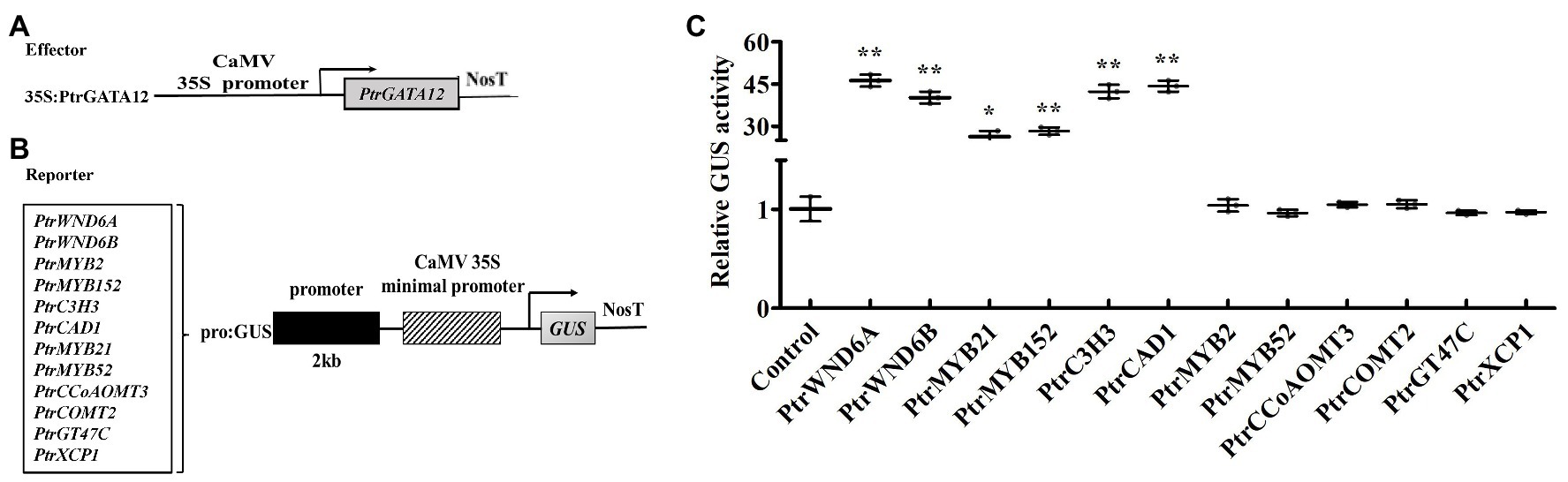
Figure 8. Activation or repression of the promoters of poplar transcription factors and secondary cell wall (SCW) pathway genes by PtrGATA12. (A,B) Diagrams of the effector and reporter constructs. (C) The expression of the β-glucuronidase (GUS) under the control of those promoters by PtrGATA12. GUS activity in tobacco leaves transfected with the reporter construct alone was used as a control and was set to 1. Error bars represent SD of three biological replicates. Asterisks indicate levels of significance of differential expression (Dunnett’s test; *, 0.01 < p < 0.05, **, p < 0.01).
The Potential of PtrGATA12 as a Higher Hierarchical TF
To test whether PtrGATA12 directly regulated these aforementioned genes through binding to the cis-acting elements in their promoters, a yeast one-hybrid assay was performed to test the potential of PtrGATA12 binding to SNBE and SMRE cis-acting elements (Supplementary Table S5), which are often present in the promoters of those genes that are directly controlled by PtrGATA12 (Supplementary Table S6) and those that are components of the currently known hierarchical transcriptional network of secondary wall formation (Ye and Zhong, 2015). The results manifested that PtrGATA12 had obvious binding affinities to these cis-acting (Figure 9), which demonstrated that PtrGATA12 as a higher hierarchical TF could bind to specific cis-acting elements to directly control TFs at low hierarchy and pathway genes during SCW biosynthesis.
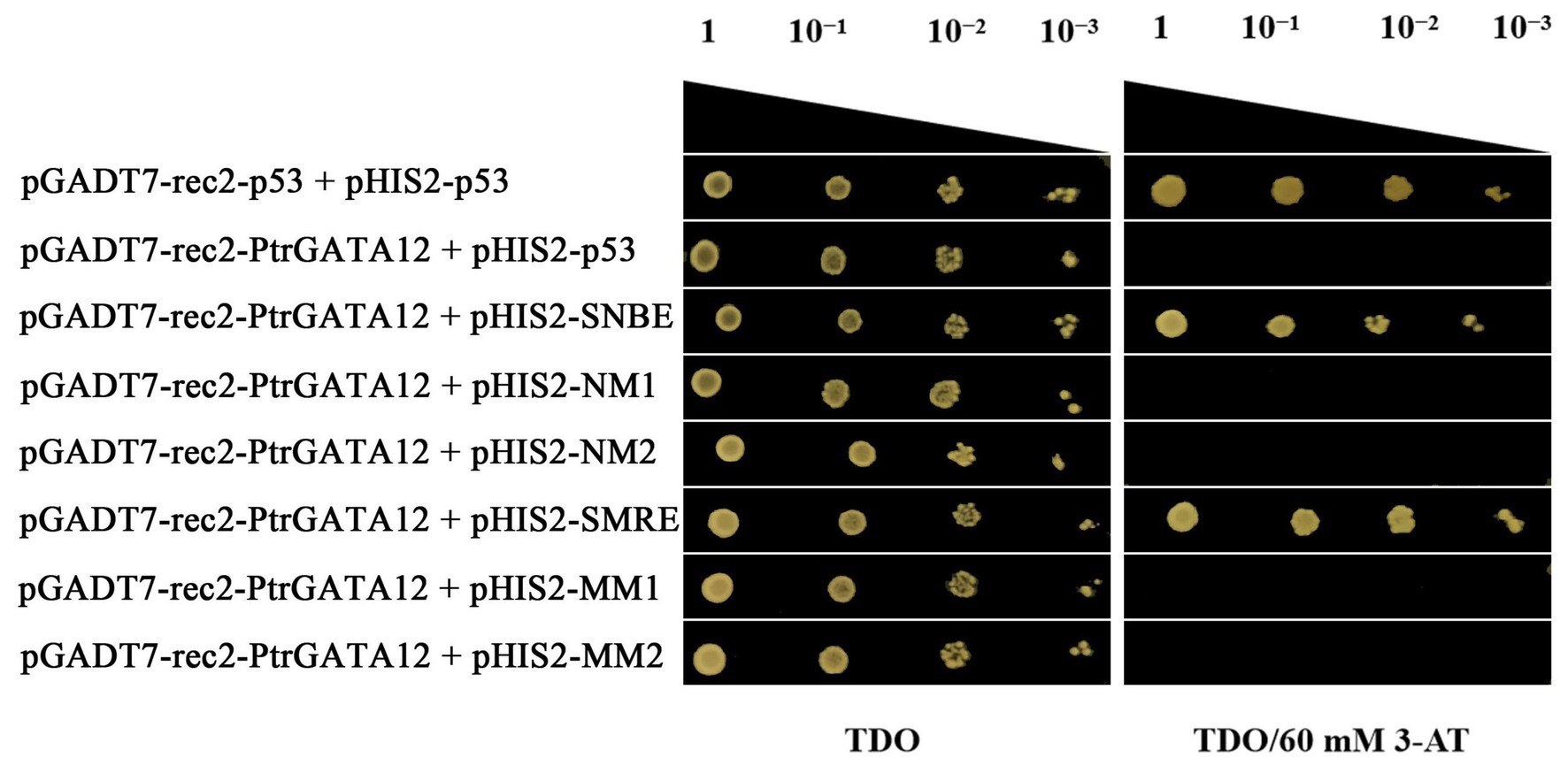
Figure 9. Yeast one-hybrid assay of PtrGATA12 binding to cis-acting elements. The pGADT7-rec2-p53/pHIS2-p53 and pGADT7-rec2-PtrGATA12/pHIS2-p53 were used as the positive and negative control, respectively. 10−1, 10−2, and 10−3 represent solution dilution ratio of transformed Y187 yeast cells.
Discussion
In the last decade, considerable studies have revealed that the SCW biosynthesis is mainly regulated by a multilayered hierarchical transcription regulation network with the regulatory edges, chains of command, pointing downward the pathway genes at the low hierarchy, which ensures a differential regulation on the formation of diverse SCW with varied components and thicknesses in various cells and across different tissues (Zhong et al., 2007, 2010a; Zhou et al., 2009; Du and Groover, 2010; Yamaguchi et al., 2010). Generally speaking, the regulators with higher hierarchy are more likely serve as master regulators or the regulatory switches exerting different regulation on multiple pathways (Balazsi et al., 2005). For example, in P. trichocapa, a well-characterized higher hierarchical regulator is PtrSND1-B1, which mediates a four-layered network (Chen et al., 2019). In this network, PtrSND1-B1 directly regulates Ptr-MYB021 and PtrMYB074, whose proteins, in turn, regulate 15 TFs, PtrNAC127, PtrMYB175, PtrWBLH3, PtrHAM3, PtrMYB059, PtrMYB090, PtrMYB161, PtrMYB174, PtrNAC123, PtrWBLH, PtrWBLH2, PtrMYB088, PtrMYB093, PtrNAC105, and PtrNAC126; these 15 TFs then directly control some lignin biosynthetic genes, PtrCAD1, PtrHCT1, PtrHCT6, PtrC4H1, PtrCOMT2, PtrCCoAOMT1,2, and PtrCAld5H1,2, and three cellulose synthases, PtrCesA4/17/18. Chen et al. (2019) also characterized the AtSND1-mediated network in A. thaliana in their study, and found that it has only three layers and is very different from PtrSND1-B1-mediated network in poplars. AtSND1 directly regulates 14 TFs (including at AtMYB46, a counterpart of PtrMYB21; Ezcurra et al., 2011) and seven cell wall biosynthetic genes, and AtMYB46 regulates 17 TFs and 12 SCW biosynthetic genes (including eight lignin pathway genes). In our study, we found that the PtrGATA12 was a regulator of PtrMYB21, a target gene of PtrSND1-B1 too, indicating that PtrGATA12 is at the same or higher hierarchical level than that of PtrSND1-B1. This is because: (1) PtrMYB21 was significantly upregulated more than five times in PtrGATA12 transgenic lines as compared to WT (Figure 7). (2) The yeast one-hybrid assay also confirmed that PtrGATA12 could bind the two SMRE motifs (ACCAACT) and SNBE motif (AACCTTGAGTTTGAAGGAT; Figure 9) and activated the reporter genes in yeast. Both motifs are present in in the 2 kb promoter of PtrMYB21 (Supplementary Table S6). (3) The overexpression of PtrGATA12 using the transient GUS-activation system in tobacco lead to a significant upregulation of PtrMYB21, which may indicate that PtrGATA12 could directly activate PtrMYB21. Collectively, these results demonstrate and suggest that PtrGATA12 is higher hierarchical regulator than PtrMYB21.
In addition to PtrMYB21, our study using GUS activation system also demonstrated that PtrWND6A and PtrWND6B are target genes of PtrGATA12 and their promoter regions (2 kb) contain both SNBE and SMRE motifs (Supplementary Table S6). These two genes are most phylogenetically related to VND7 (Ye and Zhong, 2015), which is a higher hierarchical regulator/switch regulating xylem vessel differentiation of protoxylem vessels in Arabidopsis (Endo et al., 2015). PtrWNDs have been indicated to serve as higher hierarchical regulators of not only wood associated MYB master switches (e.g., PtrMYB2, 3, 20, and 21), but also TFs activating all three SCW biosynthesis pathways (e.g., PtrMYB128, PtrNAC150, PtrMYB75, and PtrMYB18; Ye and Zhong, 2015). Our transient GUS-activation system in tobacco and yeast one-hybrid assays analysis revealed that PtrGATA12 could directly activate PtrWND6A/B, PtrMYB21, PtrMYB152, PtrC3H3, and PtrCAD1 through binding to SNBE, and SMRE cis-elements present in their promoter regions (Figures 8C, 9). These results explicitly suggest that PtrGATA12 modulate SCW biosynthesis pathways through directly regulating some high hierarchical regulators including PtrWND6A, PtrWND6B, and PtrMYB21, and indirectly regulating well-known wood-associated TFs and pathway genes that are elements of transcription regulation network of poplar SCW formation. In line with alternations of SCW components, the expression of pathway genes related to lignin and xylan biosynthesis and PCD exhibited significant changes in PtrGATA12 poplar transgenic lines (Figure 7). Besides these, the expressions of TFs in different layers of hierarchical transcription regulation network, such as PtrMYB28, PtrMYB157, PtrMYB221, PtrMYB52, and PtrMYB54 (Mellerowicz and Sundberg, 2008; Zhong et al., 2008, 2010a, 2011; Zhong and Ye, 2010; Taylor-Teeples et al., 2015; Rao and Dixon, 2018), also exhibited obvious alternations in PtrGATA12 poplar transgenic lines (Figure 7). As a result, PtrGATA12 switched on hemicellulose and lignin pathway while maintaining the cellulose biosynthesis largely unchanged.
Previous study also showed that A. thaliana TF, SHINE2 (SHN2), downregulates the lignin biosynthesis while upregulating the cellulose and hemicellulose biosynthesis in rice (Ambavaram et al., 2011). This is largely opposite to the biological function of PtrGATA12 in SCW formation. Based on the experimental evidence, a hypothetical model was established assuming that SHN2 serves as a hierarchical regulator that downregulate lignin biosynthetic genes through repressing MYB58/63, whereas upregulate the cellulose biosynthesis through activating MYB20/43. We previously overexpressed Populus simonii × Populus nigra PsnSHN2 in tobacco, leading to the altered the SCW characteristics, of which the most significant changes were the contents of cellulose and hemicellulose that increased 37 and 28%, respectively, whereas the content of lignin that decreased 34% (Liu et al., 2017). But, unfortunately, we did not test if PtrGATA12 regulate these of PsnSHN2’s target TFs in this study. Our study showed that PtrGATA12 regulated SCW pathways through possibly regulating PtrWND6A, PtrWND6B, PtrMYB21, and PtrMYB152, whose family members also showed in PsnSHN2 down-stream target TFs, for example, PsnWND1A, PsnWND3A, PsnMYB3, and PsnMYB152. These evidences and the differential regulation of PtrGATA12 and PsnSHN2 on SCW component biosynthetic pathways indicate that PtrGATA12 is just another very valuable high hierarchical regulator like PsnSHN2, and can be used alone or in conjunction with PsnSHN2 in genetic engineering of woody plants to produce the wood with altered SCW components to meet a variety of needs.
Besides alternations of SCW characteristics, the PtrGATA12 transgenic lines exhibited arrested growths of most phenotype traits and biomass accumulation (Figures 3C, 4), which was consistent with previous reports that the growth of plants often has strong inverse correlation with lignin accumulation (Novaes et al., 2009, 2010). In addition, the breaking forces of PtrGATA12 transgenic lines became stronger than those of WT (Figure 4H). The previous studies have revealed that the tensile or bending strength of plant stems is positively correlated with the cellulose content (Dhugga, 2007; Voelker et al., 2011; Liu et al., 2017), which did not exhibit significant increases in PtrGATA12 transgenic lines than in WT (Figure 6H), and thus was not the major contributor to the increased breaking forces in PtrGATA12 transgenic lines. We have noticed that the PtrGATA12 transgenic lines exhibited significant increases in diameters of stems (Figure 4C), SCW thickness of fibers and vessels (Figure 5), and hemicellulose content of SCW (Figure 6I), which might be responsible for the breaking forces of stem increased in PtrGATA12 transgenic lines. These changes also support that PtrGATA12, as a higher hierarchical regulator, had pleiotropic effects on SCW biosynthesis and vegetative growth in poplars through regulating multiple biological pathways in wood development.
In summary, our study demonstrated that PtrGATA12, as a higher hierarchical regulator, differentially modulates multiple SCW component biosynthetic pathways through possibly regulating multiple master regulators including PtrWND6A, PtrWND6B, PtrMYB21, and PtrMYB152. It can be specifically used for augmenting lignin and hemicellulose biosynthesis without reducing cellulose.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
MR and YZ conducted most experiments and data analysis. CL and YL participated in the histochemical staining. ST and HC participated in the vegetative propagation of Populus trichocarpa plantlets and phenotype analysis. HZ reviewed the manuscript. HW performed the data analysis and wrote the manuscript. ZW designed the experiments and wrote the manuscript. All authors read and approved the final version of the manuscript.
Funding
The experimental design and implementation, RNA/DNA extraction, data preprocessing analysis as well as interpretation were supported by the Fundamental Research Funds of Chinese Academy of Forestry (CAFYBB2020ZA005). MR was financially supported by Fundamental Research Funds of Chinese Academy of Forestry (CAFYBB2018ZY001-5). YZ, CL, YL, ST, and HC were financially supported by National Natural Science Foundation of China (31770640).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.657787/full#supplementary-material
Footnotes
References
Ambavaram, M. M., Krishnan, A., Trijatmiko, K. R., and Pereira, A. (2011). Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol. 155, 916–931. doi: 10.1104/pp.110.168641
An, Y., Han, X., Tang, S., Xia, X., and Yin, W. (2014). Poplar GATA transcription factor PdGNC is capable of regulating chloroplast ultrastructure, photosynthesis, and vegetative growth in Arabidopsis under varying nitrogen levels. Plant Cell Tissue Organ Cult. 119, 313–327. doi: 10.1007/s11240-014-0536-y
An, Y., Zhou, Y. Y., Han, X., Shen, C., Wang, S., Liu, C., et al. (2020). The GATA transcription factor GNC plays an important role in photosynthesis and growth in poplar. J. Exp. Bot. 71, 1969–1984. doi: 10.1093/jxb/erz564
Balazsi, G., Barabasi, A. L., and Oltvai, Z. N. (2005). Topological units of environmental signal processing in the transcriptional regulatory network of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 102, 7841–7846. doi: 10.1073/pnas.0500365102
Balmant, K. M., Noble, J. D., Alves, F. C., Dervinis, C., Conde, D., Schmidt, H. W., et al. (2020). Xylem systems genetics analysis reveals a key regulator of lignin biosynthesis in Populus deltoides. Genome Res. 30, 1131–1143. doi: 10.1101/gr.261438.120
Behringer, C., Bastakis, E., Ranftl, Q. L., Mayer, K. F., and Schwechheimer, C. (2014). Functional diversification within the family of B-GATA transcription factors through the leucine-leucine-methionine domain. Plant Physiol. 166, 293–305. doi: 10.1104/pp.114.246660
Behringer, C., and Schwechheimer, C. (2015). B-GATA transcription factors—insights into their structure, regulation, and role in plant development. Front. Plant Sci. 6:90. doi: 10.3389/fpls.2015.00090
Chen, H. F., Shao, H. X., Li, K., Zhang, D., Fan, S., Li, Y. M., et al. (2017). Genome-wide identification, evolution, and expression analysis of GATA transcription factors in apple (Malus x domestica Borkh.). Gene 627, 460–472. doi: 10.1016/j.gene.2017.06.049
Chen, H., Wang, J. P., Liu, H., Li, H., Lin, Y. J., Shi, R., et al. (2019). Hierarchical transcription factor and chromatin binding network for wood formation in black cottonwood (Populus trichocarpa). Plant Cell 31, 602–626. doi: 10.1105/tpc.18.00620
Cubria-Radio, M., and Nowack, M. K. (2019). Transcriptional networks orchestrating programmed cell death during plant development. Curr. Top. Dev. Biol. 131, 161–184. doi: 10.1016/bs.ctdb.2018.10.006
Dhugga, K. S. (2007). Maize biomass yield and composition for biofuels. Crop Sci. 47, 2211–2227. doi: 10.2135/cropsci2007.05.0299
Du, J., and Groover, A. (2010). Transcriptional regulation of secondary growth and wood formation. J. Integr. Plant Biol. 52, 17–27. doi: 10.1111/j.1744-7909.2010.00901.x
Endo, H., Yamaguchi, M., Tamura, T., Nakano, Y., Nishikubo, N., Yoneda, A., et al. (2015). Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 56, 242–254. doi: 10.1093/pcp/pcu134
Ezcurra, I., Johansson, C., Tamizhselvan, P., Winzell, A., and Aspeborg, H. (2011). An AC-type element mediates transactivation of secondary cell wall carbohydrate-active enzymes by PttMYB021, the Populus MYB46 orthologue. BMC Proc. 5(Suppl. 7):O40. doi: 10.1186/1753-6561-5-s7-o40
Gunasekara, C., Subramanian, A., Avvari, J. V., Li, B., Chen, S., and Wei, H. (2016). ExactSearch: a web-based plant motif search tool. Plant Methods 12:26. doi: 10.1186/s13007-016-0126-6
Hussey, S. G., Mizrachi, E., Creux, N. M., and Myburg, A. A. (2013). Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front. Plant Sci. 4:325. doi: 10.3389/fpls.2013.00325
Ji, X. Y., Zheng, L., Liu, Y. J., Nie, X. G., Liu, S. N., and Wang, Y. C. (2014). A transient transformation system for the functional characterization of genes involved in stress response. Plant Mol. Biol. Report. 32, 732–739. doi: 10.1007/s11105-013-0683-z
Ko, J. H., Kim, W. C., Kim, J. Y., Ahn, S. J., and Han, K. H. (2012). MYB46-mediated transcriptional regulation of secondary wall biosynthesis. Mol. Plant 5, 961–963. doi: 10.1093/mp/sss076
Kobayashi, K., and Masuda, T. (2016). Transcriptional regulation of tetrapyrrole biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 7:1811. doi: 10.3389/fpls.2016.01811
Kobayashi, K., Ohnishi, A., Sasaki, D., Fujii, S., Iwase, A., Sugimoto, K., et al. (2017). Shoot removal induces chloroplast development in roots via cytokinin signaling. Plant Physiol. 173, 2340–2355. doi: 10.1104/pp.16.01368
Kolosova, N., Miller, B., Ralph, S., Ellis, B. E., Douglas, C., Ritland, K., et al. (2004). Isolation of high-quality RNA from gymnosperm and angiosperm trees. BioTechniques 36, 821–824. doi: 10.2144/04365ST06
Li, S. F., Zhang, Y. X., Xin, X. B., Ding, C. J., Lv, F. L., Mo, W. J., et al. (2020). The Osmotin-Like protein gene PdOLP1 is involved in secondary cell wall biosynthesis during wood formation in poplar. Int. J. Mol. Sci. 21:3993. doi: 10.3390/ijms21113993
Li, S., Zhen, C., Xu, W., Wang, C., and Cheng, Y. (2017). Simple, rapid and efficient transformation of genotype Nisqually-1: a basic tool for the first sequenced model tree. Sci. Rep. 7:2638. doi: 10.1038/s41598-017-02651-x
Liao, Z., Chen, M., Guo, L., Gong, Y., Tang, F., Sun, X., et al. (2004). Rapid isolation of high-quality total RNA from taxus and ginkgo. Prep. Biochem. Biotechnol. 34, 209–214. doi: 10.1081/PB-200026790
Liu, Y., Wei, M., Hou, C., Lu, T., Liu, L., Wei, H., et al. (2017). Functional characterization of Populus PsnSHN2 in coordinated regulation of secondary wall components in tobacco. Sci. Rep. 7:42. doi: 10.1038/s41598-017-00093-z
Lowry, J. A., and Atchley, W. R. (2000). Molecular evolution of the GATA family of transcription factors: conservation within the DNA-binding domain. J. Mol. Evol. 50, 103–115. doi: 10.1007/s002399910012
McCarthy, R. L., Zhong, R., Fowler, S., Lyskowski, D., Piyasena, H., Carleton, K., et al. (2010). The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 51, 1084–1090. doi: 10.1093/pcp/pcq064
McCarthy, R. L., Zhong, R., and Ye, Z. H. (2009). MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 50, 1950–1964. doi: 10.1093/pcp/pcp139
McCarthy, R. L., Zhong, R., and Ye, Z. H. (2011). Secondary wall NAC binding element (SNBE), a key cis-acting element required for target gene activation by secondary wall NAC master switches. Plant Signal. Behav. 6, 1282–1285. doi: 10.4161/psb.6.9.16402
Mellerowicz, E. J., and Sundberg, B. (2008). Wood cell walls: biosynthesis, developmental dynamics and their implications for wood properties. Curr. Opin. Plant Biol. 11, 293–300. doi: 10.1016/j.pbi.2008.03.003
Novaes, E., Kirst, M., Chiang, V., Winter-Sederoff, H., and Sederoff, R. (2010). Lignin and biomass: a negative correlation for wood formation and lignin content in trees. Plant Physiol. 154, 555–561. doi: 10.1104/pp.110.161281
Novaes, E., Osorio, L., Drost, D. R., Miles, B. L., Boaventura-Novaes, C. R., Benedict, C., et al. (2009). Quantitative genetic analysis of biomass and wood chemistry of Populus under different nitrogen levels. New Phytol. 182, 878–890. doi: 10.1111/j.1469-8137.2009.02785.x
Ohashi-Ito, K., Oda, Y., and Fukuda, H. (2010). Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22, 3461–3473. doi: 10.1105/tpc.110.075036
Ohlrogge, J., Allen, D., Berguson, B., Dellapenna, D., Shachar-Hill, Y., and Stymne, S. (2009). Energy. Driving on biomass. Science 324, 1019–1020. doi: 10.1126/science.1171740
Rao, X., and Dixon, R. A. (2018). Current models for transcriptional regulation of secondary cell wall biosynthesis in grasses. Front. Plant Sci. 9:399. doi: 10.3389/fpls.2018.00399
Reyes, J. C., Muro-Pastor, M. I., and Florencio, F. J. (2004). The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 134, 1718–1732. doi: 10.1104/pp.103.037788
Rueda-Lopez, M., Pascual, M. B., Pallero, M., Henao, L. M., Lasa, B., Jauregui, I., et al. (2017). Overexpression of a pine Dof transcription factor in hybrid poplars: a comparative study in trees growing under controlled and natural conditions. PLoS One 12:e0174748. doi: 10.1371/journal.pone.0174748
Shikata, M., Matsuda, Y., Ando, K., Nishii, A., Takemura, M., Yokota, A., et al. (2004). Characterization of Arabidopsis ZIM, a member of a novel plant-specific GATA factor gene family. J. Exp. Bot. 55, 631–639. doi: 10.1093/jxb/erh078
Suzuki, S., Li, L., Sun, Y. H., and Chiang, V. L. (2006). The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol. 142, 1233–1245. doi: 10.1104/pp.106.086678
Taylor, S. C., Nadeau, K., Abbasi, M., Lachance, C., Nguyen, M., and Fenrich, J. (2019). The ultimate qPCR experiment: producing publication quality, reproducible data the first time. Trends Biotechnol. 37, 761–774. doi: 10.1016/j.tibtech.2018.12.002
Taylor-Teeples, M., Lin, L., de Lucas, M., Turco, G., Toal, T. W., Gaudinier, A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. doi: 10.1038/nature14099
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Voelker, S. L., Lachenbruch, B., Meinzer, F. C., and Strauss, S. H. (2011). Reduced wood stiffness and strength, and altered stem form, in young antisense 4CL transgenic poplars with reduced lignin contents. New Phytol. 189, 1096–1109. doi: 10.1111/j.1469-8137.2010.03572.x
Yamaguchi, M., Goue, N., Igarashi, H., Ohtani, M., Nakano, Y., Mortimer, J. C., et al. (2010). VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 153, 906–914. doi: 10.1104/pp.110.154013
Ye, Z. H., and Zhong, R. (2015). Molecular control of wood formation in trees. J. Exp. Bot. 66, 4119–4131. doi: 10.1093/jxb/erv081
Yoshizumi, T., Nagata, N., Shimada, H., and Matsui, M. (1999). An Arabidopsis cell cycle-dependent kinase-related gene, CDC2b, plays a role in regulating seedling growth in darkness. Plant Cell 11, 1883–1896. doi: 10.1105/tpc.11.10.1883
Zhang, C., Hou, Y., Hao, Q., Chen, H., Chen, L., Yuan, S., et al. (2015). Genome-wide survey of the soybean GATA transcription factor gene family and expression analysis under low nitrogen stress. PLoS One 10:e0125174. doi: 10.1371/journal.pone.0145988
Zhang, Z., Ren, C., Zou, L., Wang, Y., Li, S., and Liang, Z. (2018a). Characterization of the GATA gene family in Vitis vinifera: genome-wide analysis, expression profiles, and involvement in light and phytohormone response. Genome 61, 713–723. doi: 10.1139/gen-2018-0042
Zhang, J., Tuskan, G. A., Tschaplinski, T. J., Muchero, W., and Chen, J. G. (2020). Transcriptional and post-transcriptional regulation of lignin biosynthesis pathway genes in Populus. Front. Plant Sci. 11:616977. doi: 10.3389/fpls.2020.616977
Zhang, J., Xie, M., Tuskan, G. A., Muchero, W., and Chen, J. G. (2018b). Recent advances in the transcriptional regulation of secondary cell wall biosynthesis in the woody plants. Front. Plant Sci. 9:1535. doi: 10.3389/fpls.2018.01535
Zhang, Z., Zou, X. Y., Huang, Z., Fan, S. M., Qun, G., Liu, A. Y., et al. (2019). Genome-wide identification and analysis of the evolution and expression patterns of the GATA transcription factors in three species of Gossypium genus. Gene 680, 72–83. doi: 10.1016/j.gene.2018.09.039
Zhong, R., Lee, C., and Ye, Z. H. (2010a). Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol. 152, 1044–1055. doi: 10.1104/pp.109.148270
Zhong, R., Lee, C., and Ye, Z. H. (2010b). Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant 3, 1087–1103. doi: 10.1093/mp/ssq062
Zhong, R., Lee, C., Zhou, J., McCarthy, R. L., and Ye, Z. H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782. doi: 10.1105/tpc.108.061325
Zhong, R., McCarthy, R. L., Lee, C., and Ye, Z. H. (2011). Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol. 157, 1452–1468. doi: 10.1104/pp.111.181354
Zhong, R., Richardson, E. A., and Ye, Z. H. (2007). The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19, 2776–2792. doi: 10.1105/tpc.107.053678
Zhong, R., and Ye, Z. H. (2010). The poplar PtrWNDs are transcriptional activators of secondary cell wall biosynthesis. Plant Signal. Behav. 5, 469–472. doi: 10.4161/psb.5.4.11400
Zhou, J., Lee, C., Zhong, R., and Ye, Z. H. (2009). MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21, 248–266. doi: 10.1105/tpc.108.063321
Zhou, G. K., Zhong, R. Q., Himmelsbach, D. S., McPhail, B. T., and Ye, Z. H. (2007). Molecular characterization of PoGT8D and PoGT43B, two secondary wall-associated glycosyltransferases in poplar. Plant Cell Physiol. 48, 689–699. doi: 10.1093/pcp/pcm037
Keywords: PtrGATA12, hierarchical regulator, biological characterization, secondary cell wall, Populus trichocarpa, coordinated regulation
Citation: Ren M, Zhang Y, Liu C, Liu Y, Tian S, Cheng H, Zhang H, Wei H and Wei Z (2021) Characterization of a High Hierarchical Regulator, PtrGATA12, Functioning in Differentially Regulating Secondary Wall Component Biosynthesis in Populus trichocarpa . Front. Plant Sci. 12:657787. doi: 10.3389/fpls.2021.657787
Edited by:
Jin-Gui Chen, Oak Ridge National Laboratory (DOE), United StatesReviewed by:
Steven James Burgess, University of Illinois at Urbana-Champaign, United StatesFei Zhou, Huazhong Agricultural University, China
Copyright © 2021 Ren, Zhang, Liu, Liu, Tian, Cheng, Zhang, Wei and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhigang Wei, zhigangwei1973@163.com
†These authors have contributed equally to this work
 Mengxuan Ren
Mengxuan Ren Yang Zhang2†
Yang Zhang2† Hairong Wei
Hairong Wei