- 1State Key Laboratory of Crop Stress Biology for Arid Areas and College of Agronomy, Northwest A&F University, Yangling, China
- 2Shaanxi Research Station of Crop Gene Resources and Germplasm Enhancement, Ministry of Agriculture, Yangling, China
Thinopyrum ponticum (2n = 10x = 70), a member of the tertiary gene pool of wheat (Triticum aestivum L.), harbors many biotic and abiotic stress resistance genes. CH10A5, a novel disomic substitution line from a cross of T. aestivum cv. 7182 and Th. ponticum, was characterized by cytogenetic identification, in situ hybridization, molecular marker analysis, and morphological investigation of agronomic traits and disease resistance. Cytological observations showed that CH10A5 contained 42 chromosomes and formed 21 bivalents at meiotic metaphase I. Genome in situ hybridization (GISH) analysis indicated that two of its chromosomes came from the Js genome of Th. ponticum, and wheat 15K array mapping and fluorescence in situ hybridization (FISH) revealed that chromosome 1D was absent from CH10A5. Polymorphic analysis of molecular markers indicated that the pair of alien chromosomes belonged to homoeologous group one, designated as 1Js. Thus, CH10A5 was a wheat–Th. ponticum 1Js (1D) disomic substitution line. Field disease resistance trials demonstrated that the introduced Th. ponticum chromosome 1Js was probably responsible for resistance to both stripe rust and powdery mildew at the adult stage. Based on specific-locus amplified fragment sequencing (SLAF-seq), 507 STS molecular markers were developed to distinguish chromosome 1Js genetic material from that of wheat. Of these, 49 STS markers could be used to specifically identify the genetic material of Th. ponticum. CH10A5 will increase the resistance gene diversity of wheat breeding materials, and the markers developed here will permit further tracing of heterosomal chromosome fragments in the future.
Introduction
Epidemics of stripe rust, caused by Puccinia striiformis f. sp. tritici (Pst), occur frequently in areas with cool conditions during the wheat growing season. Powdery mildew is caused by Blumeria graminis (DC.) E.O.f. sp. tritici (Bgt) (An et al., 2019). These two major diseases can dramatically reduce crop yield during prevalent years (Bokore et al., 2017). Numerous powdery mildew and stripe rust resistance genes have been identified in wheat and its relatives, and these genes have played a fundamental role in wheat breeding (Liu et al., 2019). Nevertheless, most resistance genes are quickly overcome due to rapid virulence changes in pathogen populations (Zhan et al., 2015). Therefore, it is extremely important to identify new resistance genes, especially broad-spectrum resistance genes, from wheat-related species.
Thinopyrum ponticum (2n = 10x = 70) often produces large amount of biomass and is tolerant of biotic and abiotic stresses such as drought and salt stress. It is also highly resistant to wheat leaf rust, stem rust, stripe rust, powdery mildew, barley yellow dwarf virus (BYDV), and Fusarium head blight (FHB) (Ceoloni et al., 2014; Kroupin et al., 2019), and recently, Fhb7 was successfully cloned by Wang et al. (2020a) from Th. elongatum. Many stripe rust and powdery mildew resistance genes are derived from Th. ponticum and have been introduced to common wheat by additions, substitutions, translocations, and introgression of chromosomes. Despite the development of numerous wheat–Th. ponticum chromosome lines, addition or substitution lines of homoeologous group one are rarely reported (Mo et al., 2017; Zhu et al., 2017; Mago et al., 2019; Wang et al., 2019), and only a few stripe rust and powdery mildew resistance genes have been introgressed into cultivated wheat varieties (Li et al., 2003; Zhan et al., 2014; Hou et al., 2016). The resistance genes of Th. ponticum have not been fully characterized. Therefore, the creation and identification of new wheat–Th. ponticum derivative lines carrying new resistance genes are important for the expansion of wheat genetic resources (Ceoloni et al., 2017). During wheat distant hybridization, rapid tracing and accurate identification of alien fragments directly affect the development of new wheat varieties and the process of breeding (Ceoloni et al., 2014).
Molecular marker analysis is the most convenient means for identifying exogenous chromosomes or chromosome segments. Wild triticeae grasses have revealed the presence of multiple homoeologous gene copies and repetitive DNA compared with common wheat, which were helpful to molecular marker development and also chromosome identification (Li et al., 2016). Nonetheless, existing markers do not meet the demand for Th. ponticum chromatin detection (Liu et al., 2018b), and it is therefore particularly important to develop chromosome-specific markers for the detection of Th. ponticum genetic materials. At present, next generation sequencing is the most important technology for molecular marker development and includes such techniques as RNA-seq (Okada et al., 2018), whole genome sequencing (Tiwari et al., 2015), genotyping-by-sequencing (Elshire et al., 2011), and specific-locus amplified fragment sequencing (SLAF-seq) (Sun et al., 2013). SLAF-seq refers to a sequencing strategy in which restriction enzymesare used to fragment genomic DNA and high-throughput sequencing is performed on a number of specific fragments in order to fully represent the genomic information of the target species. SLAF-seq plays a fundamental role in molecular breeding, gene location, and germplasm resource identification. It has been used to develop specific markers in wheat-related species with high efficiencies, up to 66% (Chen et al., 2013). SLAF-seq can be employed to obtain sufficient markers for the construction of a high-density genetic map and permit the exploitation of useful genes for wheat breeding applications.
In this study, a novel wheat–Th. ponticum 1Js (1D) disomic substitution line with resistance to stripe rust and powdery mildew was developed from the F6 progeny of a wheat–Th. ponticum hybrid and named CH10A5. We used cytogenetic methods to assess the stability of CH10A5 through observations of chromosome pairing. Its chromosomal composition was determined using fluorescence in situ hybridization (FISH), simple sequence repeats (SSR) markers, expressed sequence tag sequence-tagged sites (EST-STS) markers, PCR-based landmark unique gene (PLUG) markers, and a single-nucleotide polymorphism (SNP) array. Its agronomic and disease resistance characteristics were also assessed. This is the first report of stripe rust and powdery mildew resistance on chromosome 1Js. Specific molecular markers of 1Js were developed using SLAF-seq and will play a very important role in reducing the length of the breeding cycle and improving selection efficiency.
Materials and Methods
Plant Materials and Isolation Scheme of the CH10A5 Line
The wheat-related species used in this study included Thinopyrum ponticum (2n = 10x = 70, EeEeEbEbExExStStStSt/JJJJJJJsJsJsJs), Th. bessarabicum (2 = 2x = 14, EbEb/JJ), Th. elongatum (2n = 2x = 14, EeEe/EE), tetraploid Pseudoroegeria spicata (2n = 4x = 28, StStStSt), Th. intermedium (2n = 6x = 42, EeEeEbEbStSt/JJJsJsStSt), Leymus mollis (Trin.) Pilger, (2n = 4x = 28, NsNsXmXm), Psathyrostachys huashanica (2n = 2x = 14, NsNs), Secale cereale L. (2n = 2x = 14, RR), Aegilops geniculata Roth. (2n = 4x = 28, UUMM), Triticum urartu Thum. (2n = 2x = 14, AA), and Ae. tauschii Coss. (2n = 2x = 14, DD).
The common wheat cultivar 7182 (2n = 6x = 42, AABBDD) was used as the male parent. Shanyou225 (SY225) was used as the susceptible control for Bgt, and Huixianhong (HXH) and Mingxian169 (MX169) were used as the susceptible controls for Pst. Chinese Spring (CS) was used as a blocker in GISH analysis, and nulli-tetrasomic materials (CSN1AT1B, CSN1BT1D, and CSN1DT1B) were used in molecular marker detection.
The isolation scheme of the CH10A5 line began with hybridization of the female parent, Th. ponticum, with 7182 in 2009. The resulting F1 progeny were backcrossed with 7182, then 18 lines with good morphologically characteristics were selected in the year of 2011, such as long spikes, strong resistance to stripe rust and powdery mildew, and so on. The chromosomes of root tip cells and their pairing in pollen mother cells (PMCs) was observed in each generation. After 6 years of strict self-crossing, a total of 375 lines were identified in BCIF6 generation, in which 136 lines had 42 chromosomes in root tip cells, accounting for 36%. CH10A5 was finally selected in BCIF6 generation (June 2016). All plant materials were preserved in our laboratory at the College of Agronomy, Northwest A&F University, China.
Cytological Stability Assessment
Root tip cells and PMCs of CH10A5 from the same plant were observed for three growing seasons (2016–2019) in a total of 34 plants. Root tip processing procedures were performed as described in Zheng et al. (2006), using a mixture of cellulase (R-10, Yakult Japan) and pectinase (Y-23, Yakult Japan) for cell enzymatic hydrolysis (Zhu et al., 2017). Young spikes (Zadoks stage 45) were sampled, and anthers at meiosis metaphase I (MI) were fixed in Carnoy fixative (a 6:3:1 ethanol-chloroform-acetic acid mixture) for at least 2 days and stained with a 1% (w/v) aceto-carmine solution prior to in situ hybridization of the PMCs as described in Yang et al. (2014). Chromosomes were fixed for 60s at an ultraviolet intensity of 125,000μJ/cm2 by UV irradiation (Spectrolinker™ XL-1500, USA). Cells were examined with an Olympus BX43 microscope (Japan) equipped with a Photometrics SenSys CCD camera.
In Situ Hybridization of CH10A5
The total genomic DNA was extracted from fresh leaves using a modified CTAB method (Allen et al., 2006) with one additional purification step to obtain high-quality DNA. The total genomic DNA of Th. ponticum was labeled with Red-5-dUTP (Invitrogen) by nick translation (Nick Translation Mix, Roche). Genomic DNA of CS was fixed in boiling water for 5 min and used as a blocker at a ratio of 1:300. The GISH procedure was performed as described in Fu et al. (2012). For multicolor GISH (mc-GISH) analysis, Th. bessarabicum DNA was labeled with Alexa Fluor 488-5-dUTP (Invitrogen), and Ps. spicata DNA was labeled with Texas Red-5-dUTP (Invitrogen) (Wang et al., 2019).
The oligonucleotide probes used to identify wheat and Th. ponticum chromosomes by non-denaturing FISH (ND-FISH) analysis included Oligo-pSc119.2 (6-FAM-5′), Oligo-pTa535 (Tamra-5′) (Tang et al., 2014), Oligo-44 (6-FAM-5′), and Oligo-D (6-FAM-5′) (Tang et al., 2018), all of which were synthesized by Shanghai Invitrogen Biotechnology Co. Ltd. (Shanghai, China). Chromosomes were counterstained with 4,6-diamidino-2-phenylindole (DAPI). Fluorescent signals were scanned and photographed with an Olympus BX53 microscope equipped with a Photometrics SenSys CCD DP80 camera (Japan) (Wang et al., 2016).
Homoeologous Group Identification
Wheat SNP Array Analysis
Genomic DNA of CH10A5 and its parents was hybridized to wheat 15K SNP genotyping arrays by China Golden Marker Biotechnology Company (Beijing, China). There were 13,947 microchip probes per chip, including a large number of diploid markers. Based on IWGSC-RefSeq-v1.0 (https://wheat-urgi.versailles.inra.fr/Seq Repository/), a total of 13,199 markers had explicit physical location information, evenly covering the entire wheat genome. Data processing and analysis of the 15K array were performed to analyze polymorphic markers using SigmaPlot V12.5 (SYSTAT software, Inc. USA) as described in Li et al. (2019).
Molecular Markers and Nulli-Tetrasomic Analysis
SSR markers, EST-STS markers (http://wheat.pw.usda.gov/SNP/new/pcr_primers.shtml) and PLUG markers for wheat homoeologous groups one to seven were synthesized by AuGCT DNA-SYN Biotechnology Co., Ltd of Beijing (Ishikawa et al., 2009). A total of 674 markers were used to identify the homoeologous group of the exogenous chromosomes carried by CH10A5. These included 467 SSR markers, 72 EST markers, and 135 PLUG markers (Supplementary Table 1). The polymerase chain reaction (PCR) amplification system contained: 10×PCR Buffer (Mg2+) 1.0 μl, 0.02 mM/μl dNTPs, 0.05 μM/μl primers, 0.05 U/μl Taq DNA polymerase (Takara), 10 ng/μl DNA, and ddH2O up to 10.0 µl. The PCR products were then digested with Taq Ⅰ (Takara) and Hae Ⅲ (Takara) (Zhu et al., 2017). The reagent for the PCR reaction was purchased from Takara Biomedical Technology (Beijing) Co., Ltd.
Disease Resistance Assessment
Stripe rust races CYR23, CYR29, CYR31, CYR 32, CYR33, and CYR 34 were kindly donated by the College of Plant Protection, Northwest A&F University, China. Disease resistance was investigated three times each year, and the most severe reaction type observed in a given year was considered to be the final disease resistance result.
Seedling stripe rust resistance trials were performed in an incubator. Each of five Pst races (CYR23, CYR29, CYR31, CYR32, and CYR34) was mixed with talcum powder at a 1:30 ratio using the shaking powder method. When seedlings had grown to the two-leaf stage, 10 plants in a pot were inoculated with one of the powder mixtures, and each treatment was replicated three times. The temperature in the incubator was 17°C during the day and 11°C at night. Day length was 16 h (6000 lux), and the relative humidity was 70%. When the susceptible control had become fully affected, we began to assess disease resistance every 4 days until 21 days after inoculation.
Adult stripe rust resistance trials were performed under field conditions over three growing seasons (2016–2019) using mixtures of Pst races from Shaanxi Province, including CYR31, CYR32, CYR33, and CYR34 (Zhan et al., 2015). The races were inoculated by using the shaking powder method at the jointing stage with the mixed talcum powder at a 1:200 ratio after raining. HXH was planted every meter in the aisle to serve as an inoculum spreader. The field trials were performed at the using a completely random experimental design. At the booting stage, 20 plants each of common wheat 7182, line CH10A5, the susceptible control HXH, as well as a cluster of Th. ponticum, were evaluated three times in the field at the College of Agronomy, Northwest A&F University. Stripe rust resistance was quantified by immunity types (ITs) using a 0–4 scale in which an IT of 0–2 indicated resistance and an IT of 3–4 indicated susceptibility (Wang et al., 2020b).
At the adult stage, powdery mildew resistance trials were performed in the nursery by spontaneous inoculation over three growing seasons (2016–2019). SY225 was planted every meter in the aisle to serve as an induction bed. Twenty plants each of 7182, CH10A5, and the susceptible control SY225, as well as a cluster of Th. ponticum, were assessed. Plants were investigated three times for infection symptoms until 20 days after flowering (Zhuang et al., 2015). Powdery mildew resistance was evaluated on a 0–9 scale, which an IT of 0–4 indicated resistance and an IT of 5–9 indicated susceptibility (Sheng and Duan, 1991).
Molecular Marker Development
SLAF-seq was performed by the Beijing Biomarker Technologies Corporation (Sun et al., 2013). Quality-checked genomic DNA was digested by the restriction enzyme HaeⅢ. The Illumina HiSeq platform was used for sequencing after the libraries had passed quality inspection. Dual indexing was used to ensure accurate demultiplexing of the sequence reads for each sample. The data filtering steps were as follows: (1) reads with adapter sequence were removed, (2) reads with an N content exceeding 10% were removed, and (3) reads in which more than 50% of the bases had quality scores less than 10 were removed. Sequence quality and data volume were evaluated after filtering the sequence reads (Chen et al., 2013).
To obtain sequences specific to 1Js, sequences with 0% similarity to CS (IWGSC-RefSeq-v1.0) were selected using the Burrows-Wheeler Alignment (BWA) tool, and from these, sequences with at least 95% similarity to Th. ponticum were selected. Primers for these sequences were developed using Primer3Plus (http://www.primer3plus.com/). After PCR, specific bands were detected on a 1% agarose gel (Liu et al., 2018b). A Venn diagram and an Upset plot were constructed using TBtools (http://mdr.tbtools.org/).
Results
Stability Evaluation of CH10A5
The CH10A5 line was derived from the F7 progeny of a 7182/Th. ponticum//7182 cross. The morphology of CH10A5 was similar to that of common wheat, with a compact plant shape and full grain (Supplementary Figure 1). Somatic cell chromosomes from mitotic root tip cells were counted. Of the 281 root tip cells observed, 277 (98.57%) had 42 chromosomes (Figure 1A), three cells (1.07%) had 41 chromosomes, and one (0.36%) had 43 chromosomes. A total of 177 PMCs were assessed over a 3-year period, of these 169 PMCs were observed 21 pairs of bivalents in meiotic metaphase I in (Figure 1B), up to 95.48%, and univalents were observed in 8 PMCs (4.52%), and on trivalents and quadrivalents were observed. The chromosomes were equally separated in a 21 + 21 manner at anaphase of meiosis II. A bivalent with a strong yellow-green Th. ponticum hybridization signal was detected by GISH in meiotic metaphase I (Figure 1C), and the alien chromosome separated equally in anaphase meiosis II (Figure 1D). Cytological observations therefore indicated that CH10A5 was highly stable.
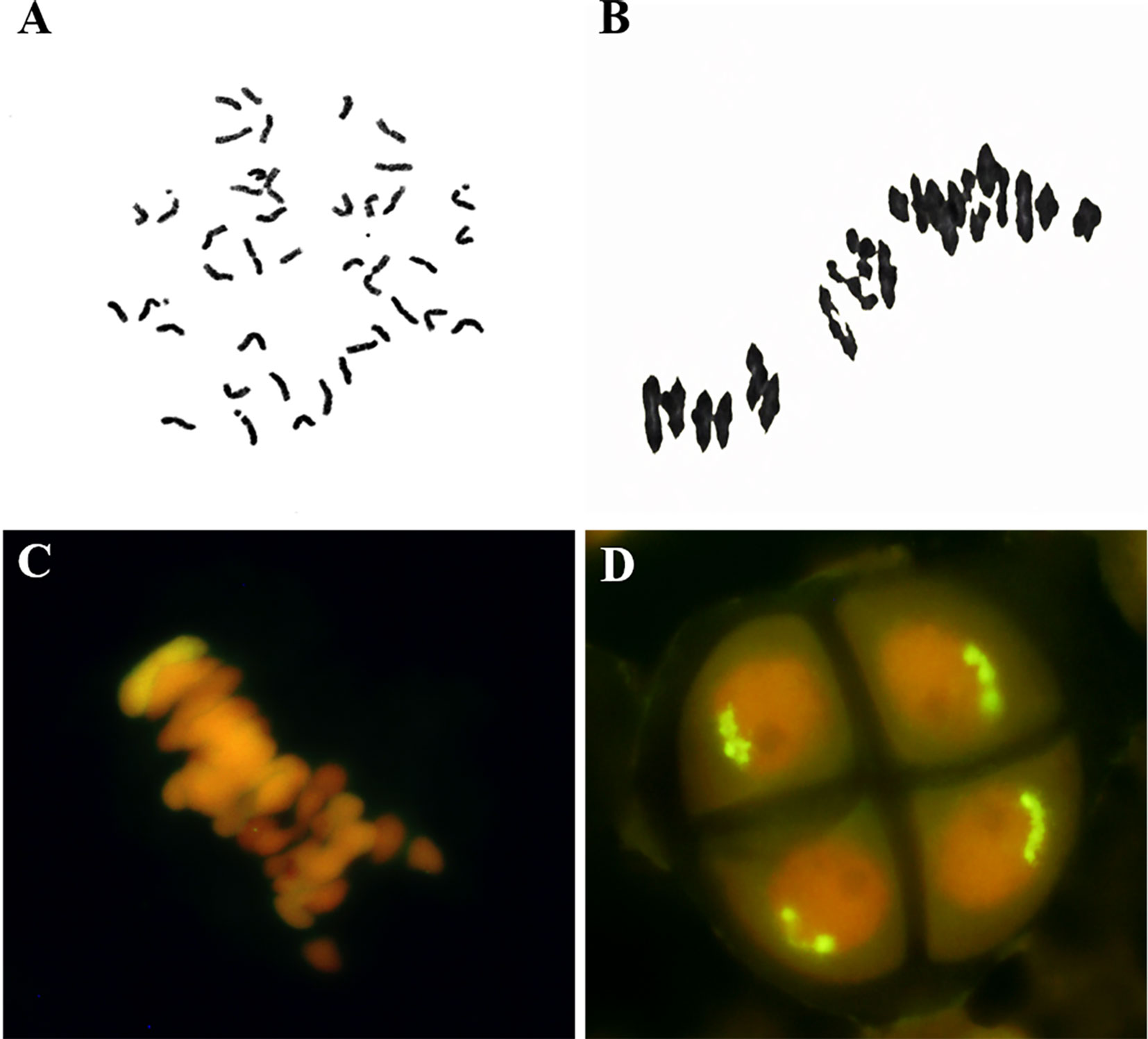
Figure 1 Cytological observations of CH10A5. (A) Mitosis stage of root tip cells, 2n = 42. (B) Meiosis stage of pollen mother cells, 2n = 21II. (C, D) GISH detection of CH10A5. The yellow-green fluorescent signals identify Th. ponticum chromosomes in meiosis metaphase. Chromosomes were counterstained with propidium iodide (red).
Chromosome Composition of CH10A5
To investigate the chromosome composition of CH10A5, the oligonucleotide probes Oligo-pTa535 (red) and Oligo-pSc119.2 (green) were used for FISH analysis of the CH10A5 chromosomes (Figure 2A). Similar to the report of Tang et al. (2014), 40 chromosomes were distinguished in CH10A5, 34 of which had the same FISH signal as CS. A pair of 1RS.1BL translocation chromosomes were observed (Jiang et al., 2017), and signal variation occurred in chromosomes 5A and 6B, and a pair of chromosomes with two arms that resembled the red Oligo-pTa535 signal were not common wheat chromosomes.
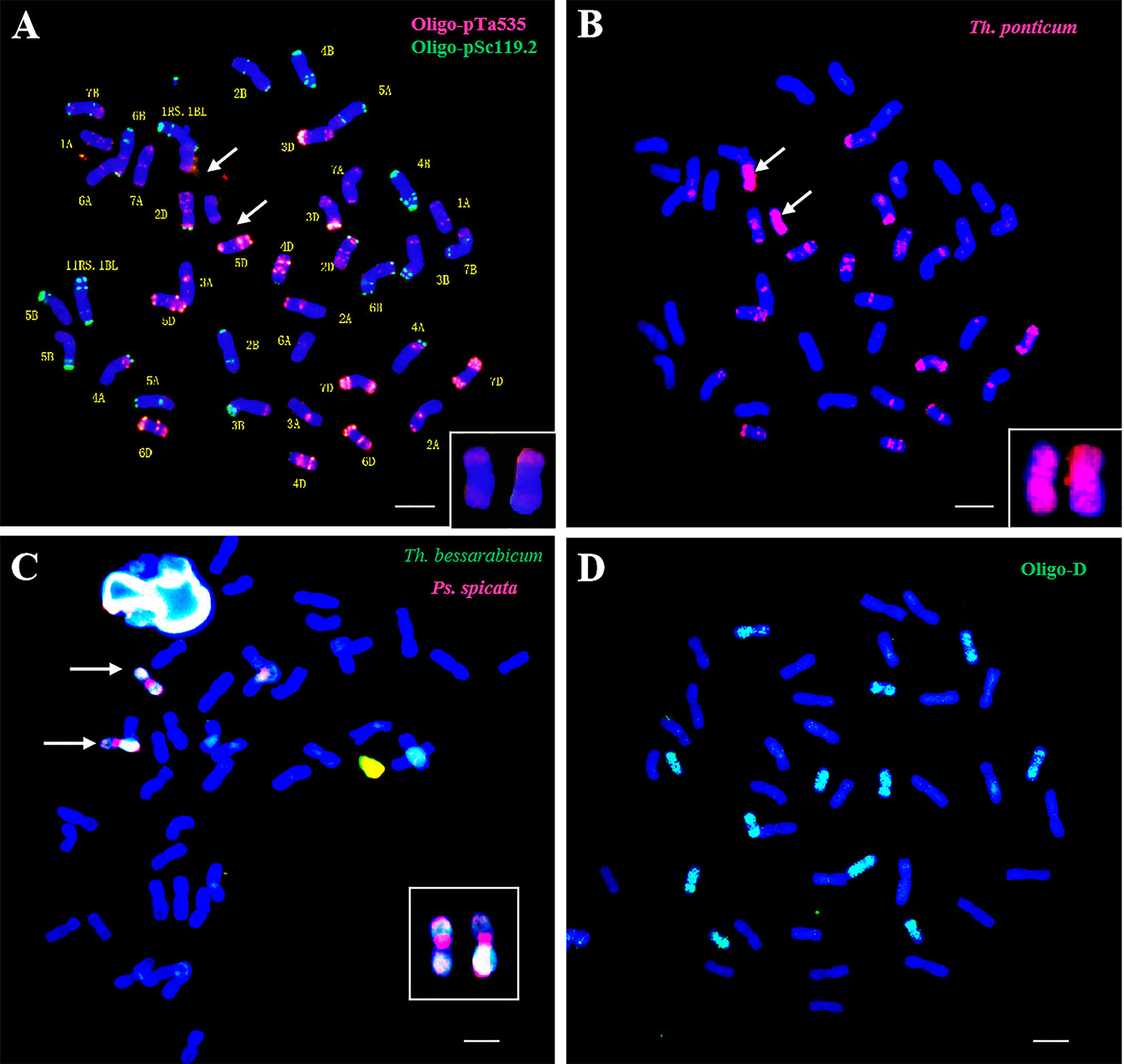
Figure 2 Cytogenetic analysis of CH10A5 by in situ hybridization. (A) mc-FISH, Oligo-pTa53 5 (red), and Oligo-pSc119.2 (green) were used to perform FISH analysis. (B) FISH-GISH, Th. ponticum genomic DNA labeled with Texas Red-5-dUTP as a probe and CS genomic DNA as a blocker. The arrows indicate the alien chromosomes in CH10A5. (C) mc-GISH, Th. bessarabicum genomic DNA (green) and Ps.spicata genomic DNA (red) were used as probes. (D) ND-FISH, Oligo-D (green) was used to detect 12 D chromosomes. The arrows referred to alien chromosomes. Chromosomes were counterstained with DAPI (blue). Scale bar = 10 μm.
Sequential FISH-GISH analysis suggested that the two chromosomes labeled with a bright red signal came from Th. ponticum (Figure 2B). In addition, mc-GISH analysis showed that two chromosomes had the red signal of Ps. spicata in the centromere region and the green signal of Th. bessarabicum on both arms. This result indicated that the alien chromosomes belonged to the Js genome (Figure 2C). Twelve D-genome chromosomes, rather than the 14 typical of common wheat, were detected in CH10A5 using Oligo-D (Figure 2D). Together, these results indicated that the CH10A5 genome was composed of 38 common wheat chromosomes (14A+12B+12D), a pair of 1RS.1BL translocation chromosomes, and two Js chromosomes from Th. ponticum.
To further characterize the chromosome variations of CH105A, the oligonucleotide probe Oligo-44 were used for ND-FISH analysis. Oligo-44 hybridizes to chromosomes 3A, 5A, 5B, 5D, and 7A, and it was therefore used to demonstrate the variation in 5A signals (Figure 3A). A sequential FISH karyotype of CH10A5 was then performed using Oligo-pTa535 and Oligo-pSc119.2 (Figure 3B). The results confirmed that the red signal on the long arm of chromosomes 5A in common wheat was changed to a green signal in CH10A5 (Figure 3C). Alien chromosomes of Th. ponticum had replaced chromosome 1D of common wheat 7182, which may cause FISH signal mutations in chromosomes 5A and 6B.
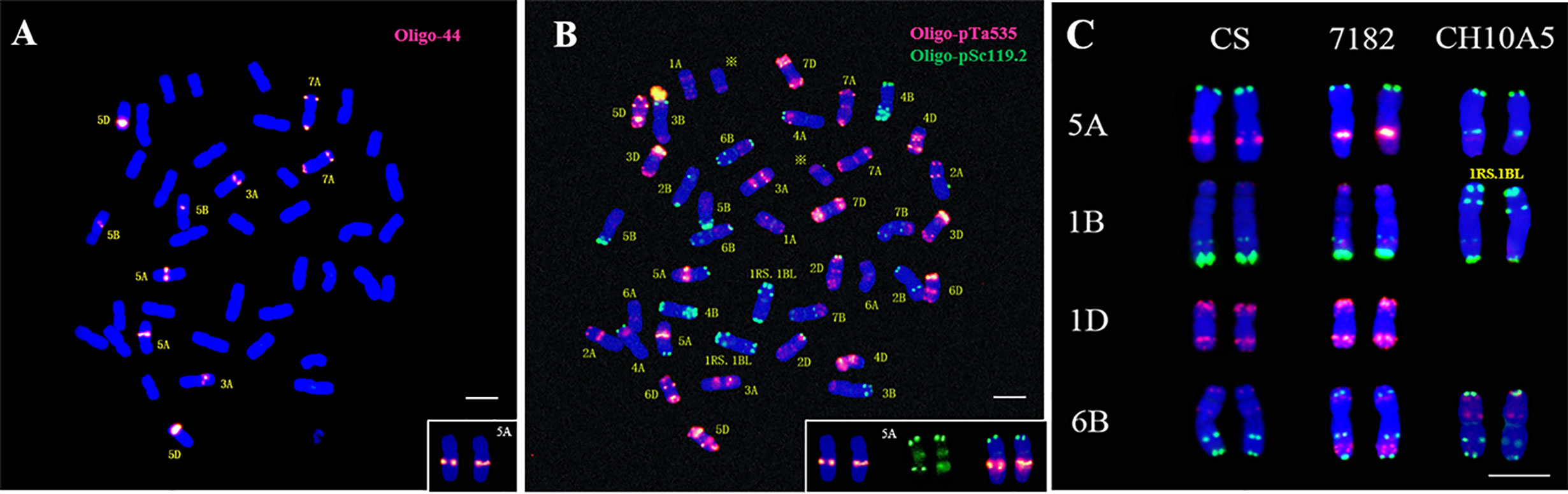
Figure 3 Chromosome variation of CH10A5. (A) ND-FISH, Oligo-44 was used to detected 3A, 5A, 5B and 7A; (B) mc-FISH, Oligo-pTa535(red), and Oligo-pSc119.2 (green) were used to attest FISH signal variation in 5A with same slides; (C) FISH signal comparison between CS, 7182, and CH10A5 on chromosomes 1B, 6B, 5A, and 1D; Chromosomes were counterstained with DAPI (blue). The “※” referred to alien chromosomes, scale bar = 10 μm.
Homoeologous Group of the Alien Chromosome
Based on wheat array mapping analysis, a total of 12,726, 13,078, and 5,916 polymorphic SNP loci were identified in CH10A5, 7182, and Th. ponticum, respectively. A total of 7,258 SNPs were polymorphic between Th. ponticum and 7182. The maximum, minimum, and mean percentages of SNP genotyping loci shared between CH10A5 and 7182 were 74.20% (on chromosome 2D), 16.62% (on 1D), and 53.27% overall. The maximum, minimum, and mean percentages of SNP genotyping loci shared between CH10A5 and Th. ponticum were 52.52% (on 1D), 11.10% (on 4A), and 19.41% overall (Supplementary Table 2). This result confirmed that a large number of wheat-origin SNPs on chromosome 1D were missing from CH10A5 and that CH10A5 and Th. ponticum shared the largest proportion of SNP loci on chromosome 1D (Figure 4A).
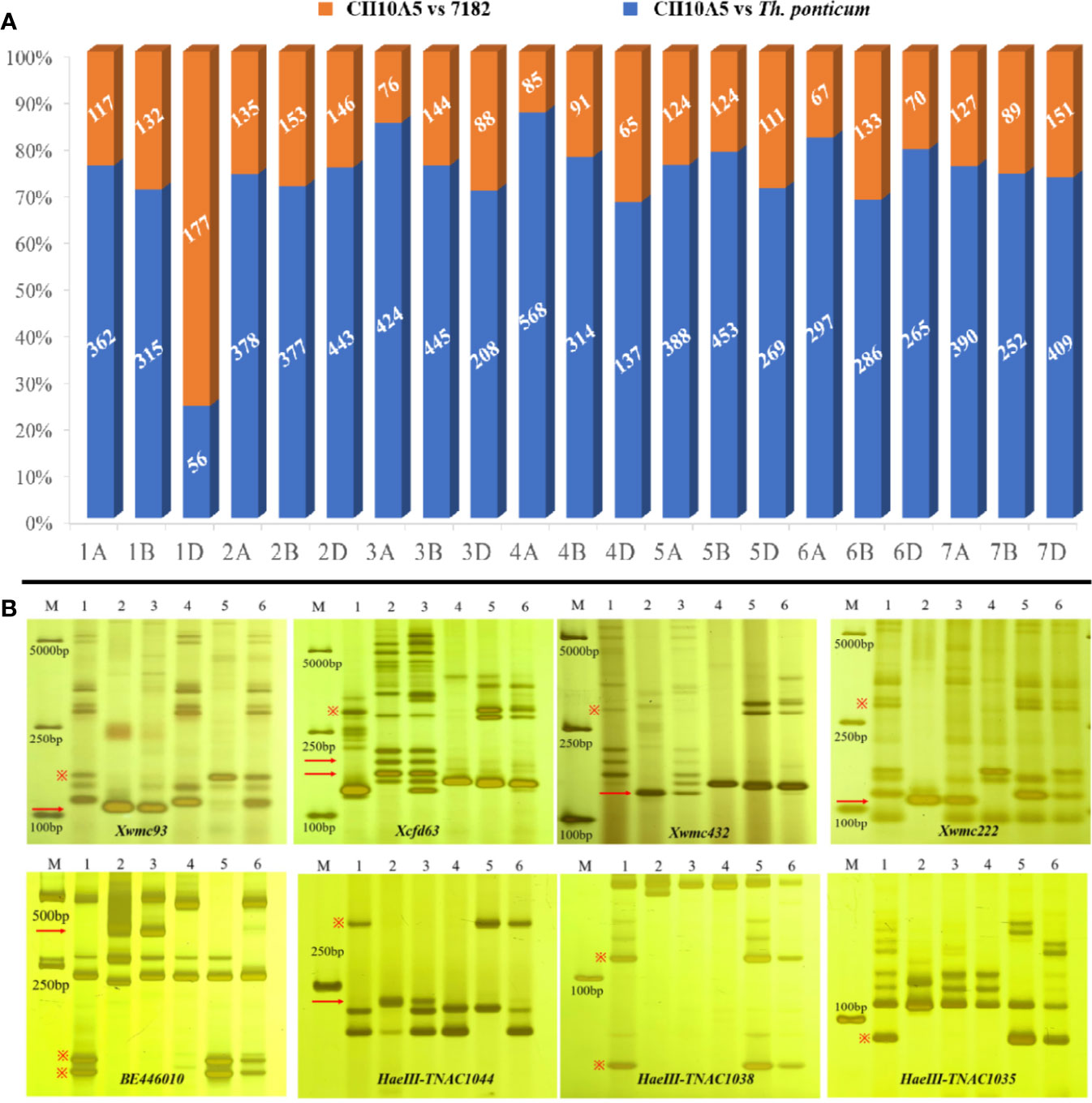
Figure 4 Molecular marker analysis of CH10A5. (A) Wheat 15K array analysis. The number refers to the same markers in CH10A5 vs. 7182 (orange) and CH10A5 vs. Th. ponticum (blue). (B) EST and PLUG marker analysis. M: DL2000 marker; 1–3 refer to 7182, Th. ponticum, and CH10A5; 4–6 refer to the CS nulli-tetrasomic materials CSN1DT1B, CSN1BT1D, and CSN1AT1B. The red arrow and “※” symbol represent the distinctive bands of Th. ponticum and chromosome 1D, respectively.
Based on SSR, EST and PLUG marker analysis, 16 markers (4 SSR markers, 4 EST markers, and 8 PLUG markers) amplified the same specific bands in CH10A5 and Th. ponticum (Table 1, Figure 4B). In addition, four SSR markers (Xwmc93, Xcfd63, Xwmc432, and Xwmc222), one EST marker (BE446010), and three PLUG markers (TNAC1038, TNAC1044, and TNAC1035) demonstrated the deletion of chromosome 1D in CH10A5. All of these markers belong to homoeologous group one.
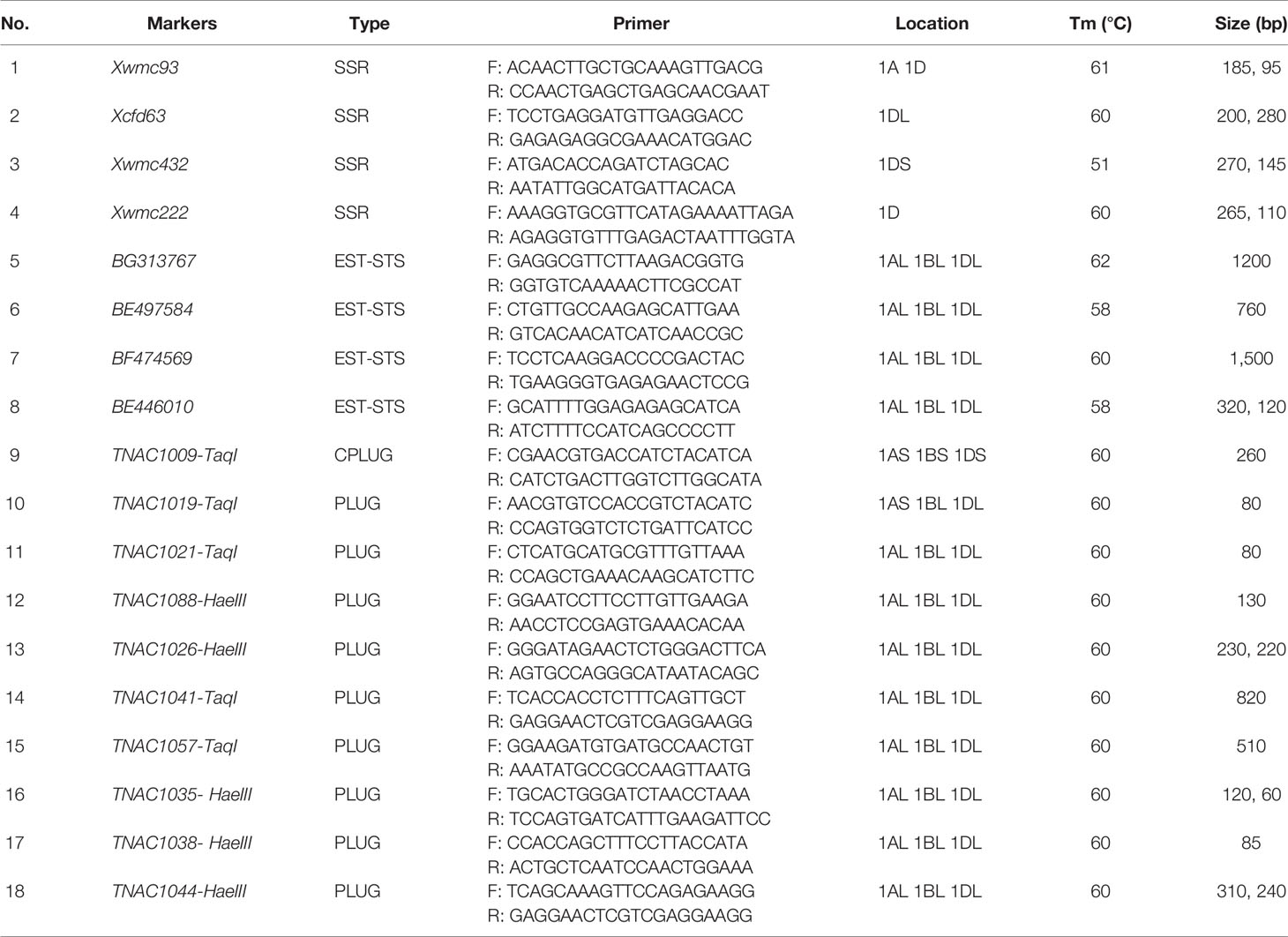
Table 1 Wheat SSR, EST-STS, and PLUG markers used to successfully analyze the chromosomal composition of CH10A5 in this study.
The molecular marker results showed that the chromosome deleted in CH10A5 was 1D and that the pair of Js chromosomes carried by CH10A5 belonged to homoeologous group one of Th. ponticum. CH10A5 was therefore a wheat–Th. ponticum 1 Js (1D) disomic substitution line.
Assessment of Disease Resistance
At the seedling stage, CH10A5 and the common wheat parent 7182 were inoculated with five Pst races (CYR23, CYR29, CYR31, CYR32, and CYR34). Ten days after inoculation, the susceptible controls HXH or MX169 had become fully infected. Three successive investigations of disease resistance showed that the immunity types of CH10A5 to CYR29, CYR31, and CYR32 were 0, indicating immunity. The immunity types of CH10A5 to CYR23 and CYR34 were 1, indicating high resistance. By contrast, 7182 was resistant to CYR23, CYR31, and CYR32 and susceptible to CYR29 and CYR34 (Table 2, Figure 5). The seedling resistance results therefore showed that CH10A5 carried new resistance gene(s) from Th. ponticum.
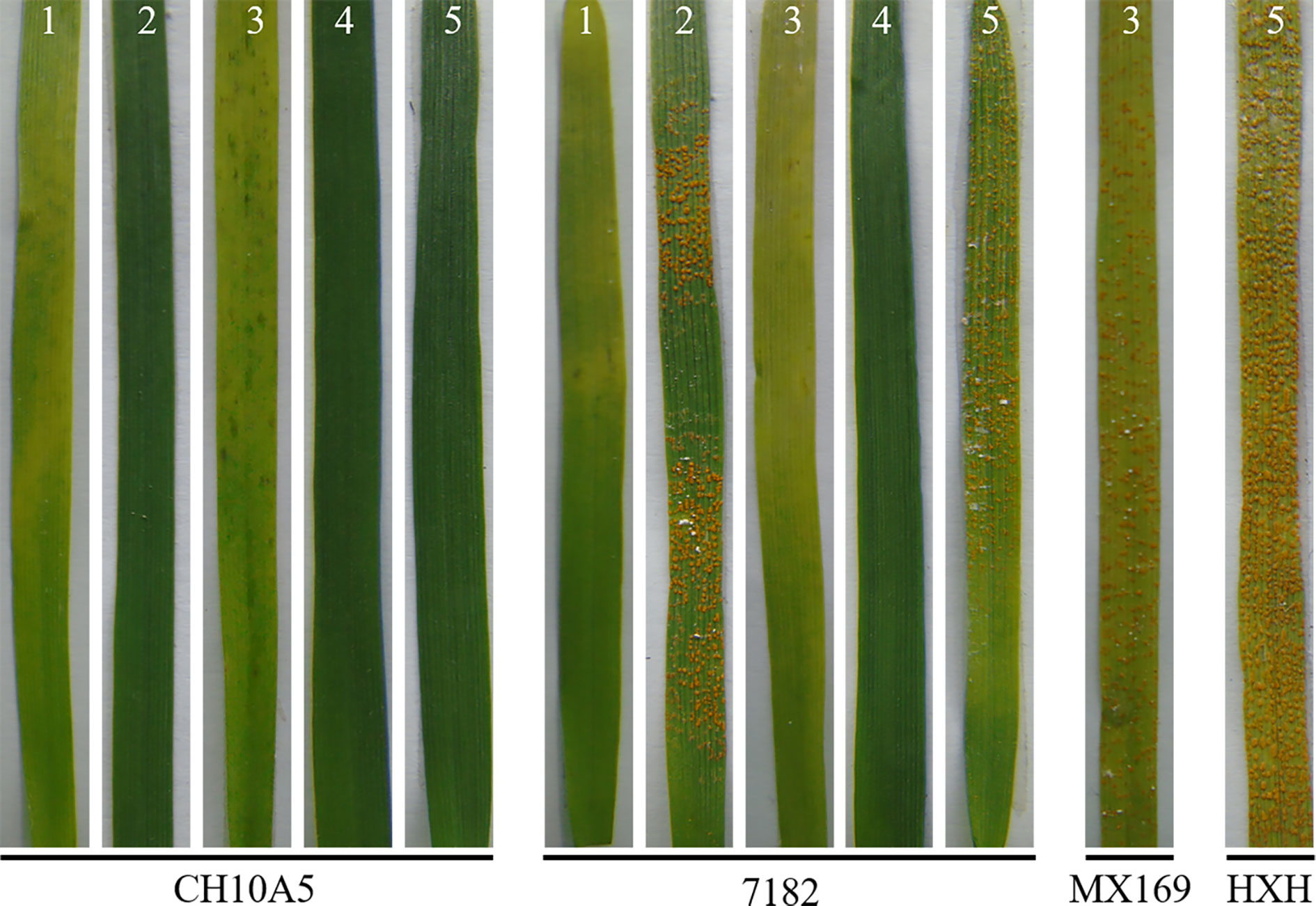
Figure 5 Stripe rust reaction of CH10A5 and 7182 to five Pst races at the seedling stage. HXH and MX169 were the susceptible controls. Numbers 1–5 refer to Pst races CYR23, CYR29, CYR31, CYR32, and CYR34, respectively.
At the adult stage, resistance to three Pst races (CYR31, CYR32, and CYR33) was assessed in CH10A5 and its parents through inoculation experiments performed over three growing seasons (2016–2019). Ten plants were randomly selected to evaluate disease resistance each year. After three consecutive years of investigation, it was found that the stripe rust resistance of each line was consistent. The IT of the susceptible control HXH was 4, indicating high sensitivity. That of CH10A5 was 0 or 1, few small uredia with distinct necrosis on leaves, indicating high resistance; that of Th. ponticum was 0, and there were no visible uredia and necrosis on leaves, indicating immunity; and that of 7182 was 3, there were a lot of medium-sized uredia, no necrosis, but with chlorosis on leaves, indicating moderate sensitivity (Table 2, Figure 6). These results suggested that the chromosome 1Js of CH10A5 might carry on a broad-spectrum resistance gene to stripe rust.

Figure 6 Stripe rust and powdery mildew resistance of CH10A5 and its parents at the adult stage. Huixianhong (HXH) was the susceptible control for Pst, and Shanyou225 (SY225) was the susceptible control for Bgt.
CH10A5 and its parents were also planted in a nursery where powdery mildew was present. When the powdery mildew spores were fully developed on the leaves of the susceptible control SY225, the ITs of the four materials were as follows: SY225, IT = 9, highly susceptible to powdery mildew; 7182, IT = 5, moderate susceptible to powdery mildew; CH10A5, IT = 3, high resistance to powdery mildew; Th. ponticum, IT = 0, immune to powdery mildew. Thus, the material with chromosomes from Th. ponticum exhibited substantial resistance to powdery mildew (Table 3, Figure 6). Together, the results of the disease resistance trials indicated that the alien chromosomes of CH10A5 carried gene(s), which probably responsible for resistance to stripe rust and powdery mildew.
Molecular Marker Development
After SLAF sequencing, a total of 11,931,086, 10,383,011, and 9,871,061 reads were obtained from CH10A5, 7182, and Th. ponticum, with an average Q30 score of 90.34% and an average GC content of 46.55%. The final SLAF numbers were 467,788, 157,996, and 500,275, and the average sequencing depth was 17.93. Using the BWA tool, 1220 reads were found to have 0% similarity to the CS wheat reference genome (IWGSC-RefSeq-v1.0). Nine hundred fifty-seven of these reads had at least 95% similarity to Th. ponticum and were considered to be specific fragments of chromosome 1Js.
To develop Th. ponticum 1Js chromosome-specific markers, 957 pairs of STS primers were designed based on these specific fragments and used to amplify sequences from CS, 7182, Th. ponticum, and CH10A5. Compared with CS and 7182, 507 STS primers were found to be specific to CH10A5 and Th. ponticum through PCR and electrophoresis detection (Supplementary Figure 2, Supplementary Table 3). The success rate for the development of markers specific to the alien chromosome using SLAF-seq was therefore reached 52.98%. Finally, 507 chromosome sequences of 1Js were obtained (unpublished data). Some chromosome sequences were related to plant disease resistance, which helped us to locate the stripe rust and powdery mildew resistance genes on chromosome 1Js.
To evaluate the polymorphisms and utility of the chromosome 1Js-specific markers, PCR was performed on 7182 and eleven wheat-related species to identify markers that amplified specific bands that distinguished 7182 from other wheat-related species. The numbers of specific markers for each species were as follows: 49 for Th. ponticum, 38 for Th. bessarabicum, 45 for Th. elongatum, 89 for Ps. spicata, and 57 for Th. intermedium (Figure 7A). In addition, Venn diagrams showed that some markers were shared by two or three species (Figure 7B). PCR of other wheat-related species also amplified specific bands. There were 70, 51, 67, 54, 17, and 43 markers for L. mollis, Ps. huashanica, S. cereale, Ae. geniculata, T. urartu, and Ae. tauschii, respectively (Supplementary Table 4).
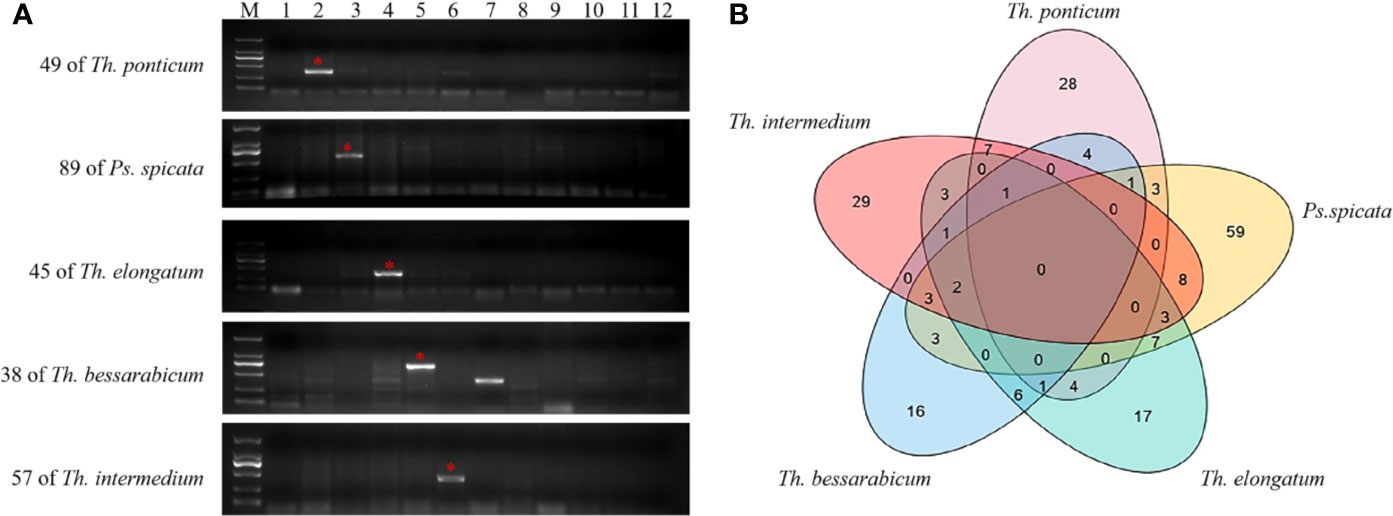
Figure 7 Molecular marker development and PCR amplification in 7182 and other wheat-related species. (A) Specific markers for Th. ponticum, Ps. spicata, Th elongatum, Th. bessarabicum, and Th. intermedium. Lanes: M, DL2000; 1–12 refer to 7182, Th. ponticum, Ps. spicata, Th. elongatum, Th. bessarabicum, Th. intermedium, L. mollis, Ps. huashanica, S. cereale, Ae. geniculata, T. urartu, and Ae. tauschii, respectively. The red stars and arrows indicate target bands. (B) Venn diagrams show the number of specific markers for Th. ponticum, Th. bessarabicum, Th. elongatum, Ps. spicata, and Th. intermedium. Overlaps represent the number of markers that were shared among two or three species.
Discussion
Since the 1960s, at least 12 genes for resistance to fungal or viral diseases have been reported from Th. ponticum in the form of chromosome translocations. Among them, two leaf rust resistance genes (Lr19 and Lr29), two stem rust resistance genes (Sr25 and Sr43), and one FHB gene (Fhb7) were located on homoeologous seven; Sr24 and Lr24 were located on homoeologous three; and Sr26 and SrB were located on homoeologous six (Li and Wang, 2009; Niu et al., 2014; Guo et al., 2015; Kuzmanovic et al., 2019; Mago et al., 2019). A wheat–Th. ponticum introgression line was resistant to powdery mildew (Pm51) and stripe rust (Yr69), and the resistance genes were all located on homoeologous two (Zhan et al., 2014; Hou et al., 2016). Likewise, resistance to eyespot and stripe rust at the adult stage was associated with homoeologous groups four and five in wheat–Th. ponticum line (Li et al., 2004; Mo et al., 2017). The cloning of Fhb7 will greatly promote the development of wheat FHB resistance breeding in China (Wang et al., 2020a). However, stable addition, substitution, and translocation wheat–Th. ponticum lines are rarely reported, particularly for homoeologous group one, and it is therefore important to explore potential resistance genes in Th. ponticum. In this study, the wheat–Th. ponticum 1Js (1D) disomic substitution line CH10A5 was probably resistant to both stripe rust and powdery mildew. This is first report of stripe rust and powdery mildew resistance probably associated with chromosome 1Js. By comparing the resistance response of CH10A5 to that of 7182, we demonstrated that chromosomes 1Js of Th. ponticum might possess new resistance gene(s) for stripe rust and powdery mildew. CH10A5 is therefore an important material for the exploration of potential resistance genes from Th. ponticum. For further use of this 1Js (1D) disomic substitution line, small fragment translocation lines will be developed using radiation and the Ph1b gene (Zheng et al., 2006; Dai et al., 2020) to explore the segments of resistance genes and identify new useful traits.
FISH analysis can easily and efficiently detect different structural rearrangements and polymorphic blocks in wheat chromosomes (Huang et al., 2018). The introduction of an alien chromosome may lead to structural changes or altered gene expression of common wheat chromosomes (Rey et al., 2018). Previous studies have indicated that structural alterations of wheat chromosomes occur randomly (Yang et al., 2016), but Huang et al. (2018) have suggested that chromosomes 1B, 1D, 4A, and 5B undergo more rearrangements. Previous studies have shown that different wheat–Th. ponticum addition or substitution lines exhibit random chromosomal variations (Chen et al., 2012; Zheng et al., 2014; Pei et al., 2018; Wang et al., 2019). The 1RS.1BL translocation was successfully identified by using Oligo-pSc119.2 (6-FAM-5’) and Oligo-pTa535 (Tamra-5’) (Jiang et al., 2017), and a pair of 1RS.1BL translocation chromosomes was detected in CH10A5, which may occurred the cross pollination of wheat during breeding process. In this study, the FISH signals of chromosomes 5A and 6B were changed in CH10A5 (see Figure 3), but no change was observed in the FISH signals of the D genome, indicating that chromosome Js caused chromosomal rearrangement of the A and B genomes. This result confirms that the St or E genome is more closely related to the D genome than to the A and B genomes, consistent with previous studies (Liu et al., 2007). The researchers have constructed a FISH-based karyotype of wheat relatives using oligonucleotide probes, such as Th. bessarabicum, Th. elongatum (Grewal et al., 2018; Li et al., 2018), and Th. intermedium (Cui et al., 2018). We concluded that the chromosome 1E of Th. elongatum could be hybridized with strong signal using Oligo-pSc119.2 (green) and Oligo-pTa535 (red). Chromosomes 1J of Th. bessarabicum had Oligo-pSc119.2 signals (green) on terminal regions of both arms, and the FISH pattern of chromosome Th. intermedium was not very clear, but the GISH pattern was different from the chromosomes 1Js in CH10A5. In our study, the FISH pattern of 1Js was hybridized with weak signal using Oligo- pTa535 (red), but not introducted the signal of Oligo-pSc119.2. In conclusion, the results suggested that chromosomes 1Js in CH10A5 derive from Th. ponticum.
Molecular markers play an important role in tracking alien genetic material in a wheat background and are widely used in genetic breeding, genomic mapping (Zhou et al., 2018), gene localization (Zhang et al., 2018), identification of species relatedness (Wang et al., 2017; Cseh et al., 2019), gene bank construction (Zhang et al., 2002), gene cloning, and other applications (Ma and Tomita, 2013). The lack of rich and reliable molecular markers and the shortage of high-density linkage maps are significant constraints for research work. Some specific molecular markers have been obtained from wheat-related species by SLAF-seq (Chen et al., 2013), and SLAF-seq technology is widely used in molecular marker development for wheat and related species. Chen et al. (2013) developed a large number of Th. elongatum 7E chromosome-specific molecular markers. Liu et al. (2018a) constructed a physical map of chromosome 4Ag from Th. ponticum and obtained 89 blue-grain-related molecular markers; 67 markers specific to Th. ponticum were developed in a separate study (Liu et al., 2018b). All of these markers were developed using SLAF-seq. In this study, some markers were associated with resistance to disease, making them using for the identification and study of related genes. Here, specific markers for Th. ponticum (49 markers), Th. bessarabicum (38), Th. elongatum (45), Ps. spicata (89), and Th. intermedium (57) were also obtained using SLAF-seq, and a number of markers had polymorphisms in two or all five species. These results indicate that chromosome 1Js in CH10A5 may undergo chromosome rearrangements between the J and St genomes, consistent with previous studie (Kruppa and Molnar-Lang, 2016; He et al., 2017). These molecular markers could help us further study the relationships among wheat-related species and detect the introgression of foreign genes.
Conclusions
CH10A5 is a novel 1Js (1D) disomic substitution line derived from a cross between common wheat 7182 and Th. ponticum. The chromosome composition of CH10A5 is 14A+12B+12D+1RS.1BL+1Js. Moreover, it possesses resistance gene(s) to both wheat stripe rust and powdery mildew at the adult stage. In addition, 507 chromosome-specific markers for 1Js were obtained based on SLAF-seq, of which 49 were helpful for characterizing the genetic material of Th. ponticum. All these specific markers will be useful for revealing the genetic diversity among wheat-related species and accelerating the process of molecular marker-assisted breeding. The novel disomic substitution line CH10A5 can be exploited as a promising germplasm for wheat resistance breeding and chromosome engineering.
Data Availability Statement
The datasets for this article are not publicly available because: a large number of verification experiments are still under way. Requests to access the datasets should be directed to WJ, jiwanquan2008@126.com.
Author Contributions
WJ, CW, and HZ designed the research. YanW, QC, JZ, and SW performed the research. CC and CW contributed to the development of the material. YanW, CQ, and JZ collected the samples for DNA extraction and performed the genotyping. YanW analyzed the data. HZ, CW, and YajW contributed to the writing of the article.
Funding
This work was supported by the National Key Research and Development Program of China (No.2016YFD0102000).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Jiachuang Li for GISH of PMCs and Dr. Wei for conducting assessment of the reactions to stripe rust. Thank you for all your help and support. We would also like to thank TopEdit (www.topeditsci.com) for English language editing of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01282/full#supplementary-material
Supplementary Figure1 | Agronomic performance of CH10A5 and its parents. Letters (A–C) refer to plant, spike, and spikelets; numbers 1, 2, and 3 stands for CH10A5, 7182, and Th. ponticum, respectively.
Supplementary Figure 2 | Molecular marker development of CH10A5. Lanes 1, 2, 3 and 4 correspond to CS, 7182, Th. ponticum, and CH10A5, respectively. “*” represents the specific bands.
Supplementary Table 1 | All 72 wheat EST-STS and 135 of PLUG markers used to analyze the chromosomal composition of CH10A5.
Supplementary Table 2 | Genotype data from wheat 15K SNP arrays for CH10A5 and its parents.
Supplementary Table 3 | 507 specific primers for Th. ponticum.
Supplementary Table 4 | The number of specific markers for each wheat relative species.
References
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
An, D., Ma, P., Zheng, Q., Fu, S., Li, L., Han, F., et al. (2019). Development and molecular cytogenetic identification of a new wheat-rye 4R chromosome disomic addition line with resistances to powdery mildew, stripe rust and sharp eyespot. Theor. Appl. Genet. 132, 257–272. doi: 10.1007/s00122-018-3214-3
Bokore, F. E., Cuthbert, R. D., Knox, R. E., Randhawa, H. S., Hiebert, C. W., DePauw, R. M., et al. (2017). Quantitative trait loci for resistance to stripe rust of wheat revealed using global field nurseries and opportunities for stacking resistance genes. Theor. Appl. Genet. 130, 2617–2635. doi: 10.1007/s00122-017-2980-7
Ceoloni, C., Kuzmanović, L., Gennaro, A., Forte, P., Giorgi, D., Rosaria Grossi, M., et al. (2014). “Genomes, Chromosomes and Genes of the Wheatgrass Genus Thinopyrum: the Value of their Transfer into Wheat for Gains in Cytogenomic Knowledge and Sustainable Breeding,” in Genomics of Plant Genetic Resources. Eds. Tuberosa, R., Graner, A., Frison, E. (Dordrecht: Springer Netherlands), 333–358.
Ceoloni, C., Forte, P., Kuzmanovic, L., Tundo, S., Moscetti, I., De Vita, P., et al. (2017). Cytogenetic mapping of a major locus for resistance to Fusarium head blight and crown rot of wheat on Thinopyrum elongatum 7EL and its pyramiding with valuable genes from a Th. ponticum homoeologous arm onto bread wheat 7DL. Theor. Appl. 130, 2005–2024. doi: 10.1007/s00122-017-2939-8
Chen, G., Zheng, Q., Bao, Y., Liu, S., Wang, H., Li, X. (2012). Molecular cytogenetic identification of a novel dwarf wheat line with introgressed Thinopyrum ponticum chromatin. J. Biosci. 37, 149–155. doi: 10.1007/s12038-011-9175-1
Chen, S., Huang, Z., Dai, Y., Qin, S., Gao, Y., Zhang, L., et al. (2013). The development of 7E chromosome-specific molecular markers for Thinopyrum elongatum based on SLAF-seq technology. PLoS One 8, e65122. doi: 10.1371/journal.pone.0065122
Cseh, A., Yang, C., Hubbart-Edwards, S., Scholefield, D., Ashling, S. S., Burridge, A. J., et al. (2019). Development and validation of an exome-based SNP marker set for identification of the St Jr and Jvs genomes of Thinopyrym intermedium in a wheat background. Theor. Appl. Genet. 132, 1555–1570. doi: 10.1007/s00122-019-03300-9
Cui, Y., Zhang, Y., Qi, J., Wang, H., Wang, R. R., Bao, Y., et al. (2018). Identification of chromosomes in Thinopyrum intermedium and wheat-Th. intermedium amphiploids based on multiplex oligonucleotide probes. Genome 61, 515–521. doi: 10.1139/gen-2018-0019
Dai, K., Zhao, R., Shi, M., Xiao, J., Yu, Z., Jia, Q., et al. (2019). Dissection and cytological mapping of chromosome arm 4VS by the development of wheat-Haynaldia villosa structural aberration library. Theor. Appl. Genet. 133, 217–226. doi: 10.1007/s00122-019-03452-8
Dai, K., Zhao, R., Shi, M., Xiao, J., Yu, Z., Jia, Q., et al. (2020). Dissection and cytological mapping of chromosome arm 4VS by the development of wheat-Haynaldia villosa structural aberration library. Theor. Appl. Genet. 133, 217–226. doi: 10.1007/s00122-019-03452-8
Elshire, R. J., Glaubitz, J. C., Sun, Q., Poland, J. A., Kawamoto, K., Buckler, E. S., et al. (2011). A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6, e19379. doi: 10.1371/journal.pone.0019379
Fu, S., Lv, Z., Qi, B., Guo, X., Li, J., Liu, B., et al. (2012). Molecular cytogenetic characterization of wheat-Thinopyrum elongatum addition, substitution and translocation lines with a novel source of resistance to wheat Fusarium Head Blight. J. Genet. Genomics 39, 103–110. doi: 10.1016/j.jgg.2011.11.008
Grewal, S., Yang, C., Edwards, S. H., Scholefield, D., Ashling, S., Burridge, A. J., et al. (2018). Characterisation of Thinopyrum bessarabicum chromosomes through genome-wide introgressions into wheat. Theor. Appl. Genet. 131, 389–406. doi: 10.1007/s00122-017-3009-y
Guo, J., Zhang, X., Hou, Y., Cai, J., Shen, X., Zhou, T., et al. (2015). High-density mapping of the major FHB resistance gene Fhb7 derived from Thinopyrum ponticum and its pyramiding with Fhb1 by marker-assisted selection. Theor. Appl. Genet. 128, 2301–2316. doi: 10.1007/s00122-015-2586-x
He, F., Wang, Y., Bao, Y., Ma, Y., Wang, X., Li, X., et al. (2017). Chromosomal constitutions of five wheat - Elytrigia elongata partial amphiploids as revealed by GISH, multicolor GISH and FISH. Comp. Cytogenet. 11, 525–540. doi: 10.3897/CompCytogen.v11i3.11883
Hou, L. Y., Jia, J., Zhang, X., Li, X., Yang, Z., Ma, J., et al. (2016). Molecular Mapping of the Stripe Rust Resistance Gene Yr69 on Wheat Chromosome 2AS. Plant Dis. 100, 1717–1724. doi: 10.1094/pdis-05-15-0555-re
Huang, X., Zhu, M., Zhuang, L., Zhang, S., Wang, J., Chen, X., et al. (2018). Structural chromosome rearrangements and polymorphisms identified in Chinese wheat cultivars by high-resolution multiplex oligonucleotide FISH. Theor. Appl. Genet. 131, 1967–1986. doi: 10.1007/s00122-018-3126-2
Ishikawa, G., Nakamura, T., Ashida, T., Saito, M., Nasuda, S., Endo, T. R., et al. (2009). Localization of anchor loci representing five hundred annotated rice genes to wheat chromosomes using PLUG markers. Theor. Appl. Genet. 118, 499–514. doi: 10.1007/s00122-008-0916-y
Jiang, M., Xaio, Z. Q., Fu, S. L., Tang, Z. X. (2017). FISH Karyotype of 85 Common Wheat Cultivars/Lines Displayed by ND-FISH Using Oligonucleotide Probes. Cereal Res. Commun. 45, 549–563. doi: 10.1556/0806.45.2017.049
Kroupin, P. Y., Kuznetsova, V. M., Nikitina, E. A., Martirosyan, Y. T., Karlov, G. I., Divashuk, M. G. (2019). Development of new cytogenetic markers for Thinopyrum ponticum (Podp.) Z.-W. Liu & R.-C. Wang. Comp. Cytogenet. 13, 231–243. doi: 10.3897/CompCytogen.v13i3.36112
Kruppa, K., Molnar-Lang, M. (2016). Simultaneous visualization of different genomes (J, JSt and St) in a Thinopyrum intermedium x Thinopyrum ponticum synthetic hybrid (Poaceae) and in its parental species by multicolour genomic in situ hybridization (mcGISH). Comp. Cytogenet. 10, 283–293. doi: 10.3897/CompCytogen.v10i2.7305
Kuzmanovic, L., Mandala, G., Tundo, S., Ciorba, R., Frangella, M., Ruggeri, R., et al. (2019). Equipping Durum Wheat-Thinopyrum ponticum Recombinant Lines With a Thinopyrum elongatum Major QTL for Resistance to Fusarium Diseases Through a Cytogenetic Strategy. Front. Plant Sci. 10, 1324. doi: 10.3389/fpls.2019.01324
Li, H., Wang, X. (2009). Thinopyrum ponticum and Th. intermedium: the promising source of resistance to fungal and viral diseases of wheat. J. Genet. Genomics 36, 557–565. doi: 10.1016/S1673-8527(08)60147-2
Li, H., Chen, Q., Conner, R. L., Guo, B., Zhang, Y., Graf, R. J., et al. (2003). Molecular characterization of a wheat-Thinopyrum ponticum partial amphiploid and its derivatives for resistance to leaf rust. Genome 46, 906–913. doi: 10.1139/g03-053
Li, H. J., Arterburn, M., Jones, S. S., Murray, T. D. (2004). A New Source of Resistance to Tapesia yallundae Associated with a Homoeologous Group 4 Chromosome in Thinopyrum ponticum. Phytopathology 94, 932–937. doi: 10.1094/PHYTO.2004.94.9.932
Li, G., Wang, H., Lang, T., Li, J., La, S., Yang, E., et al. (2016). New molecular markers and cytogenetic probes enable chromosome identification of wheat-Thinopyrum intermedium introgression lines for improving protein and gluten contents. Planta 244, 865–876. doi: 10.1007/s00425-016-2554-y
Li, D., Li, T., Wu, Y., Zhang, X., Zhu, W., Wang, Y., et al. (2018). FISH-Based Markers Enable Identification of Chromosomes Derived From Tetraploid Thinopyrum elongatum in Hybrid Lines. Front. Plant Sci. 9 (526). doi: 10.3389/fpls.2018.00526
Li, J. C., Yao, X. N., Yang, Z. J., Cheng, X. N., Yuan, F. P., Liu, Y., et al. (2019). Molecular cytogenetic characterization of a novel wheat-Psathyrostachys huashanica Keng 5Ns (5D) disomic substitution line with stripe rust resistance. Mol. Breed. 39, 109–123. doi: 10.1007/s11032-019-1014-3
Liu, Z., Li, D., Zhang, X. (2007). Genetic relationships among five basic genomes St, E, A, B and D in Triticeae revealed by genomic southern and in situ hybridization. J. Integr. Plant Biol. 49, 1080–1086. doi: 10.1111/j.1672-9072.2007.00462.x
Liu, L., Luo, Q., Li, H., Li, B., Li, Z., Zheng, Q. (2018a). Physical mapping of the blue-grained gene from Thinopyrum ponticum chromosome 4Ag and development of blue-grain-related molecular markers and a FISH probe based on SLAF-seq technology. Theor. Appl. Genet. 131, 2359–2370. doi: 10.1007/s00122-018-3158-7
Liu, L., Luo, Q., Teng, W., Li, B., Li, H., Li, Y., et al. (2018b). Development of Thinopyrum ponticum-specific molecular markers and FISH probes based on SLAF-seq technology. Planta 247, 1099–1108. doi: 10.1007/s00425-018-2845-6
Liu, L., Yuan, C. Y., Wang, M. N., See, D. R., Zemetra, R. S., Chen, X. M. (2019). QTL analysis of durable stripe rust resistance in the North American winter wheat cultivar Skiles. Theor. Appl. Genet. 132, 1677–1691. doi: 10.1007/s00122-019-03307-2
Ma, Y. Z., Tomita, M. (2013). Thinopyrum 7Ai-1-derived small chromatin with Barley Yellow Dwarf Virus (BYDV) resistance gene integrated into the wheat genome with retrotransposon. Cytol. Genet. 47, 1–7. doi: 10.3103/S0095452713010064
Mago, R., Zhang, P., Xia, X., Zhang, J., Hoxha, S., Lagudah, E., et al. (2019). Transfer of stem rust resistance gene SrB from Thinopyrum ponticum into wheat and development of a closely linked PCR-based marker. Theor. Appl. Genet. 132, 371–382. doi: 10.1007/s00122-018-3224-1
Mo, Q., Wang, C. Y., Chen, C. H., Wang, Y. J., Zhang, H., Liu, X. L., et al. (2017). Molecular cytogenetic identification of a wheat-Thinopyrum ponticum substitution line with stripe rust resistance. Cereal Res. Commun. 45, 564–573. doi: 10.1556/0806.45.2017.037
Niu, Z., Klindworth, D. L., Yu, G., L Friesen, T., Chao, S., Jin, Y., et al. (2014). Development and characterization of wheat lines carrying stem rust resistance gene Sr43 derived from Thinopyrum ponticum. Theor. Appl. Genet. 127, 969–980. doi: 10.1007/s00122-014-2272-4
Okada, M., Yoshida, K., Nishijima, R., Michikawa, A., Motoi, Y., Sato, K., et al. (2018). RNA-seq analysis reveals considerable genetic diversity and provides genetic markers saturating all chromosomes in the diploid wild wheat relative Aegilops umbellulata. BMC Plant Biol. 18, 271. doi: 10.1186/s12870-018-1498-8
Pei, Y., Cui, Y., Zhang, Y., Wang, H., Bao, Y., Li, X. (2018). Molecular cytogenetic identification of three rust-resistant wheat-Thinopyrum ponticum partial amphiploids. Mol. Cytogenet. 11, 27. doi: 10.1186/s13039-018-0378-0
Rey, E., Abrouk, M., Keeble-Gagnere, G., Karafiatova, M., Vrana, J., Balzergue, S., et al. (2018). Transcriptome reprogramming due to the introduction of a barley telosome into bread wheat affects more barley genes than wheat. Plant Biotechnol. J. 16, 1767–1777. doi: 10.1111/pbi.12913
Sheng, B., Duan, X. (1991). Improvement of scale 0–9 method for scoring adult plant resistance to powdery mildew of wheat (Beijing Agricultural Sciences).
Sun, X., Liu, D., Zhang, X., Li, W., Liu, H., Hong, W., et al. (2013). SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS One 8, e58700. doi: 10.1371/journal.pone.0058700
Tang, Z., Yang, Z., Fu, S. (2014). Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55, 313–318. doi: 10.1007/s13353-014-0215-z
Tang, S., Tang, Z., Qiu, L., Yang, Z., Li, G., Lang, T., et al. (2018). Developing New Oligo Probes to Distinguish Specific Chromosomal Segments and the A, B, D Genomes of Wheat (Triticum aestivum L.) Using ND-FISH. Front. Plant Sci. 9, 1104. doi: 10.3389/fpls.2018.01104
Tiwari, V. K., Wang, S., Danilova, T., Koo, D. H., Vrana, J., Kubalakova, M., et al. (2015). Exploring the tertiary gene pool of bread wheat: sequence assembly and analysis of chromosome 5M(g) of Aegilops geniculata. Plant J. 84, 733–746. doi: 10.1111/tpj.13036
Wang, Y., Quan, W., Peng, N., Wang, C., Yang, X., Liu, X., et al. (2016). Molecular cytogenetic identification of a wheat–Aegilops geniculata Roth 7Mg disomic addition line with powdery mildew resistance. Mol. Breed. 36, 143. doi: 10.1007/s11032-016-0463-1
Wang, R. R., Larson, S. R., Jensen, K. B. (2017). Differential transferability of EST-SSR primers developed from the diploid species Pseudoroegneria spicata, Thinopyrum bessarabicum, and Thinopyrum elongatum. Genome 60, 530–536. doi: 10.1139/gen-2016-0157
Wang, S. W., Wang, C. Y., Wang, Y. Z., Wang, Y. J., Chen, C. H., Ji, W. Q. (2019). Molecular cytogenetic identification of two wheat–Thinopyrum ponticum substitution lines conferring stripe rust resistance. Mol. Breed. 39, 143–153. doi: 10.1007/s11032-019-1053-9
Wang, H. W., Sun, S., Ge, W., Zhao, L., Hou, B., Wang, K., et al. (2020a). Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 368, eaba5435. doi: 10.1126/science.aba5435
Wang, H., Zou, S., Li, Y., Lin, F., Tang, D. (2020b). An ankyrin-repeat and WRKY-domain-containing immune receptor confers stripe rust resistance in wheat. Nat. Commun. 11, 1353. doi: 10.1038/s41467-020-15139-6
Yang, X., Wang, C., Chen, C., Zhang, H., Tian, Z., Li, X., et al. (2014). Chromosome constitution and origin analysis in three derivatives of Triticum aestivum-Leymus mollis by molecular cytogenetic identification. Genome 57, 583–591. doi: 10.1139/gen-2014-0161
Yang, W., Wang, C., Chen, C., Wang, Y., Zhang, H., Liu, X., et al. (2016). Molecular cytogenetic identification of a wheat-rye 1R addition line with multiple spikelets and resistance to powdery mildew. Genome 59, 277–288. doi: 10.1139/gen-2015-0129
Zhan, H., Li, G., Zhang, X., Li, X., Guo, H., Gong, W., et al. (2014). Chromosomal location and comparative genomics analysis of powdery mildew resistance gene Pm51 in a putative wheat-Thinopyrum ponticum introgression line. PLoS One 9, e113455. doi: 10.1371/journal.pone.0113455
Zhan, H., Zhang, X., Li, G., Pan, Z., Hu, J., Li, X., et al. (2015). Molecular characterization of a new wheat-Thinopyrum intermedium translocation line with resistance to powdery mildew and stripe rust. Int. J. Mol. Sci. 16, 2162–2173. doi: 10.3390/ijms16012162
Zhang, Z. Y., Wang, L. L., Xin, Z. Y., Lin, Z. S. (2002). Development of new PCR markers specific to a Thinopyrum intermedium chromosome 2Ai-2 and cloning of the St-specific sequences. Dasypyrum Villosum 29, 627–633.
Zhang, R., Fan, Y., Kong, L., Wang, Z., Wu, J., Xing, L., et al. (2018). Pm62, an adult-plant powdery mildew resistance gene introgressed from Dasypyrum villosum chromosome arm 2VL into wheat. Theor. Appl. Genet. 131, 2613–2620. doi: 10.1007/s00122-018-3176-5
Zheng, Q., Li, B., Zhang, X., Mu, S., Zhou, H., Li, Z. (2006). Molecular cytogenetic characterization of wheat-Thinopyrum ponticum translocations bearing blue-grained gene(s) induced by r-ray. Euphytica 152, 51–60. doi: 10.1007/s10681-006-9176-6
Zheng, Q., Lv, Z., Niu, Z., Li, B., Li, H., Xu, S. S., et al. (2014). Molecular cytogenetic characterization and stem rust resistance of five wheat-Thinopyrum ponticum partial amphiploids. J. Genet. Genomics 41, 591–599. doi: 10.1016/j.jgg.2014.06.003
Zhou, S., Zhang, J., Che, Y., Liu, W., Lu, Y., Yang, X., et al. (2018). Construction of Agropyron Gaertn. genetic linkage maps using a wheat 660K SNP array reveals a homoeologous relationship with the wheat genome. Plant Biotechnol. J. 16, 818–827. doi: 10.1111/pbi.12831
Zhu, C., Wang, Y., Chen, C., Wang, C., Zhang, A., Peng, N., et al. (2017). Molecular cytogenetic identification of a wheat-Thinopyrum ponticum substitution line with stripe rust resistance. Genome 60, 860–867. doi: 10.1139/gen-2017-0099
Keywords: Thinopyrum ponticum, wide hybridization, wheat 15K array, Pst, Bgt SLAF-seq
Citation: Wang Y, Cao Q, Zhang J, Wang S, Chen C, Wang C, Zhang H, Wang Y and Ji W (2020) Cytogenetic Analysis and Molecular Marker Development for a New Wheat–Thinopyrum ponticum 1Js (1D) Disomic Substitution Line With Resistance to Stripe Rust and Powdery Mildew. Front. Plant Sci. 11:1282. doi: 10.3389/fpls.2020.01282
Received: 06 March 2020; Accepted: 06 August 2020;
Published: 21 August 2020.
Edited by:
Luigi Cattivelli, Council for Agricultural and Economics Research, ItalyReviewed by:
Steve Richard Larson, United States Department of Agriculture, United StatesZujun Yang, University of Electronic Science and Technology of China, China
Copyright © 2020 Wang, Cao, Zhang, Wang, Chen, Wang, Zhang, Wang and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanquan Ji, jiwanquan2008@126.com
 Yanzhen Wang
Yanzhen Wang Qiang Cao1,2
Qiang Cao1,2 Hong Zhang
Hong Zhang Wanquan Ji
Wanquan Ji