- 1Department of Horticulture, Plant Biotechnology Resource and Outreach Center, Michigan State University, East Lansing, MI, United States
- 2Biotechnology Laboratory, La Platina Station, Instituto de Investigaciones Agropecuarias, Santiago de Chile, Chile
- 3Citrus Research and Education Center, Institute of Food and Agricultural Sciences (IFAS), University of Florida, Lake Alfred, FL, United States
Genetic engineering based on Agrobacterium-mediated transformation has been a desirable tool to manipulate single or multiple genes of existing genotypes of woody fruit crops, for which conventional breeding is a difficult and lengthy process due to heterozygosity, sexual incompatibility, juvenility, or a lack of natural sources. To date, successful transformation has been reported for many fruit crops. We review the major progress in genetic transformation of these fruit crops made in the past 5 years, emphasizing reproducible transformation protocols as well as the strategies that have been tested in fruit crops. While direct transformation of scion cultivars was mostly used for fruit quality improvement, biotic and abiotic tolerance, and functional gene analysis, transgrafting on genetically modified (GM) rootstocks showed a potential to produce non-GM fruit products. More recently, genome editing technology has demonstrated a potential for gene(s) manipulation of several fruit crops. However, substantial efforts are still needed to produce plants from gene-edited cells, for which tremendous challenge remains in the context of either cell’s recalcitrance to regeneration or inefficient gene-editing due to their polyploidy. We propose that effective transient transformation and efficient regeneration are the key for future utilization of genome editing technologies for improvement of fruit crops.
Introduction
Fruits and nuts (F&N) provide essential nutrients for human growth and health. There are 494 culinary fruits (excluding fruit vegetables such as watermelons and tomatoes) and 14 nuts listed as of submission in Wikipedia. The increasing consumer awareness regarding health benefits and growing population worldwide are boosting a market for more and higher-quality fruits and nuts. For example, the total global production of fresh fruit increased from 13.6 million metric tons in 1996 to 33.3 million metric tons in 2016 (FAO). On the other hand, many adverse impacts, including emerging diseases (e.g., citrus greening and papaya ringspot virus), abiotic stress (e.g., salt, drought, and extreme temperatures) due to global warming, and natural resource depletion (e.g., land and water) due to the growing population, have threatened fruit yield and quality of F&N crops (Gottwald, 2010; Castillo et al., 2011; Fiore et al., 2018). Thus, the major task for F&N breeders is to develop new cultivars with improved resistance to diseases and abiotic stress, and higher productivity. Conventional breeding for many F&N crops often takes a few to over 10 years due to their long juvenile periods and asexual propagation nature (Janick, 2005). Utilization of biotechnology in breeding is an efficient alternative allowing for the manipulation of gene(s) of interest (GOI) through genetic engineering in shorter period of time, relative to conventional breeding. The efficiency of biotechnology application in breeding has become evident with the successes achieved via Agrobacterium-mediated transformation (Figure 1 and Table 1), and carries a great potential with increased availability of sequenced genomes of F&N crops that can be used by technical advances such as gene editing for trait improvement (Table 2).
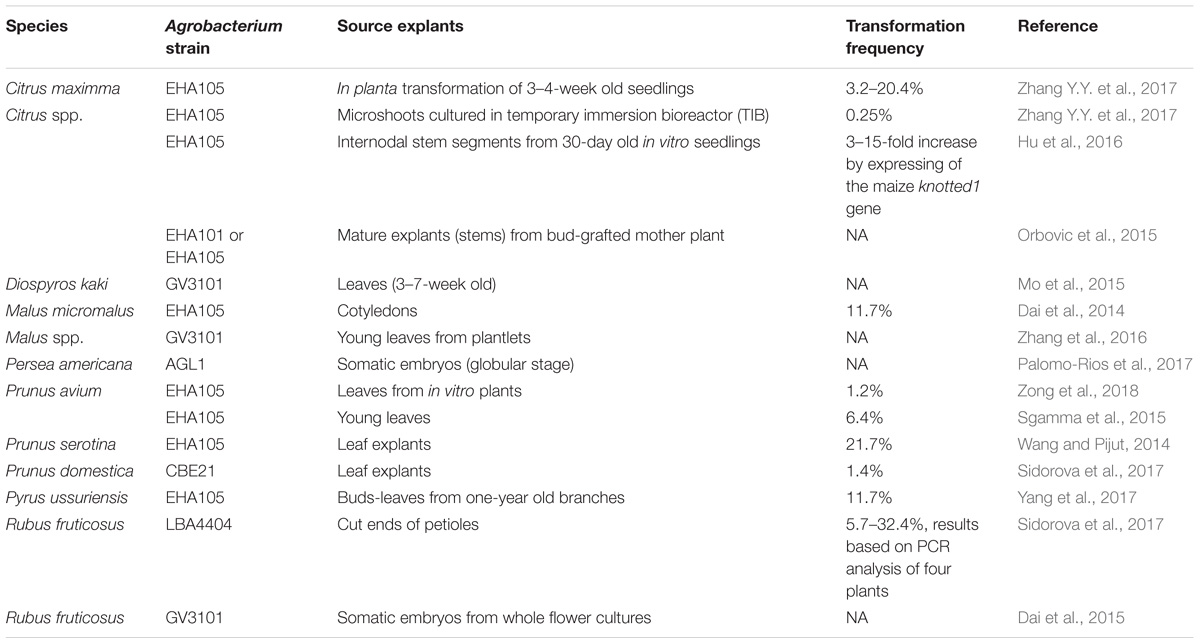
Table 1. Recent successes (2014-present) in Agrobacterium tumefaciens-mediated transformation of F&N crops for transformation technology development.
New biotechnological tools revolutionized plant breeding and offered new and effective ways for plant breeders to manipulate traits at the levels of individual gene(s) or gene blocks (Gelvin, 2012; Hiei et al., 2014; Nester, 2014). Except for the widely commercialized virus-resistant papaya produced in 1992 through biolistic-mediated transformation (Fitch et al., 1992), virus-resistant plum (Ravelonandro et al., 1997; Scorza et al., 2001, 2007) and non-browning apples (Waltz, 2015) have been both produced by Agrobacterium-mediated transformation. The transgenic plum is on the horizon to be commercially released and the non-browning apples are in the stores. Agrobacterium-mediated transformation protocols for stable and transient expression remain a major platform for gene editing technologies in F&N species (Table 1). In this review, we summarize the availability of reliable transformation protocols and discuss recent progress, current constraints, and future perspectives of application of Agrobacterium-mediated transformation for the improvement of woody F&N crops.
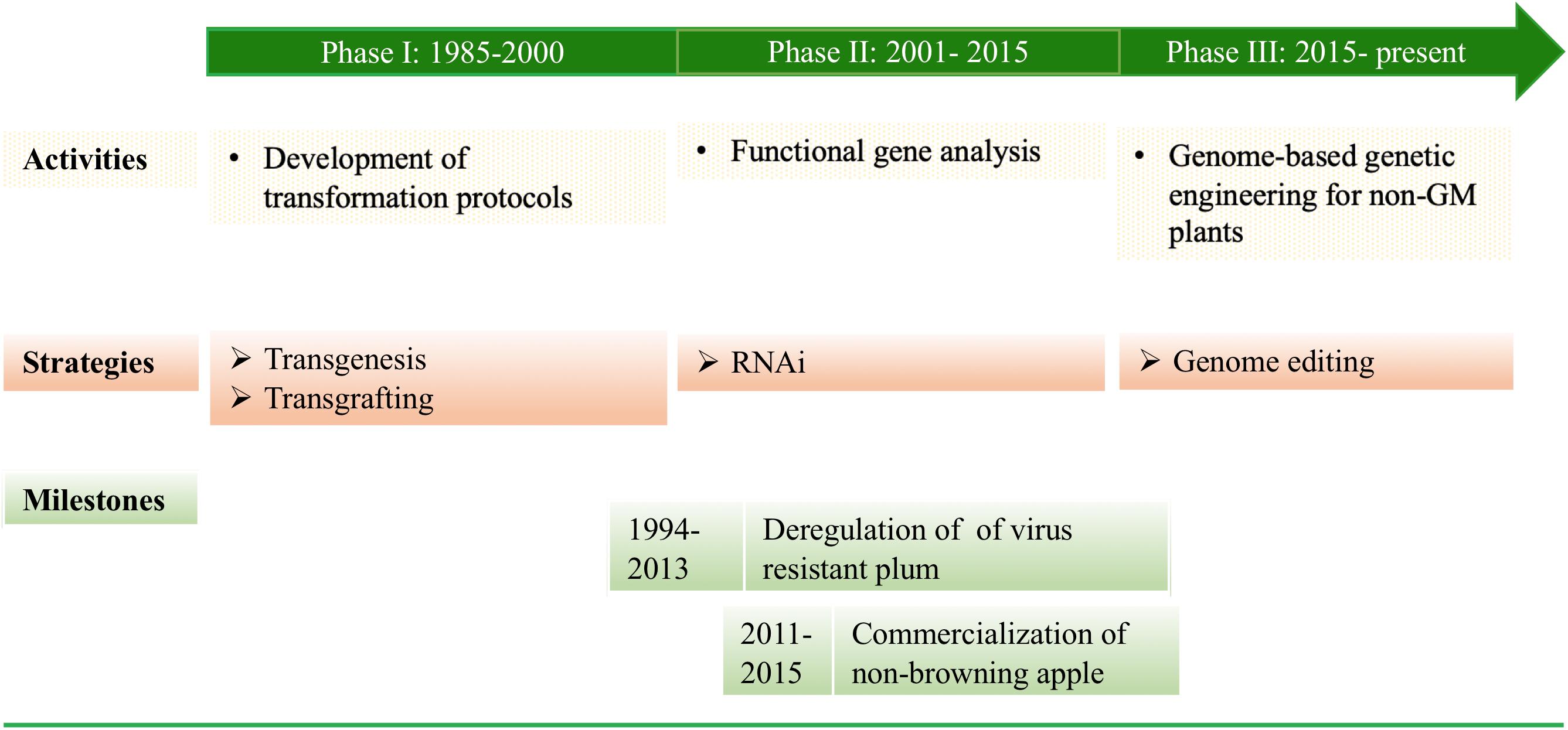
History of Genetic Transformation of F&N Crops (Figure 1)
Phase I (1985–2000)
Development of transformation protocols. After success in Agrobacterium tumefaciens-mediated stable transformation of tobacco plants (Horsch et al., 1985), substantial effort was made to develop A. tumefaciens-mediated transformation protocols for F&N crops (Kole and Hall, 2008), such as apple (Yao et al., 1995), pear (Mourgues et al., 1996), plum (Mante et al., 1991), cherry rootstock (Gutierrez-Pesce et al., 1998), grapes (Mullins et al., 1990), walnuts (Mcgranahan et al., 1988), kiwifruits (Uematsu et al., 1991), citrus (Cervera et al., 1998), and European chestnuts (Seabra and Pais, 1998).
Phase II (2001–2015)
RNA interference (RNAi) technologies. The efforts to develop/improve transformation protocols for more F&N crops or cultivars continued and as a result blueberry (Song and Sink, 2004) and sour cherry (Song and Sink, 2006) were transformed. RNAi was used to suppress either virus RNAs or plant endogenous RNAs in plum (Scorza et al., 2004, 2013), cherry (Song et al., 2013), and apple (Saurabh et al., 2014; Waltz, 2015). On the other hand, driven by advances in sequencing, cloning, and RNAi technologies, functional gene analysis became the major focus for the F&N crops, and workable transformation protocols had been developed, including transient expression systems (Scorza et al., 2013). The recombination/excision systems [e.g., Cre/LoxP and Flp-FRT (flippase recognition target)] have been demonstrated to be effective in producing selectable marker gene (SMG)-free apple (Kost et al., 2015; Krens et al., 2015), apricot (Petri et al., 2012), and citrus (Zou et al., 2013). The most significant progress at this stage include: (1) Deregulation of transgenic plum with plum pox virus (PPV) resistance (Scorza et al., 2007, 2013); and (2) Commercialization of non-browning apples (Waltz, 2015).
Phase III (2015–Present)
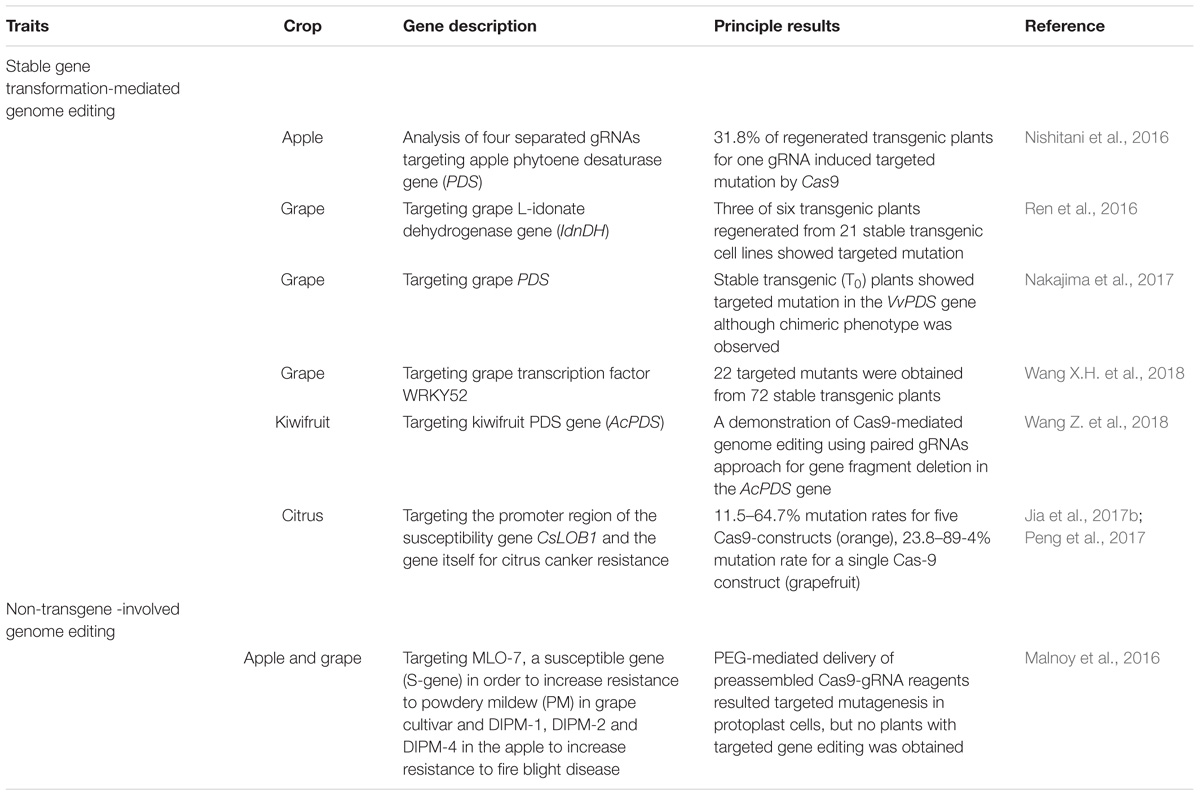
Precision breeding. Gene editing technologies have become powerful tools to precisely manipulate nucleic acids in a plant cell. The very first attempts of these technologies in apple (Nishitani et al., 2016), grape (Ren et al., 2016; Nakajima et al., 2017; Wang X.H. et al., 2018), sweet orange and grapefruit (Jia and Wang, 2014; Zhang F. et al., 2017), and kiwifruit (Wang Z. et al., 2018) have relied on the use of Agrobacterium to produce stable transgenic plants expressing either editing reagents or small RNAs inducers. Ideally, transient expression of editing reagents leading to stable editing of a GOI or a regulatory DNA sequence, similar to those demonstrated in annual crops (Svitashev et al., 2016; Liang et al., 2018), will be the next step for F&N plants.
Transformation Protocols for Woody Fruit and Nut Crops
The current transformation protocols rely on procedures mainly developed between 1990 and 2000. Within the group of F&N species, the majority (over 95%) are still recalcitrant for transformation, and most of the transgenic F&N crops were produced using A. tumefaciens-mediated transformation (Singh and Sansavini, 1998; Kole and Hall, 2008). The scarce availability of regenerable explant sources and the time required to produce transgenic individuals are the major constraints (Figure 2). For example, only 16 papers on developing or optimizing Agrobacterium-mediated transformation of F&N crops have been published since 2014, but regardless of these excellent efforts, little break-through in terms of transformed species and transformation frequency has been made (Table 1).
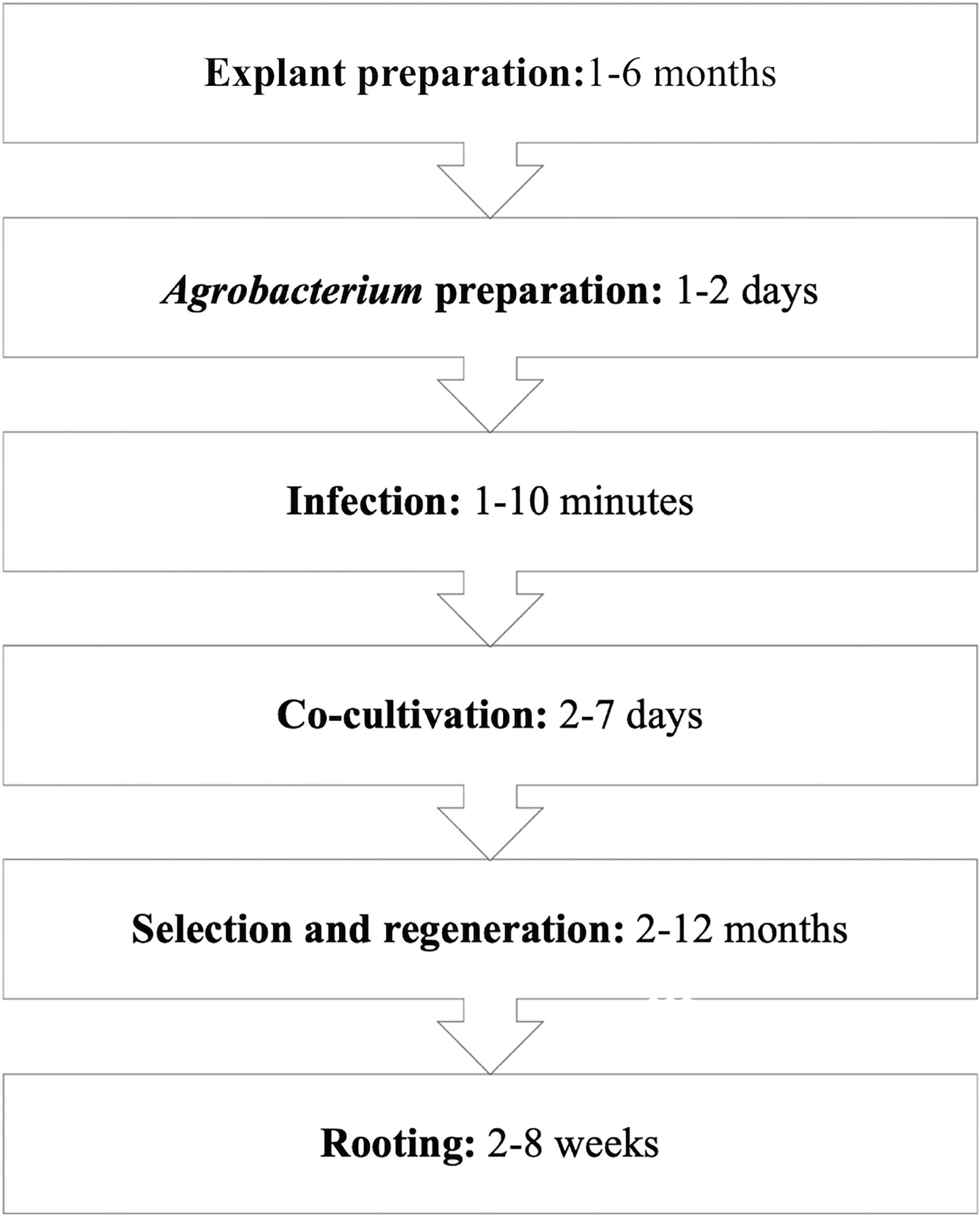
Figure 2. General protocols for Agrobacterium tumefaciens-mediated transformation of F&N crops (e.g., blueberry) (Song, 2015).
Key Limitations for Transformation of F&N Crops
Efficient protocols for transformation rely on effective gene delivery, easy but accurate selection, and prolific regeneration from treated explants (Figure 2).
Agrobacterium-mediated gene transfer is a powerful tool for delivery of transgenes although optimal conditions for efficient gene delivery vary for different species, genotypes and strains of Agrobacterium (Wang, 2015). Transfer DNA (T-DNA) has been shown to be a consistent carrier for a considerable variety of cargoes ranging from conventional expression cassettes used for GOIs, to the current RNA hairpin inducers (Song et al., 2013) or shuttle vectors for secondary DNA-replicons used in gene editing (Baltes et al., 2014). A. tumefaciens is preferable to biolistic guns for stable transformation of F&N crops due mainly to its low cost in operation and the high potential in producing transformations with a low-copy number of the inserted sequence (such as GOI) (Gelvin, 2012). A. tumefaciens with ACC deaminase activity has been developed to improve transformation frequency of annual plants through reducing ethylene levels in plants (Nonaka and Ezura, 2014), although it has not been tested in F&N crops. Up to now, gene delivery is not a key limitation for transformation of F&N crops, A. tumefaciens-mediated transformation remains the major approach (Tables 1–3).
Within the protocols for transformation of F&N crops, the effective selection is achieved mainly by using a SMG conferring resistance to the plant against either an antibiotic or an herbicide (Miki and McHugh, 2004). Three major SMG for plant transformation include the neomycin phosphotransferase II (nptII) gene that confers kanamycin resistance, the hygromycin-B-phosphotransferase gene (hph) conferring hygromycin B resistance, and the bar gene conferring resistance to phosphinothricin, a glutamine synthetase inhibitor produced by plants from the herbicide bialophos (Miki and McHugh, 2004). The nptII is, to date, the most widely used SMG for the transformation of F&N crops (Kole and Hall, 2008). However, effective selection of transformed cells without assistance of SMGs remains a key limitation for plant transformation.
In general, plants with high regeneration capability from non-meristem containing explants are amenable for genetic transformation. Unlike annual crops (e.g., wheat, maize, rice, and soybean), the cultivated F&N crops, for example, blueberries (Song and Hancock, 2012) and cherries (Song, 2014; Zong et al., 2018), are often clonally propagated; to maintain genetic stability, regeneration from explants of clonally propagated tissues is preferable. Lack of available explants sources (e.g., seedlings) makes plant regeneration studies for F&N crops a lengthy process (Figure 3), because in many instances an efficient micropropagation system has to be established for obtaining regenerable explants (e.g., leaves and petioles) prior to regeneration studies (Kole and Hall, 2008). Optimal conditions for both micropropagation and regeneration are determined by many factors (e.g., blueberries) (Liu et al., 2010; Figure 3). In addition, little has been documented on the potential impacts of plant-Agrobacterium interaction on plant regeneration. Finally, for some extremely recalcitrant species in the group (e.g., Prunus persica) species, regeneration can be achieved mostly on the basis of the use of seed explants (Song, 2014; Petri et al., 2018; Zong et al., 2018). In terms of using transgenes to improve regeneration efficiency, one report demonstrated that constitutive expression of the class I KNOX gene of maize increased production of adventitious shoots from leaf explants of plum (Srinivasan et al., 2011). More recently, overexpression of morphogenic regulators, i.e., maize Baby boom and maize Wuschel2, has been demonstrated to be effective in improving monocot transformation (Artlip et al., 2016); and the potential for use of these genes for F&N crops has been recently demonstrated in citrus (Miki and McHugh, 2004).
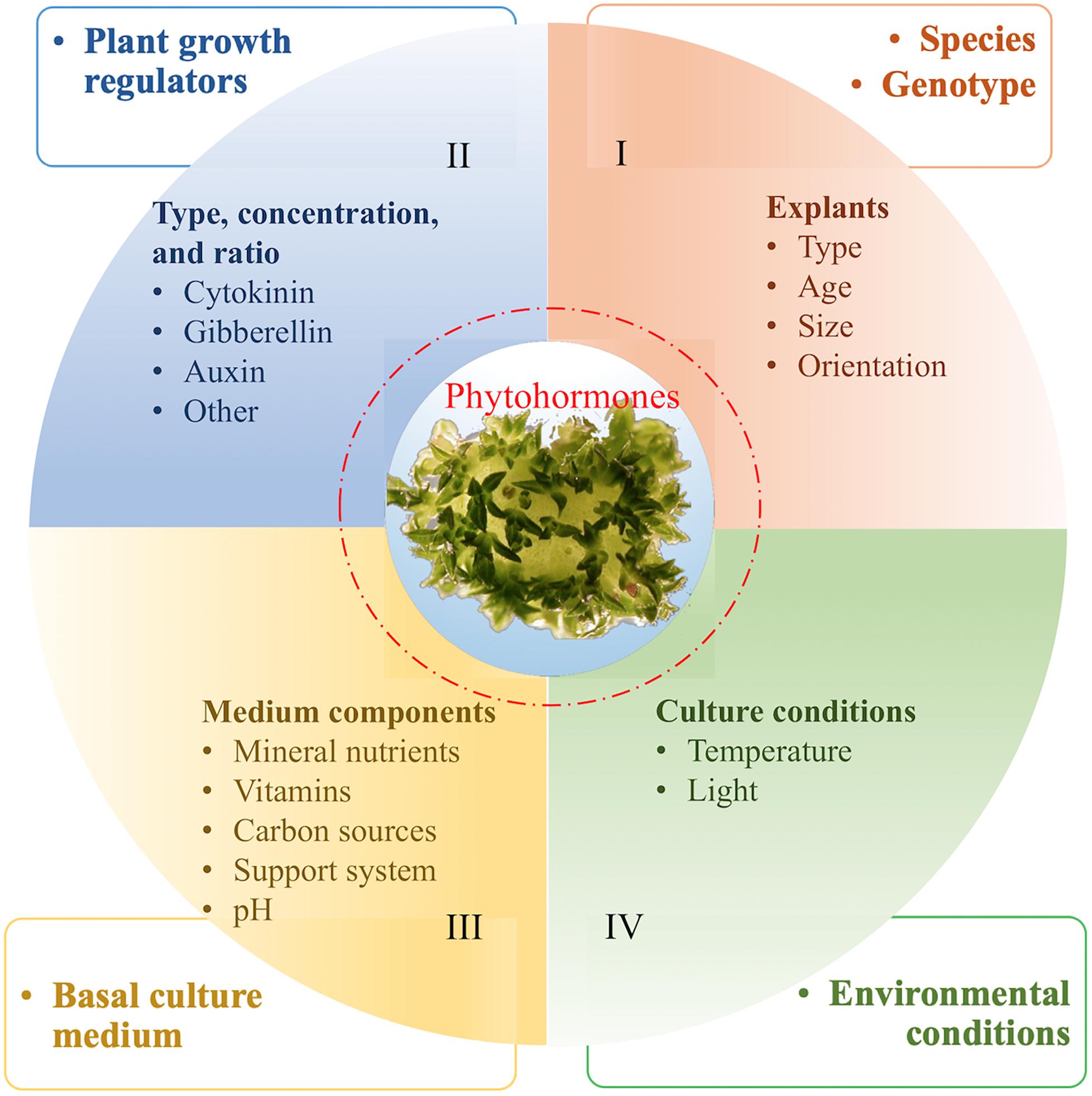
Figure 3. Factors affecting regeneration of plant cells [An example of shoot organogenesis from leaf explants of blueberries (Song and Sink, 2004; Liu et al., 2010)]. We hypothesize that regeneration capability is determined by endogenous phytohormone levels, which are genetically controlled (I) and can be affected by II–IV.
Recent Developments in Genetic Engineering of F&N Crops
Genetic transformation of F&N crops might be necessary when a breeding goal is not easily achievable through traditional breeding approaches (Singh and Sansavini, 1998). Of many approaches developed, gene addition and gene subtraction are two basic strategies for GM trait development (Pena and Seguin, 2001).
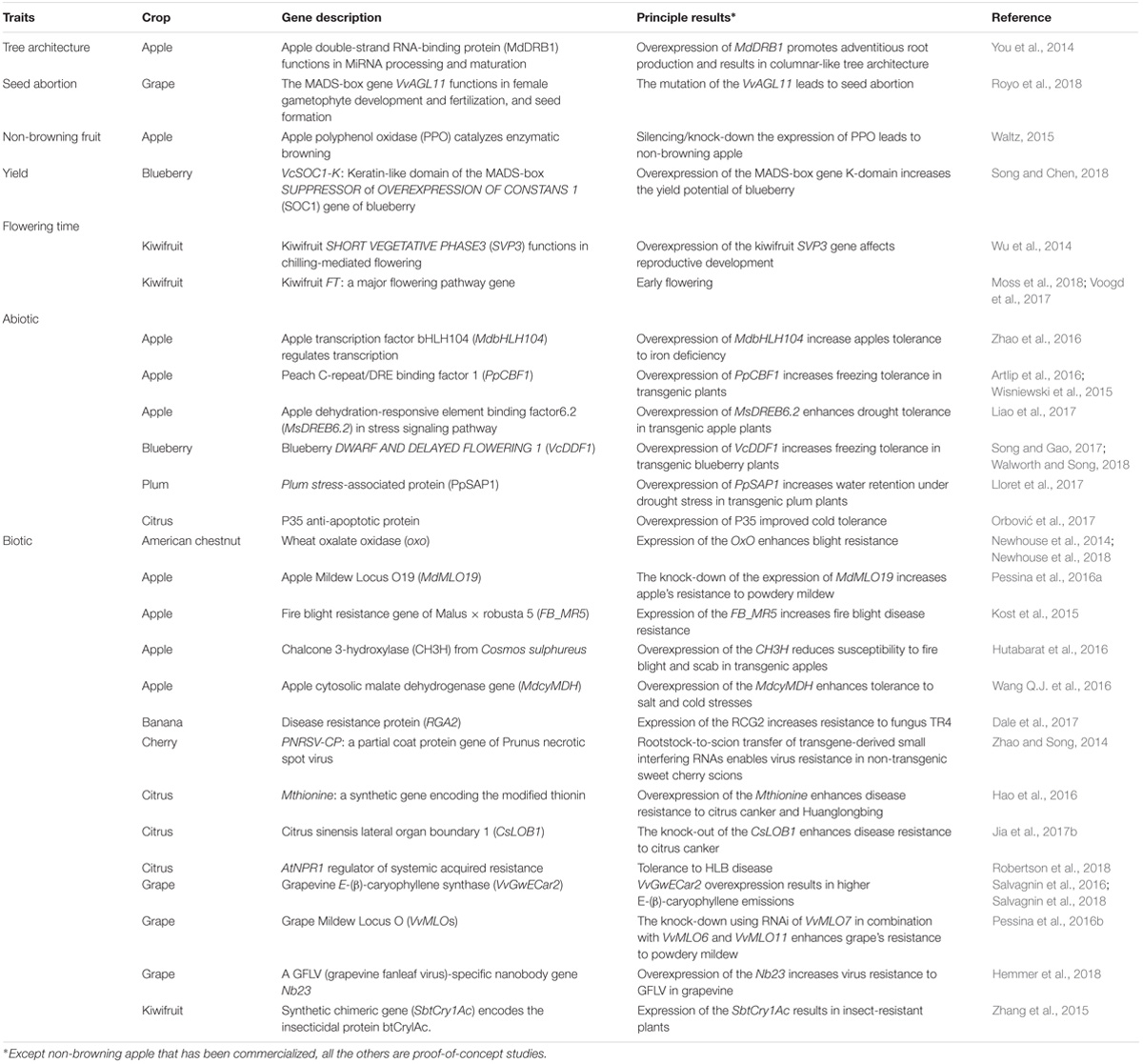
Table 3. Recent successes (2014-present) in A. tumefaciens-mediated transformation of F&N crops for agronomic trait improvement.
Transgrafting
Grafting by artificially conjoining different vascular systems (i.e., rootstock and scion) is a widely used agricultural practice with over 3000-year history for horticultural crops, especially for F&N crops (Jensen et al., 2012; Nawaz et al., 2016; Chitarra et al., 2017). Traditionally, grafting is used to produce plants for asexual propagation, altered plant vigor and architecture, increased tolerance to biotic/abiotic stresses, precocity, and higher yield. The term transgrafting was introduced when GE rootstocks were used in grafting in 1990 (Haroldsen et al., 2012; Figure 4). To date, long-distance transportation of transgene-derived small interfering RNAs (siRNA) from rootstock to non-transgenic sweet cherry scions have been verified by small RNA sequencing (Zhao and Song, 2014). For large mRNA molecules, qRT-PCR analysis detected short-distance transported mRNAs of the reporter of the red fluorescent protein gene (DsRED) from transgenic rootstock to non-transgenic scion of walnut (Juglans regia) (Liu et al., 2017); in contrast, for long-distance (>1 m) transportation, the transgene (e.g., the SMG nptII gene) was not detected in non-transgenic sweet cherry scions grafted on transgenic rootstock through RT-PCR (Zhao and Song, 2014). More recently, a new study has suggested that cell-to-cell movement of mRNAs is selective (Luo et al., 2018). In fact, interaction of transgenic rootstocks and non-transgenic scions in transgrafted plants through either the mobile transgenic products or immobile transgenic products has been demonstrated to be effective in facilitating changes in non-transgenic scions directly or indirectly (Smolka et al., 2010; Song et al., 2015; Artlip et al., 2016); this is also well-supported by recent studies of grafting in non-transgenic rootstocks and scions (Jensen et al., 2012; Chitarra et al., 2017). Using transgenic apple rootstock expressing the root-inducing rolB gene of Agrobacterium rhizogenes T-DNA reduced the vegetative growth of nontransgenic scions (Smolka et al., 2010). Thus, transgenic rootstocks have the potential to expand the use of transgenesis for production of non-transgenic F&N crops (Haroldsen et al., 2012).
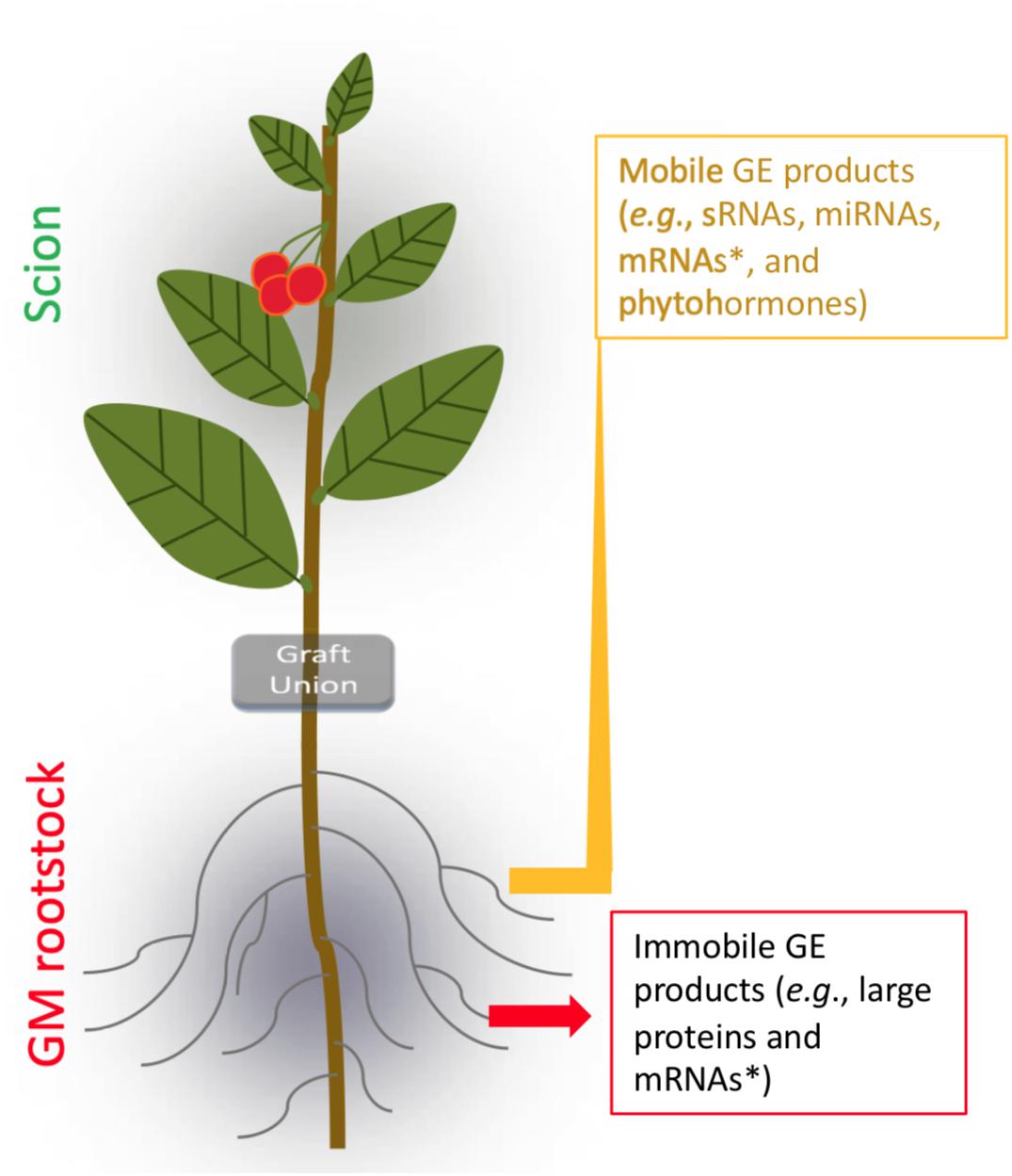
Figure 4. Transgrafting of F&N crops. GE, genetic engineering; SRNAs, small RNAs; miRNAs, micro RNAs. ∗Selective targeting movement (Luo et al., 2018).
FastTrack Breeding
The long juvenile phase exhibited by several F&N crops can severely limit the traditional breeding efforts which are dependent on the ability to make genetic crosses (Janick, 2005). “FastTrack breeding” as demonstrated in many recent publications is done through the manipulation of flowering pathway genes to hasten flowering (Figure 5 and Table 3; Petri et al., 2018). Stable transformation of elite cultivars by either overexpression of flower promoting genes (e.g., FLOWERING LOCUS T, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1, LEAFY, and APETALA1) or repression of flower repressing genes (e.g., TERMINAL FLOWER 1) is an effective approach to enable a fast introduction of genes of interest from wild germplasm through the early flowering seedlings from both crosses and backcrosses (Figure 5A). This approach relies on efficient transformation systems for elite cultivars and low cross-incompatibility between the transformed elite cultivars and donor plants. Transgrafting on transgenic rootstocks can affect flowering in non-transgenic scions (Artlip et al., 2016), suggesting that there is a potential to use a rootstock overexpressing FT to promote early flowering of the scions from juvenile seedlings, although this potential has not yet successful been demonstrated for trees (Zhang et al., 2010; Srinivasan et al., 2012; Figure 5B). Alternatively, transient transformation using virus vectors either to enhance expression of flower promoting genes expression or to repress flower repressing genes has showed a potential in promoting flowering of juvenile plants (Yamagishi et al., 2014; Velazquez et al., 2016).
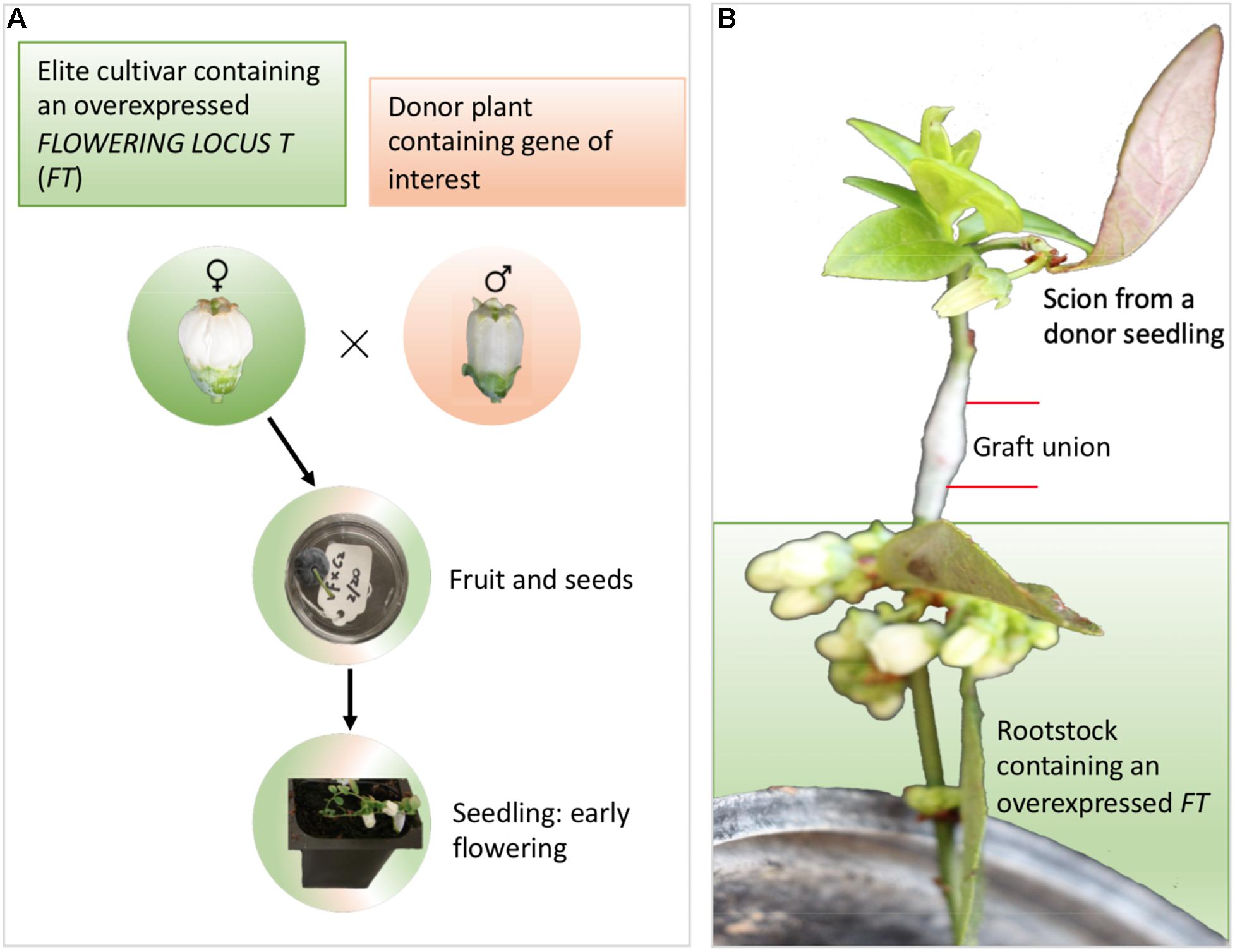
Figure 5. Different approaches for FastTrack breeding – an example for blueberries. (A) Stable gene transformation into elite cultivars for overexpression of flower-promoting genes (e.g., FT, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1, LEAFY, and APETALA1) or repression of flower-repressing genes (e.g., TERMINAL FLOWER 1). The early flowering seedlings enable blueberry to skip 2–3 years of juvenile period for each cross. (B) Transgrafting on FT-overexpressing rootstock promotes early flowering of the scion.
Selectable Marker Gene-Free Plants and Intragenesis
The negative perception of transgenic plants by consumers spurred research to develop either plants modified with plant derived sequences (intragenesis) or transgenic plants from which the SMG was removed (Figure 6). While these strategies have the potential to allay consumer fears, both approaches are currently regulated as transgenic plants worldwide (Holme et al., 2013).
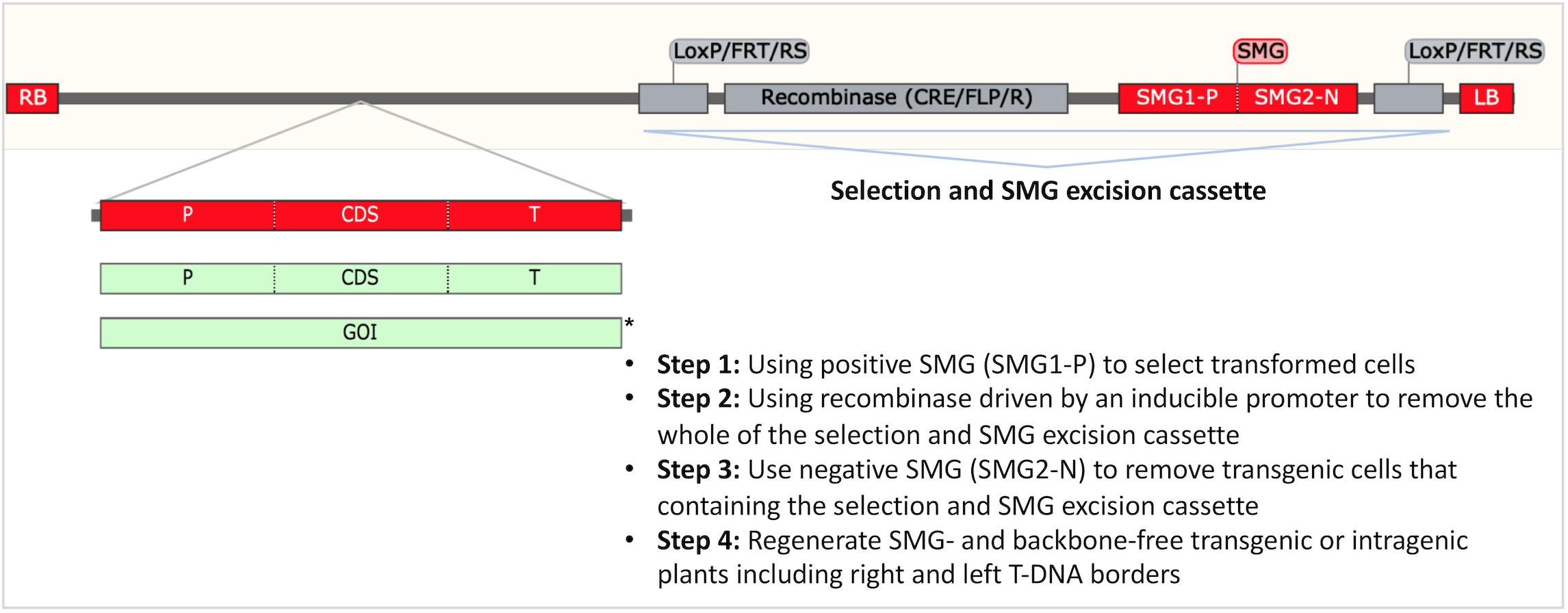
Figure 6. Transgenesis without a SMG and intragenesis. A strategy to obtain SMG free and backbone free transgenic and intragenic F&N crops. The T-DNA region is drawn according to the construct used for intragenic apple (De Paepe et al., 2009; Kost et al., 2015; Collier et al., 2018). This construct allows production of SMG-free T0 transformants. RB: right T-DNA border. LB: left T-DNA border. P: promoter. T: terminator. CDS: coding sequence. GOI: gene of interest. Green boxes show plant-derived intragenic/cisgenic components. ∗Cisgenesis has not been demonstrated in A. tumefaciens-mediated transformation of plants because RB and LB are difficult to replace or remove.
Transgenesis without a SMG or with a plant-derived SMG is preferable to SMG-containing transgenic products by the public (Miki and McHugh, 2004). To date, plant-derived reporters, for example, the genes regulating anthocyanin biosynthesis have been successfully used as alternative to non-plant-derived reporters [i.e., green fluorescent protein (GFP), β-glucuronidase (GUS)] for transformation (Krens et al., 2015; Kandel et al., 2016; Dutt et al., 2018). Co-transformation enables production of SMG-free GM crops through the crosses for the segregation of SMG-removal transformants in next generation, but this is not desirable for asexually propagated F&N crops. The recombination/excision systems [e.g., Cre/LoxP and Flp-FRT (flippase recognition target)] are effective in generating SMG-free apple (Kost et al., 2015; Krens et al., 2015), apricot (Petri et al., 2012), grape (Dalla Costa et al., 2016), and citrus (Zou et al., 2013).
Intragenesis is desirable for food crops because it relies on the gene pool for conventional breeding. Technically, intragenesis is more challenging than SMG-assisted transgenesis (Figure 6). Previous studies have demonstrated the potential of production of intragenic apples and citrus (An et al., 2013; Kost et al., 2015; Krens et al., 2015).
Genome Editing
Genome editing technologies provide powerful tools for precise manipulation of targeted genome sequence(s) for crop improvement (Arora and Narula, 2017; Limera et al., 2017). Remarkably, these technologies make it possible to edit or excise a specific gene in a genome without introduction of any extra DNA.
Programmable DNA binding proteins such zinc finger (ZF) and transcription activator-like effector (TALE) emerged as the first generation engineered nucleases to create targeted mutagenesis, which is an alternative to classical protocols for random mutagenesis. These tools have a recognition capability of specific target DNA sequences based on customized arrangements of one (TALE) or three (ZF) nucleotides, and in such way bringing to these places a nuclease (for instance C-terminal domain of FokI) which disrupts DNA adjacent to the recognition zones. Both ZF- and TALE-nucleases, require two effectors (left and right) in order to define the nuclease cutting site (Gaj et al., 2013). Prior to the wide application of these technologies for F&N crops, a more powerful gene editing tool -The clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 mediated gene editing technology was developed (Jinek et al., 2012). The CRISPR/Cas9 has been a revolutionary molecular tool since its discovery as an adaptive line of defense against viral infection in Archaea (Mojica et al., 2000). This system operates through guide RNAs (gRNAs) that contain specific sequences designed according to their targets in the genome. The Cas nuclease (commonly Cas9), when directed by the gRNA generates a double strand break adjacent to the gRNA’s annealing location allowing for a target-specific mutagenesis. More recently, CRISPR/Cpf1, another CRISPR/Cas system that overcomes some of the CRISPR/Cas9 system limitations, has been found more efficient at DNA editing (Ledford, 2015; Zetsche et al., 2015; Fonfara et al., 2016; Jia et al., 2017a).
Delivery of CRISPR/Cas9 components into the plant cell has been achieved by either transgenic or non-transgenic approaches (Table 2). To date, as proof-of-concept, CRISPR/Cas9 guided DNA editing through stable transformation has been demonstrated in four major fruit crops [i.e., apple (Nishitani et al., 2016), grape (Ren et al., 2016; Nakajima et al., 2017; Wang X.H. et al., 2018), sweet orange and grapefruit (Jia and Wang, 2014; Zhang F. et al., 2017), and kiwifruit (Wang Z. et al., 2018)], suggesting that the CRISPR/Cas9 is suitable for precise gene knockout. The main constraint using stable transformation is that unlike annual crops, from which gene editing associated transgenes can be effectively segregated post editing through traditional crosses, breeding out the transgenes through crosses for F&N crops is undesirable and will eliminate the identity of the clonally propagated variety. Transient transformation without stable integration of the CRISPR/Cas9 components is possible and desirable for gene editing (Chen et al., 2018); whereas technically it is very challenging to identify the targeted mutant cells caused by the CRISPR/Cas9 due to the lack of SMG for transformed plant cells (Table 2). Non-transgene-involved gene editing by using guide gRNA-Cas9/Cpf1 ribonucleoprotein (RNP) complex is ideal for protoplasts of F&N crops and has been demonstrated in apple and grape cells (Malnoy et al., 2016). An alternative approach has been the use of DNA-replicon strategy (Baltes et al., 2014), based on the Bean yellow dwarf virus (BeYDV) genome structure in the absence of proteins required for its infection and mobility (i.e., disarmed virus). This allowed a high copy number in the cell without the insertion of the replicon into the plant genome (Gil-Humanes et al., 2017). Despite these improvements in the genetic engineering area, it is extremely challenging to induce plant regeneration from protoplast cells in F&N species (Table 2). Again, the main challenge for using gene editing technologies to improve the F&N crops remains the lack of efficient regeneration systems.
Genes and Traits
Regardless of the approach, genetic engineering of the F&N crops aims to modify selected traits through manipulation of targeted gene(s) or gene regulatory sequence(s). The progress made in genetic engineering for fruit crops prior to 2013 has been well-documented (Rai and Shekhawat, 2014).
More recently (2014-present), progress has been made for A. tumefaciens-mediated transformation of nine F&N crops using 24 genes for improving seven traits (Table 2). This progress has demonstrated that A. tumefaciens-mediated transformation is a powerful tool and remains a major approach for improvement of F&N crops. In this regard, the availability of genome drafts in several F&N species allow for the advance toward improved and safer application of techniques such as RNAi and CRISPR/Cas editing. Interference of target mRNAs using artificial microRNAs, or editing of on-target and effect over off-target loci can be assessed by ex-ante analyses using dedicated designers which predict these activities in an efficient rate (Doench et al., 2014; Castro et al., 2016). For instance, computing of protospacer adjacent motifs (PAMs) for Cas9 over a target genome allows for a first-dimension analysis of the putative cut sites for the nuclease upon gRNA leadership. In that way, Wang et al. (Peng et al., 2017) described the occurrence of more than 35 million PAMs in the grapevine genome and similar developments for other species are becoming available tools (Wang Y. et al., 2016; Pulido-Quetglas et al., 2017).
Concluding Remarks
Low success rate and recalcitrance of some species to transformation remain as major challenges for adoption of the new breeding techniques to fruit crops. As those challenges are overcome, several new technologies based on Agrobacterium-mediated transformation, such as transgrafting, fast-track breeding, intragenesis, and genome editing will be employed more frequently for solving problems facing tree fruit industry worldwide.
Data Availability
All datasets generated for this study are included in the manuscript and/or the supplementary files.
Author Contributions
HP wrote the gene editing part. G-qS and VO drafted the rest of the manuscript. All authors read and approved the final manuscript.
Funding
VO was supported by the grant 15-033C from the Citrus Research and Development Foundation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely thank our reviewers for their constructive and insightful comments. We also thank Mr. Aaron Walworth for drawing the diagram in Figure 4.
References
An, C. F., Orbovic, V., and Mou, Z. L. (2013). An efficient intragenic vector for generating intragenic and cisgenic plants in citrus. Am. J. Plant Sci. 4, 2131–2137. doi: 10.4236/ajps.2013.411265
Arora, L., and Narula, A. (2017). gene editing and crop improvement using CRISPR-Cas9 system. Front. Plant Sci. 8:1932. doi: 10.3389/fpls.2017.01932
Artlip, T. S., Wisniewski, M. E., Arora, R., and Norelli, J. L. (2016). An apple rootstock overexpressing a peach CBF gene alters growth and flowering in the scion but does not impact cold hardiness or dormancy. Hortic. Res. 3:16006. doi: 10.1038/hortres.2016.6
Baltes, N. J., Gil-Humanes, J., Cermak, T., Atkins, P. A., and Voytas, D. F. (2014). DNA replicons for plant genome engineering. Plant Cell 26, 151–163. doi: 10.1105/tpc.113.119792
Castillo, X. A. O., Fermin, G., Tabima, J., Rojas, Y., Tennant, P. F., Fuchs, M., et al. (2011). Phylogeography and molecular epidemiology of Papaya ringspot virus. Virus Res. 159, 132–140. doi: 10.1016/j.virusres.2011.04.011
Castro, A., Quiroz, D., Sanchez, E., Miccono, M. D., Aguirre, C., Ramirez, A., et al. (2016). Synthesis of an artificial Vitis vinifera miRNA 319e using overlapping long primers and its application for gene silencing. J. Biotechnol. 233, 200–210. doi: 10.1016/j.jbiotec.2016.06.028
Cervera, M., Pina, J. A., Juarez, J., Navarro, L., and Pena, L. (1998). Agrobacterium-mediated transformation of citrange: factors affecting transformation and regeneration. Plant Cell Rep. 18, 271–278. doi: 10.1007/s002990050570
Chen, L., Li, W., Katin-Grazzini, L., Ding, J., Gu, X., Li, Y., et al. (2018). A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Hortic. Res. 5:13. doi: 10.1038/s41438-018-0023-4
Chitarra, W., Perrone, I., Avanzato, C. G., Minio, A., Boccacci, P., Santini, D., et al. (2017). Grapevine grafting: scion transcript profiling and defense-related metabolites induced by rootstocks. Front. Plant Sci. 8:654. doi: 10.3389/fpls.2017.00654
Collier, R., Thomson, J. G., and Thilmony, R. (2018). A versatile and robust Agrobacterium-based gene stacking system generates high-quality transgenic Arabidopsis plants. Plant J. 95, 573–583. doi: 10.1111/tpj.13992
Dai, H. Y., Li, W. R., Mao, W. J., Zhang, L., Han, G. F., Zhao, K., et al. (2014). Development of an efficient regeneration and Agrobacterium-mediated transformation system in crab apple (Malus micromalus) using cotyledons as explants. In Vitro Cell. Dev. Biol. Plant 50, 1–8. doi: 10.1007/s11627-013-9544-6
Dai, L. M., Zhou, Q., Li, R. M., Du, Y. J., He, J., Wang, D., et al. (2015). Establishment of a picloram-induced somatic embryogenesis system in Vitis vinifera cv. chardonnay and genetic transformation of a stilbene synthase gene from wild-growing Vitis species. Plant Cell Tissue Organ Cult. 121, 397–412. doi: 10.1007/s11240-015-0711-9
Dale, J., James, A., Paul, J. Y., Khanna, H., Smith, M., Peraza-Echeverria, S., et al. (2017). Transgenic Cavendish bananas with resistance to Fusarium wilt tropical race 4. Nat. Commun. 8:1496. doi: 10.1038/s41467-017-01670-6
Dalla Costa, L., Piazza, S., Campa, M., Flachowsky, H., Hanke, M. V., and Malnoy, M. (2016). Efficient heat-shock removal of the selectable marker gene in genetically modified grapevine. Plant Cell Tissue Organ Cult. 124, 471–481. doi: 10.1007/s11240-015-0907-z
De Paepe, A., De Buck, S., Hoorelbeke, K., Nolf, J., Peck, I., and Depicker, A. (2009). High frequency of single-copy T-DNA transformants produced by floral dip in CRE-expressing Arabidopsis plants. Plant J. 59, 517–527. doi: 10.1111/j.1365-313X.2009.03889.x
Doench, J. G., Hartenian, E., Graham, D. B., Tothova, Z., Hegde, M., Smith, I., et al. (2014). Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267. doi: 10.1038/nbt.3026
Dutt, M., Zambon, F. T., Erpen, L., Soriano, L., and Grosser, J. (2018). Embryo-specific expression of a visual reporter gene as a selection system for citrus transformation. PLoS One 13:e0190413. doi: 10.1371/journal.pone.0190413
Fiore, A., Lardo, E., Montanaro, G., Laterza, D., Loiudice, C., Berloco, T., et al. (2018). Mitigation of global warming impact of fresh fruit production through climate smart management. J. Clean Prod. 172, 3634–3643. doi: 10.1016/j.jclepro.2017.08.062
Fitch, M. M. M., Manshardt, R. M., Gonsalves, D., Slightom, J. L., and Sanford, J. C. (1992). Virus resistant papaya plants derived from tissues bombarded with the coat protein gene of papaya ringspot virus. Nat. Biotechnol. 10, 1466–1472. doi: 10.1038/nbt1192-1466
Fonfara, I., Richter, H., Bratovic, M., Le Rhun, A., and Charpentier, E. (2016). The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521. doi: 10.1038/nature17945
Gaj, T., Gersbach, C. A., and Barbas, C. F. III (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405. doi: 10.1016/j.tibtech.2013.04.004
Gelvin, S. B. (2012). Traversing the Cell: Agrobacterium T-DNA’s journey to the host genome. Front. Plant Sci. 3:52. doi: 10.3389/fpls.2012.00052
Gil-Humanes, J., Wang, Y. P., Liang, Z., Shan, Q. W., Ozuna, C. V., Sanchez-Leon, S., et al. (2017). High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 89, 1251–1262. doi: 10.1111/tpj.13446
Gottwald, T. R. (2010). Current epidemiological understanding of citrus huanglongbing. Annu. Rev. Phytopathol. 48, 119–139. doi: 10.1146/annurev-phyto-073009-114418
Gutierrez-Pesce, P., Taylor, K., Muleo, R., and Rugini, E. (1998). Somatic embryogenesis and shoot regeneration from transgenic roots of the cherry rootstock Colt (Prunus avium x P-pseudocerasus) mediated by pRi 1855 T-DNA of Agrobacterium rhizogenes. Plant Cell Rep. 17, 574–580. doi: 10.1007/s002990050445
Hao, G., Stover, E., and Gupta, G. (2016). Overexpression of a modified plant thionin enhances disease resistance to citrus canker and huanglongbing (HLB). Front. Plant Sci. 7:1078. doi: 10.3389/fpls.2016.01078
Haroldsen, V. M., Szczerba, M. W., Aktas, H., Lopez-Baltazar, J., Odias, M. J., Chi-Ham, C. L., et al. (2012). Mobility of transgenic nucleic acids and proteins within grafted rootstocks for agricultural improvement. Front. Plant Sci. 3:39. doi: 10.3389/fpls.2012.00039
Hemmer, C., Djennane, S., Ackerer, L., Hleibieh, K., Marmonier, A., Gersch, S., et al. (2018). Nanobody-mediated resistance to Grapevine fanleaf virus in plants. Plant Biotechnol. J. 16, 660–671. doi: 10.1111/pbi.12819
Hiei, Y., Ishida, Y., and Komari, T. (2014). Progress of cereal transformation technology mediated by Agrobacterium tumefaciens. Front. Plant Sci. 5:628. doi: 10.3389/fpls.2014.00628
Holme, I. B., Wendt, T., and Holm, P. B. (2013). Intragenesis and cisgenesis as alternatives to transgenic crop development. Plant Biotechnol. J. 11, 395–407. doi: 10.1111/pbi.12055
Horsch, R. B., Fry, J. E., Hoffmann, N. L., Eichholtz, D., Rogers, S. G., and Fraley, R. T. (1985). A simple and general-method for transferring genes into plants. Science 227, 1229–1231. doi: 10.1126/science.227.4691.1229
Hu, W., Li, W., Xie, S. X., Fagundez, S., McAvoy, R., Deng, Z. N., et al. (2016). Kn1 gene overexpression drastically improves genetic transformation efficiencies of citrus cultivars. Plant Cell Tissue Organ Cult. 125, 81–91. doi: 10.1007/s11240-015-0931-z
Hutabarat, O. S., Flachowsky, H., Regos, I., Miosic, S., Kaufmann, C., Faramarzi, S., et al. (2016). Transgenic apple plants overexpressing the chalcone 3-hydroxylase gene of Cosmos sulphureus show increased levels of 3-hydroxyphloridzin and reduced susceptibility to apple scab and fire blight. Planta 243, 1213–1224. doi: 10.1007/s00425-016-2475-9
Janick, J. (2005). The origins of fruits, fruit growing, and fruit breeding. Plant Breed. Rev. 25, 255–320. doi: 10.1002/9780470650301.ch8
Jensen, P. J., Halbrendt, N., Fazio, G., Makalowska, I., Altman, N., Praul, C., et al. (2012). Rootstock-regulated gene expression patterns associated with fire blight resistance in apple. BMC Genomics 13:9. doi: 10.1186/1471-2164-13-9
Jia, H., Xu, J., Orbovic, V., Zhang, Y., and Wang, N. (2017a). Editing citrus genome via SaCas9/sgRNA system. Front. Plant Sci. 8:2135. doi: 10.3389/fpls.2017.02135
Jia, H. G., and Wang, N. (2014). Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS One 9:e93806. doi: 10.1371/journal.pone.0093806
Jia, H., Zhang, Y. Z., Orbović, V., Xu, J., White, F. F., Jones, J. B., et al. (2017b). Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 15, 817–823. doi: 10.1111/pbi.12677
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Kandel, R., Bergey, D. R., Dutt, M., Sitther, V., Li, Z. T., Gray, D. J., et al. (2016). Evaluation of a grapevine-derived reporter gene system for precision breeding of Vitis. Plant Cell Tissue Organ Cult. 124, 599–609. doi: 10.1007/s11240-015-0918-9
Kole, C., and Hall, T. C. (2008). Transgenic Temperate Fruits and Nuts, Vol. 4, Chichester: Wiley-Blackwell.
Kost, T. D., Gessler, C., Jansch, M., Flachowsky, H., Patocchi, A., and Broggini, G. A. (2015). Development of the first cisgenic apple with increased resistance to fire blight. PLoS One 10:e0143980. doi: 10.1371/journal.pone.0143980
Krens, F. A., Schaart, J. G., van der Burgh, A. M., Tinnenbroek-Capel, I. E., Groenwold, R., Kodde, L. P., et al. (2015). Cisgenic apple trees; development, characterization, and performance. Front. Plant Sci. 6:286. doi: 10.3389/fpls.2015.00286
Ledford, H. (2015). Bacteria yield new gene cutter. Nature 526, 17–17. doi: 10.1038/nature.2015.18432
Liang, Z., Chen, K. L., Zhang, Y., Liu, J. X., Yin, K. Q., Qiu, J. L., et al. (2018). Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 &ITin vitro&IT transcripts or ribonucleoproteins. Nat. Protoc. 13, 413–430. doi: 10.1038/nprot.2017.145
Liao, X., Guo, X., Wang, Q., Wang, Y., Zhao, D., Yao, L., et al. (2017). Overexpression of MsDREB6.2 results in cytokinin-deficient developmental phenotypes and enhances drought tolerance in transgenic apple plants. Plant J. 89, 510–526. doi: 10.1111/tpj.13401
Limera, C., Sabbadini, S., Sweet, J. B., and Mezzetti, B. (2017). New biotechnological tools for the genetic improvement of major woody fruit species. Front. Plant Sci. 8:1418. doi: 10.3389/fpls.2017.01418
Liu, C., Callow, P., Rowland, L. J., Hancock, J. F., and Song, G. Q. (2010). Adventitious shoot regeneration from leaf explants of southern highbush blueberry cultivars. Plant Cell Tissue Organ Cult. 103, 137–144. doi: 10.1007/s11240-010-9755-z
Liu, X., Walawage, S. L., Leslie, C. A., Dandekar, A. M., Tricoli, D. M., Hu, H., et al. (2017). In vitro gene expression and mRNA translocation from transformed walnut (Juglans regia) rootstocks expressing DsRED fluorescent protein to wild-type scions. Plant Cell Rep. 36, 877–885. doi: 10.1007/s00299-017-2116-1
Lloret, A., Conejero, A., Leida, C., Petri, C., Gil-Munoz, F., Burgos, L., et al. (2017). Dual regulation of water retention and cell growth by a stressassociated protein (SAP) gene in Prunus. Sci. Rep. 7:332. doi: 10.1038/s41598-017-00471-7
Luo, K. R., Huang, N. C., and Yu, T. S. (2018). Selective targeting of mobile mRNAs to plasmodesmata for cell-to-cell movement. Plant Physiol. 177, 604–614. doi: 10.1104/pp.18.00107
Malnoy, M., Viola, R., Jung, M. H., Koo, O. J., Kim, S., Kim, J. S., et al. (2016). DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7:1904. doi: 10.3389/fpls.2016.01904
Mante, S., Morgens, P. H., Scorza, R., Cordts, J. M., and Callahan, A. M. (1991). Agrobacterium-mediated transformation of plum (Prunus-Domestica L) hypocotyl slices and regeneration of transgenic plants. Nat. Biotechnol. 9, 853–857. doi: 10.1038/nbt0991-853
Mcgranahan, G. H., Leslie, C. A., Uratsu, S. L., Martin, L. A., and Dandekar, A. M. (1988). Agrobacterium-mediated transformation of walnut somatic embryos and regeneration of transgenic plants. Nat. Biotechnol. 6, 800–804. doi: 10.1038/nbt0788-800
Miki, B., and McHugh, S. (2004). Selectable marker genes in transgenic plants: applications, alternatives and biosafety. J. Biotechnol. 107, 193–232. doi: 10.1016/j.jbiotec.2003.10.011
Mo, R. L., Huang, Y. M., Yang, S. C., Zhang, Q. L., and Luo, Z. R. (2015). Development of Agrobacterium-mediated transient transformation in persimmon (Diospyros kaki Thunb.). Sci. Hortic. 192, 29–37. doi: 10.1016/j.scienta.2015.05.013
Mojica, F. J., Diez-Villasenor, C., Soria, E., and Juez, G. (2000). Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 36, 244–246. doi: 10.1046/j.1365-2958.2000.01838.x
Moss, S. M. A., Wang, T., Voogd, C., Brian, L. A., Wu, R., Hellens, R. P., et al. (2018). AcFT promotes kiwifruit in vitro flowering when overexpressed and Arabidopsis flowering when expressed inthe vasculature under its own promoter. Plant Direct 2:e00068. doi: 10.1002/pld3.68
Mourgues, F., Chevreau, E., Lambert, C., and deBondt, A. (1996). Efficient Agrobacterium-mediated transformation and recovery of transgenic plants from pear (Pyrus communis L). Plant Cell Reports 16, 245–249. doi: 10.1007/s002990050216
Mullins, M. G., Tang, F. C. A., and Facciotti, D. (1990). Agrobacterium-mediated genetic-transformation of grapevines - transgenic plants of vitis-rupestris-scheele and buds of Vitis-Vinifera L. Nat. Biotechnol. 8, 1041–1045. doi: 10.1038/nbt1190-1041
Nakajima, I., Ban, Y., Azuma, A., Onoue, N., Moriguchi, T., Yamamoto, T., et al. (2017). CRISPR/Cas9-mediated targeted mutagenesis in grape. PLoS One 12:e0177966. doi: 10.1371/journal.pone.0177966
Nawaz, M. A., Imtiaz, M., Kong, Q., Cheng, F., Ahmed, W., Huang, Y., et al. (2016). Grafting. A technique to modify ion accumulation in horticultural crops. Front. Plant Sci. 7:1457.
Nester, E. W. (2014). Agrobacterium: nature’s genetic engineer. Front. Plant Sci. 5:730. doi: 10.3389/fpls.2014.00730
Newhouse, A. E., Oakes, A. D., Pilkey, H. C., Roden, H. E., Horton, T. R., and Powell, W. A. (2018). Transgenic American chestnuts do not inhibit germination of native seeds or colonization of Mycorrhizal fungi. Front. Plant Sci. 9:1046. doi: 10.3389/fpls.2018.01046
Newhouse, A. E., Polin-McGuigan, L. D., Baier, K. A., Valletta, K. E. R., Rottmann, W. H., Tschaplinski, T. J., et al. (2014). Transgenic American chestnuts show enhanced blight resistance and transmit the trait to T1 progeny. Plant Sci. 228, 88–97. doi: 10.1016/j.plantsci.2014.04.004
Nishitani, C., Hirai, N., Komori, S., Wada, M., Okada, K., Osakabe, K., et al. (2016). Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 6:31481. doi: 10.1038/srep31481
Nonaka, S., and Ezura, H. (2014). Plant-Agrobacterium interaction mediated by ethylene and super-Agrobacterium conferring efficient gene transfer. Front. Plant Sci. 5:681. doi: 10.3389/fpls.2014.00681
Orbovic, V., Shankar, A., Peeples, M. E., Hubbard, C., and Zale, J. (2015). Citrus transformation using mature tissue explants. Methods Mol. Biol. 1224, 259–273. doi: 10.1007/978-1-4939-1658-0_21
Orbović, V., Fields, J. S., and Syvertsen, J. P. (2017). Transgenic citrus plants expressing the p35 anti-apoptotic gene have altered response to abiotic stress. Horticulture Environ. Biotechnol. 58, 303–309. doi: 10.1007/s13580-017-0196-z
Palomo-Rios, E., Cerezo, S., Mercado, J. A., and Pliego-Alfaro, F. (2017). Agrobacterium-mediated transformation of avocado (Persea americana Mill.) somatic embryos with fluorescent marker genes and optimization of transgenic plant recovery. Plant Cell Tissue Organ Cult. 128, 447–455. doi: 10.1007/s11240-016-1122-2
Pena, L., and Seguin, A. (2001). Recent advances in the genetic transformation of trees. Trends Biotechnol. 19, 500–506. doi: 10.1016/S0167-7799(01)01815-7
Peng, A. H., Chen, S. C., Lei, T. G., Xu, L. Z., He, Y. R., Wu, L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. doi: 10.1111/pbi.12733
Pessina, S., Angeli, D., Martens, S., Visser, R. G., Bai, Y., Salamini, F., et al. (2016a). The knock-down of the expression of MdMLO19 reduces susceptibility to powdery mildew (Podosphaera leucotricha) in apple (Malus domestica). Plant Biotechnol. J. 14, 2033–2044. doi: 10.1111/pbi.12562
Pessina, S., Lenzi, L., Perazzolli, M., Campa, M., Dalla Costa, L., Urso, S., et al. (2016b). Knockdown of MLO genes reduces susceptibility to powdery mildew in grapevine. Hortic. Res. 3:16016. doi: 10.1038/hortres.2016.16
Petri, C., Alburquerque, N., Faize, M., Scorza, R., and Dardick, C. (2018). Current achievements and future directions in genetic engineering of European plum (Prunus domestica L.). Transgenic Res. 27, 225–240. doi: 10.1007/s11248-018-0072-3
Petri, C., Lopez-Noguera, S., Wang, H., Garcia-Almodovar, C., Alburquerque, N., and Burgos, L. (2012). A chemical-inducible Cre-LoxP system allows for elimination of selection marker genes in transgenic apricot. Plant Cell Tissue Organ Cult. 110, 337–346. doi: 10.1007/s11240-012-0155-4
Pulido-Quetglas, C., Aparicio-Prat, E., Arnan, C., Polidori, T., Hermoso, T., Palumbo, E., et al. (2017). Scalable design of paired CRISPR guide RNAs for genomic deletion. PLoS Comput. Biol. 13:e1005341. doi: 10.1371/journal.pcbi.1005341
Rai, M. K., and Shekhawat, N. S. (2014). Recent advances in genetic engineering for improvement of fruit crops. Plant Cell Tissue Organ Cult. 116, 1–15. doi: 10.1007/s11240-013-0389-9
Ravelonandro, M., Scorza, R., Bachelier, J. C., Labonne, G., Levy, L., Damsteegt, V., et al. (1997). Resistance of transgenic Prunus domestica to plum pox virus infection. Plant Dis. 81, 1231–1235. doi: 10.1094/PDIS.1997.81.11.1231
Ren, C., Liu, X. J., Zhang, Z., Wang, Y., Duan, W., Li, S. H., et al. (2016). CRISPR/Cas9-mediated efficient targeted mutagenesis in chardonnay (Vitis vinifera L.). Sci. Rep. 6:32289. doi: 10.1038/srep32289
Robertson, C. J., Zhang, X., Gowda, S., Orbović, V., Dawson, W. O., and Mou, Z. (2018). Overexpression of the Arabidopsis NPR1 protein in citrus confers tolerance to Huanglongbing. J. Cit. Pathol. 5:38911.
Royo, C., Torres-Perez, R., Mauri, N., Diestro, N., Cabezas, J. A., Marchal, C., et al. (2018). The major origin of seedless grapes is associated with a missense mutation in the MADS-Box gene VviAGL11. Plant Physiol. 177, 1234–1253. doi: 10.1104/pp.18.00259
Salvagnin, U., Carlin, S., Angeli, S., Vrhovsek, U., Anfora, G., Malnoy, M., et al. (2016). Homologous and heterologous expression of grapevine E-(beta)-caryophyllene synthase (VvGwECar2). Phytochemistry 131, 76–83. doi: 10.1016/j.phytochem.2016.08.002
Salvagnin, U., Malnoy, M., Thoming, G., Tasin, M., Carlin, S., Martens, S., et al. (2018). Adjusting the scent ratio: using genetically modified Vitis vinifera plants to manipulate European grapevine moth behaviour. Plant Biotechnol. J., 16, 264–271. doi: 10.1111/pbi.12767
Saurabh, S., Vidyarthi, A. S., and Prasad, D. (2014). RNA interference: concept to reality in crop improvement. Planta 239, 543–564. doi: 10.1007/s00425-013-2019-5
Scorza, R., Callahan, A., Dardick, C., Ravelonandro, M., Polak, J., Malinowski, T., et al. (2013). Genetic engineering of Plum pox virus resistance: ‘HoneySweet’ plum-from concept to product. Plant Cell Tissue Organ Cult. 115, 1–12. doi: 10.1007/s11240-013-0339-6
Scorza, R., Callahan, A., Levy, L., Damsteegt, V., Webb, K., and Ravelonandro, M. (2001). Post-transcriptional gene silencing in plum pox virus resistant transgenic European plum containing the plum pox potyvirus coat protein gene. Transgenic Res. 10, 201–209. doi: 10.1023/A:1016644823203
Scorza, R., Hily, J. M., Callahan, A., Malinowski, T., Cambra, M., Capote, N., et al. (2007). Deregulation of plum pox resistant transgenic plum ‘HoneySweet’. Acta Hortic. 738, 669–673. doi: 10.17660/ActaHortic.2007.738.88
Scorza, R., Webb, K., and Ravelonandro, M. (2004). Analysis of methylation and post-transcriptional gene silencing in embryos and seedlings of C5, a transgenic Plum pox virus resistant plum Prunus domestica L. Acta Hortic. 657, 317–320. doi: 10.17660/ActaHortic.2004.657.49
Seabra, R. C., and Pais, M. S. (1998). Genetic transformation of European chestnut. Plant Cell Rep. 17, 177–182. doi: 10.1007/s002990050374
Sgamma, T., Thomas, B., and Muleo, R. (2015). Ethylene inhibitor silver nitrate enhances regeneration and genetic transformation of Prunus avium (L.) cv Stella. Plant Cell Tissue Organ Cult. 120, 79–88. doi: 10.1007/s11240-014-0581-6
Sidorova, T., Mikhailov, R., Pushin, A., Miroshnichenko, D., and Dolgov, S. (2017). A non-antibiotic selection strategy uses the phosphomannose-isomerase (PMI) gene and green fluorescent protein (GFP) gene for Agrobacterium-mediated transformation of Prunus domestica L. leaf explants. Plant Cell Tissue Organ Cult. 128, 197–209. doi: 10.1007/s11240-016-1100-8
Singh, Z., and Sansavini, S. (1998). “Genetic transformation and fruit crop improvement,” in Plant Breeding Reviews, Vol. 16, ed. J. Janick (Hoboken, NJ: John Wiley & Sons), 87–134.
Smolka, A., Li, X. Y., Heikelt, C., Welander, M., and Zhu, L. H. (2010). Effects of transgenic rootstocks on growth and development of non-transgenic scion cultivars in apple. Transgenic Res. 19, 933–948. doi: 10.1007/s11248-010-9370-0
Song, G. Q. (2014). Recent Advances and Opportunities in Cherry Biotechnology. Acta Hortic. 1020, 89–97. doi: 10.17660/ActaHortic.2014.1020.11
Song, G. Q. (2015). Blueberry (Vaccinium corymbosum L.). Methods Mol. Biol. 1224, 121–131. doi: 10.1007/978-1-4939-1658-0_11
Song, G. Q., and Chen, Q. X. (2018). Overexpression of the MADS-box gene K-domain increases the yield potential of blueberry. Plant Sci. 276, 22–31. doi: 10.1016/j.plantsci.2018.07.018
Song, G. Q., and Gao, X. (2017). Transcriptomic changes reveal gene networks responding to the overexpression of a blueberry DWARF AND DELAYED FLOWERING 1 gene in transgenic blueberry plants. BMC Plant Biol. 17:106. doi: 10.1186/s12870-017-1053-z
Song, G.-Q., and Hancock, J. F. (2012). Recent advances in blueberry transformation. Int. J. Fruit Sci. 12, 316–332. doi: 10.1080/15538362.2011.623083
Song, G. Q., and Sink, K. C. (2004). Agrobacterium tumefaciens-mediated transformation of blueberry (Vaccinium corymbosum L.). Plant Cell Rep. 23, 475–484. doi: 10.1007/s00299-004-0842-7
Song, G. Q., and Sink, K. C. (2006). Transformation of Montmorency sour cherry (Prunus cerasus L.) and Gisela 6 (P-cerasus x P-canescens) cherry rootstock mediated by Agrobacterium tumefaciens. Plant Cell Rep. 25, 117–123. doi: 10.1007/s00299-005-0038-9
Song, G. Q., Sink, K. C., Walworth, A. E., Cook, M. A., Allison, R. F., and Lang, G. A. (2013). Engineering cherry rootstocks with resistance to Prunus necrotic ring spot virus through RNAi-mediated silencing. Plant Biotechnol. J. 11, 702–708. doi: 10.1111/pbi.12060
Song, G. Q., Walworth, A. E., and Loescher, W. H. (2015). Grafting of genetically engineered plants. J. Am. Soc. Hortic. Sci. 140, 203–213. doi: 10.21273/JASHS.140.3.203
Srinivasan, C., Dardick, C., Callahan, A., and Scorza, R. (2012). Plum (Prunus domestica) trees transformed with poplar FT1 result in altered architecture, dormancy requirement, and continuous flowering. PLoS One 7:e40715. doi: 10.1371/journal.pone.0040715
Srinivasan, C., Liu, Z., and Scorza, R. (2011). Ectopic expression of class 1 KNOX genes induce adventitious shoot regeneration and alter growth and development of tobacco (Nicotiana tabacum L) and European plum (Prunus domestica L). Plant Cell Rep. 30, 655–664. doi: 10.1007/s00299-010-0993-7
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K., and Mark Cigan, A. (2016). Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat. Commun. 7:13274. doi: 10.1038/ncomms13274
Uematsu, C., Murase, M., Ichikawa, H., and Imamura, J. (1991). Agrobacterium-mediated transformation and regeneration of kiwi fruit. Plant Cell Rep. 10, 286–290. doi: 10.1007/BF00193143
Velazquez, K., Aguero, J., Vives, M. C., Aleza, P., Pina, J. A., Moreno, P., et al. (2016). Precocious flowering of juvenile citrus induced by a viral vector based on Citrus leaf blotch virus: a new tool for genetics and breeding. Plant Biotechnol. J. 14, 1976–1985. doi: 10.1111/pbi.12555
Voogd, C., Brian, L. A., Wang, T., Allan, A. C., and Varkonyi-Gasic, E. (2017). Three FT and multiple CEN and BFT genes regulate maturity, flowering, and vegetative phenology in kiwifruit. J. Exp. Bot. 68, 1539–1553. doi: 10.1093/jxb/erx044
Waltz, E. (2015). Nonbrowning GM apple cleared for market. Nat. Biotechnol. 33, 326–327. doi: 10.1038/nbt0415-326c
Walworth, A., and Song, G. Q. (2018). The cold-regulated genes of blueberry and their response to overexpression of VcDDF1 in several tissues. Int. J. Mol. Sci. 19:E1553. doi: 10.3390/ijms19061553
Wang, K. (2015). Preface. Agrobacterium protocols. Methods Mol. Biol. 1224, 7–8. doi: 10.1007/978-1-4939-1658-0
Wang, Q. J., Sun, H., Dong, Q. L., Sun, T. Y., Jin, Z. X., Hao, Y. J., et al. (2016). The enhancement of tolerance to salt and cold stresses by modifying the redox state and salicylic acid content via the cytosolic malate dehydrogenase gene in transgenic apple plants. Plant Biotechnol. J. 14, 1986–1997. doi: 10.1111/pbi.12556
Wang, X. H., Tu, M. X., Wang, D. J., Liu, J. W., Li, Y. J., Li, Z., et al. (2018). CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 16, 844–855. doi: 10.1111/pbi.12832
Wang, Y., Liu, X. J., Ren, C., Zhong, G. Y., Yang, L., Li, S. H., et al. (2016). Identification of genomic sites for CRISPR/Cas9-based genome editing in the Vitis vinifera genome. BMC Plant Biol. 16:96. doi: 10.1186/s12870-016-0787-3
Wang, Y., and Pijut, P. M. (2014). Improvement of Agrobacterium-mediated transformation and rooting of black cherry. In Vitro Cell Dev. Biol. Plant 50, 307–316. doi: 10.1007/s11627-014-9608-2
Wang, Z., Wang, S., Li, D., Zhang, Q., Li, L., Zhong, C., et al. (2018). Optimized paired-sgRNA/Cas9 cloning and expression cassette triggers high-efficiency multiplex genome editing in kiwifruit. Plant Biotechnol. J. 16, 1424–1433. doi: 10.1111/pbi.12884
Wisniewski, M., Norelli, J., and Artlip, T. (2015). Overexpression of a peach CBF gene in apple: a model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front. Plant Sci. 6:85. doi: 10.3389/fpls.2015.00085
Wu, R. M., Wang, T. C., McGie, T., Voogd, C., Allan, A. C., Hellens, R. P., et al. (2014). Overexpression of the kiwifruit SVP3 gene affects reproductive development and suppresses anthocyanin biosynthesis in petals, but has no effect on vegetative growth, dormancy, or flowering time. J. Exp. Bot. 65, 4985–4995. doi: 10.1093/jxb/eru264
Yamagishi, N., Kishigami, R., and Yoshikawa, N. (2014). Reduced generation time of apple seedlings to within a year by means of a plant virus vector: a new plant-breeding technique with no transmission of genetic modification to the next generation. Plant Biotechnol. J. 12, 60–68. doi: 10.1111/pbi.12116
Yang, Y. J., Wang, D. F., Wang, C. S., Wang, X. H., Li, J. N., and Wang, R. (2017). Construction of high efficiency regeneration and transformation systems of Pyrus ussuriensis maxim. Plant Cell Tissue Organ Cult. 131, 139–150. doi: 10.1007/s11240-017-1271-y
Yao, J. L., Cohen, D., Atkinson, R., Richardson, K., and Morris, B. (1995). Regeneration of transgenic plants from the commercial apple cultivar Royal Gala. Plant Cell Rep. 14, 407–412. doi: 10.1007/BF00234044
You, C. X., Zhao, Q., Wang, X. F., Xie, X. B., Feng, X. M., Zhao, L. L., et al. (2014). A dsRNA-binding protein MdDRB1 associated with miRNA biogenesis modifies adventitious rooting and tree architecture in apple. Plant Biotechnol. J. 12, 183–192. doi: 10.1111/pbi.12125
Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771. doi: 10.1016/j.cell.2015.09.038
Zhang, F., LeBlanc, C., Irish, V. F., and Jacob, Y. (2017). Rapid and efficient CRISPR/Cas9 gene editing in Citrus using the YAO promoter. Plant Cell Rep. 36, 1883–1887. doi: 10.1007/s00299-017-2202-4
Zhang, H., Harry, D. E., Ma, C., Yuceer, C., Hsu, C. Y., Vikram, V., et al. (2010). Precocious flowering in trees: the FLOWERING LOCUS T gene as a research and breeding tool in Populus. J. Exp. Bot. 61, 2549–2560. doi: 10.1093/jxb/erq092
Zhang, H. Y., Liu, H. M., and Liu, X. Z. (2015). Production of transgenic kiwifruit plants harboring the SbtCry1Ac gene. Genet. Mol. Res. 14, 8483–8489. doi: 10.4238/2015.July.28.16
Zhang, J., Tian, J., Tai, D. Q., Li, K. T., Zhu, Y. J., and Yao, Y. C. (2016). An optimized TRV-based virus-induced gene silencing protocol for Malus crabapple. Plant Cell Tissue Organ Cult. 126, 499–509. doi: 10.1007/s11240-016-1019-0
Zhang, Y. Y., Zhang, D. M., Zhong, Y., Chang, X. J., Hu, M. L., and Cheng, C. Z. (2017). A simple and efficient in planta transformation method for pommelo (Citrus maxima) using Agrobacterium tumefaciens. Sci. Hortic. 214, 174–179. doi: 10.1016/j.scienta.2016.11.033
Zhao, D., and Song, G. Q. (2014). Rootstock-to-scion transfer of transgene-derived small interfering RNAs and their effect on virus resistance in nontransgenic sweet cherry. Plant Biotechnol. J. 12, 1319–1328. doi: 10.1111/pbi.12243
Zhao, Q., Ren, Y. R., Wang, Q. J., Yao, Y. X., You, C. X., and Hao, Y. J. (2016). Overexpression of MdbHLH104 gene enhances the tolerance to iron deficiency in apple. Plant Biotechnol. J. 14, 1633–1645. doi: 10.1111/pbi.12526
Zong, X., Chen, Q., Nagaty, M. A., Kang, Y., Lang, G. A., and Song, G.-Q. (2018). Adventitious shoot regeneration and Agrobacterium tumefaciens -mediated transformation of leaf explants of sweet cherry (Prunus avium L.). J. Hortic. Sci. Biotechnol. 94, 229–236. doi: 10.1080/14620316.2018.1470908
Keywords: Agrobacterium, genome editing, genetic engineering, genetic transformation, woody fruit and nut crops
Citation: Song G-q, Prieto H and Orbovic V (2019) Agrobacterium-Mediated Transformation of Tree Fruit Crops: Methods, Progress, and Challenges. Front. Plant Sci. 10:226. doi: 10.3389/fpls.2019.00226
Received: 04 October 2018; Accepted: 11 February 2019;
Published: 01 March 2019.
Edited by:
Stanton B. Gelvin, Purdue University, United StatesReviewed by:
Ray Collier, Wisconsin Crop Innovation Center (WCIC), United StatesSteven Henry Strauss, Oregon State University, United States
Copyright © 2019 Song, Prieto and Orbovic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guo-qing Song, songg@msu.edu Vladimir Orbovic, orbovic@ufl.edu
 Guo-qing Song
Guo-qing Song Humberto Prieto
Humberto Prieto Vladimir Orbovic
Vladimir Orbovic