- 1Department of Agricultural Biotechnology, Research Institute of Agriculture and Life Sciences, Seoul National University, Seoul, South Korea
- 2Graduate School of International Agricultural Technology and Crop Biotechnology Institute, GreenBio Science and Technology, Seoul National University, Pyeongchang, South Korea
- 3School of Biological Sciences and Technology, Chonnam National University, Gwangju, South Korea
The jasmonic acid (JA) and gibberellic acid (GA) signaling pathways interact to coordinate stress responses and developmental processes. This coordination affects plant growth and yield, and is mediated by interactions between the repressors of each pathway, the JASMONATE ZIM-DOMAIN PROTEIN (JAZ) and DELLA proteins. In this study we attempted to identify rice (Oryza sativa) JAZs that interact with rice DELLAs such as SLENDER RICE 1 (SLR1). Analysis of protein–protein interactions showed that OsJAZ8 and OsJAZ9 interact with SLR1; OsJAZ9 also interacted with the SLR1-LIKE (SLRL) protein SLRL2. Based on this broader interaction, we explored the function of OsJAZ9 in JA and GA responses by analyzing transcript levels of the JA-responsive gene OsbHLH148 and the GA-responsive gene OsPIL14 in OsJAZ9-overexpressing (OsJAZ9-Ox) and osjaz9 mutant plants. OsbHLH148 and OsPIL14 encode key transcription factors controlling JA and GA responses, respectively, and JA and GA antagonistically regulate their expression. In OsJAZ9-Ox, the expression of OsbHLH148 was downregulated and the expression of OsPIL14 was upregulated. By contrast, in osjaz9 mutants, the expression of OsbHLH148 was upregulated and the expression of OsPIL14 was downregulated. These observations indicated that OsJAZ9 regulates both JA and GA responses in rice, and this finding was supported by the opposite expression patterns of OsDREB1s, downstream targets of OsbHLH148 and OsPIL14, in the OsJAZ9-Ox and osjaz9 plants. Together, these findings indicate that OsJAZ9 suppresses JA responses and promotes GA responses in rice, and the protein–protein interaction between OsJAZ9 and SLR1 is involved in the antagonistic interplay between JA and GA.
Introduction
Plants coordinate their defenses and growth in response to environmental conditions; this process strongly affects growth and productivity in many crops, including rice, Oryza sativa (Hou et al., 2010; Yang et al., 2012; Jang et al., 2017). Previous studies showed that jasmonic acid (JA) or JA-dependent environmental stress responses affect the expression of genes involved in gibberellic acid (GA) responses (Navarro et al., 2008; Yang et al., 2012; Wild et al., 2012; Heinrich et al., 2013; Wild and Achard, 2013). These findings suggest that the interaction between JA and GA affects the coordination of defense responses and plant growth, which may have implications for developing crops with high-yield traits via manipulating the regulatory interactions between JA and GA.
JA is a key phytohormone that mediates plant responses to abiotic and biotic stresses and JA is synthesized from linolenic acid through the octadecanoid pathway (Creelman and Mullet, 1997; Stintzi, 2000; Wasternack and Hause, 2002; Farmer et al., 2003). In plant cells, JA is further metabolized into to a bioactive conjugated form, JA-isoleucine (JA-Ile), by the activity of JASMONATE RESISTANT 1 (JAR1) (Staswick and Tiryaki, 2004). In Arabidopsis thaliana, the interaction between JA-Ile and its receptor, CORONATINE INSENSITIVE1 (COI1), promotes JA signaling by provoking 26S proteasome-mediated degradation of the JASMONATE ZIM-DOMAIN (JAZ) repressor proteins (Xie et al., 1998; Thines et al., 2007; Fonseca et al., 2009). The proteolysis of JAZs leads to the release of transcription factors, such as MYC2 in Arabidopsis and OsbHLH148 in rice; these transcription factors induce the expression of JA-responsive genes. The Arabidopsis thaliana genome encodes 12 JAZ proteins (Lorenzo and Solano, 2005; Chini et al., 2007) and previous studies using overexpression and knock-down systems showed the essential role of JAZs in JA responses. For example, Shyu et al. (2012) showed that overexpression of AtJAZ8 reduces the JA response (Shyu et al., 2012), and Yang et al. (2012) showed that overexpression of JAZs such as AtJAZ1, AtJAZ3, AtJAZ9, and AtJAZ10 mimics the phenotype of coi1 mutants, which have defects in JA signaling (Yang et al., 2012). By contrast, AtJAZ1 or AtJAZ10 knock-down plants were hypersensitive to JA (Yan et al., 2007; Grunewald et al., 2009). These observations indicate the important roles of JAZs in the JA response in Arabidopsis.
GA plays pivotal roles in many aspects of plant development such as plant height, leaf sheath growth, stem elongation, leaf expansion, flower development, and seed germination (Matsukura et al., 1998; Ikeda et al., 2001; Fu and Harberd, 2003; Cheng et al., 2004; Piskurewicz et al., 2008; Ubeda-Tomás et al., 2008; Achard et al., 2009; Qi et al., 2014). Similar to the JA signaling pathway, in the GA signaling pathway, proteolysis of the DELLA repressor proteins, is critical for the regulation of GA responses. In Arabidopsis, the interaction between GA and the GA INSENSITIVE DWARF1 (GID1) receptor provokes degradation of DELLAs through the 26S proteasome (Sasaki et al., 2003; Sun, 2011). This degradation leads to the release of transcription factors, such as PHYTOCHROME INTERACTING FACTORS (PIFs) 3 and 4 in Arabidopsis and PIF-LIKE (PIL) 13 and 14 in rice, which are responsible for inducing the expression of GA-responsive genes (Nakamura et al., 2007; De Lucas et al., 2008; Feng et al., 2008; Todaka et al., 2012; Cordeiro et al., 2016). The Arabidopsis thaliana genome contains 5 DELLAs, including the REPRESSOR OF GA1-3 (RGA) gene. Plants that overexpress RGA show a reduced GA response and mutants lacking RGA exhibit enhanced GA responses, suggesting a crucial role of RGA in the GA response in Arabidopsis (Silverstone et al., 1998; Dill and Sun, 2001; King et al., 2001; Lee et al., 2002; Cheng et al., 2004).
The JA and GA signals interact synergistically or antagonistically in various aspects of plant development and defense. In Arabidopsis, JA and GA synergistically interact to regulate the development of stamen filaments and trichomes (Cheng et al., 2009; Qi et al., 2014), and a interaction between JAZs/DELLAs and the WD-repeat/bHLH/MYB complex is involved in the synergistic interaction between JA and GA in trichome development (Qi et al., 2014). JA interacts with GA antagonistically in hypocotyl elongation and root development (Hou et al., 2010; Yang et al., 2012; Hou et al., 2013). The interaction between JAZs and DELLAs attenuates their functions as signaling repressors. These reports indicate that the inhibitory interaction between JAZs and DELLAs plays a role in the coordination of defense and growth in Arabidopsis.
JA and GA signals also interact antagonistically in rice. For example, the suppression of plant height by JA is not observed in the slender rice 1 (slr1) mutant, which lacks the activity of SLR1, a rice DELLA. This indicates that an antagonistic interaction between JA and GA strongly affects rice growth, and that SLR1 is involved in this process (Yang et al., 2012). The expression pattern of OsDREB1s, which act downstream of the JA-responsive gene OsbHLH148 and the GA-responsive gene OsPIL14, supports the antagonistic interaction between JA and GA, as OsbHLH148 promotes OsDREB1s expression and OsPIL14 suppresses OsDREB1s expression (Seo et al., 2011; Cordeiro et al., 2016). However, the molecular mechanism controlling the antagonistic interaction between JA and GA remains largely unknown in rice.
The Oryza sativa genome encodes 12 JAZs and 3 DELLAs, SLR1, SLRL1, and SLRL2 (Ikeda et al., 2001; Itoh et al., 2002; Tian et al., 2004; Ueguchi-Tanaka et al., 2007; Fukao and Bailey-Serres, 2008; Hirano et al., 2010). In this study, we found that OsJAZ8 and OsJAZ9 interact with SLRs, including SLR1. OsJAZ9 interacts with more SLRs than OsJAZ8, so we further investigated the role of OsJAZ9 in the regulation of JA and GA responses. Analysis of OsJAZ9-overexpressing and osjaz9 mutant plants revealed that OsJAZ9 differently regulates expression of OsbHLH148 and OsPIL14, whose transcript levels are antagonistically regulated by JA and GA. These observations indicated that OsJAZ9 is involved in antagonistic regulation of JA and GA responses in rice, and this finding was supported by the phenotype of OsJAZ9-overexpressing transgenic and osjaz9 mutant plants. Taking these results together with the finding that OsJAZ9 interacts with SLR1, we propose that OsJAZ9 mediates the antagonistic interaction between stress-responsive JA signals and growth-promoting GA signals through interaction with SLR1 in rice.
Materials and Methods
Plant Materials, Growth Conditions, and Chemical Treatments
Oryza sativa L. cv. Dongjin was used as the wild-type rice. Rice seeds were germinated in one-half strength Murashige and Skoog (1/2 MS) agar medium in a growth chamber in the dark at 28°C for 3 days. After 3 days of germination, plants were grown in long-day conditions of 16-h light/8-h dark at 28°C for 14 days. For the GA and methyl jasmonate (MeJA) treatments, 8-week-old plants grown in soil were treated with 10 μM GA3 or 10 μM MeJA for the indicated times. The osjaz9 knockout mutants were obtained by targeted CRISPR/Cas9 mutagenesis as previously described in Jang et al. (2016). The homozygous mutant osjaz9-1, which corresponds to line 2–3 and has a single nucleotide insertion at +194 bp from the start codon, and osjaz9-2, which corresponds to line 6–27 and has a single nucleotide insertion at +192 bp, were used in this study.
Construction of Recombinant DNA Plasmids
For the construction of the PGD1::OsJAZ9 and PGD1::SLR1 plasmids, full-length OsJAZ9 and SLR1 cDNAs were amplified from total RNA by RT-PCR. The OsJAZ9 and SLR1 cDNAs were inserted into BamHI/SmaI and XbaI/SmaI-digested pPZP200, respectively; this vector contains the constitutively overexpressing PHOSPHOGLUCONATE DEHYDROGENASE1 (PGD1) promoter (Park et al., 2010). The recombinant plasmid was verified by sequencing and then introduced into Agrobacterium tumefaciens LBA4404 for Agrobacterium-mediated rice transformation. Primer sequences are listed in Supplementary Table 1.
Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
RT-qPCR analyses were performed using total RNA extracted from the indicated plants. Extraction of total RNA was carried out using TRIzol (Invitrogen). For cDNA synthesis, 20 μL reactions were performed using 2 μg of total RNA, Superscript III reverse transcriptase, and oligo dT primers (Invitrogen). For quantitative PCR, a LightCycler 480 Instrument II Real-Time PCR machine with SYBR GREEN I Master Mix (Roche) was used. PCR conditions were programmed according to the manufacturer’s instructions (initial denaturation at 95°C for 5 min followed by 45 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 10 s, and extension at 72°C for 10 s). OsTubA2 (Os11g0247300) was used as an internal control. Three technical replicates of the RT-qPCRs were performed using three biological replicates. Primer sequences are listed in Supplementary Table 1.
Yeast Two-Hybrid Assay
The Matchmaker Gold Yeast Two-Hybrid System (Clontech) was used to test the JAZ–DELLA interactions. Full-length cDNAs of JAZ and DELLA genes were amplified by RT-PCR from total RNA extracted from 14-day-old rice (Dongjin) or 2-week-old Arabidopsis (Col-0) plants. For construction of the bait plasmids, the JAZs were inserted into the Y2H bait vector pGBKT7 (Clontech), which contains a tryptophan biosynthesis gene, TRP1, for selection. For construction of the prey plasmid, the DELLA genes were inserted into the Y2H prey vector pGADT7 (Clontech), which contains a leucine biosynthesis gene, LEU2, for selection. The cDNAs of the JAZs and DELLAs were inserted using multiple enzyme sites (NdeI, EcoRI, and BamHI) of the pGBKT7 and PGADT7 vectors. The recombinant plasmids were co-transformed into the Y2H Gold yeast strain with the Aureobasidin A antibiotic resistance gene to test for protein–protein interactions. Co-transformed yeasts were selected in minimal yeast growth media without tryptophan and leucine (double dropout media, DDO). To test for the interactions between JAZs and DELLAs, co-transformed yeast (OD600 = 0.01) were placed on the DDO media including 250 ng/mL Aureobasidin A. After a 5-day incubation in the dark at 30°C, the growth of the yeast was captured using a Coolpix p300 (Nikon) digital camera. Sequences of the primers used for the bait and prey plasmids are provided in Supplementary Table 1.
Protoplast Transformation and Co-immunoprecipitation (Co-IP) Assay
To analyze the interaction between OsJAZ9 and SLR1, OsJAZ9 and OsbHLH148 or SLR1 and OsPIL14, pHBT-6xMyc-OsJAZ9, pHBT-GFP-SLR1, pHBT-GFP-OsbHLH148, and pHBT-6xMyc-OsPIL14 were generated by introducing amplified cDNAs of OsJAZ9, SLR1, OsbHLH148, and OsPIL14 into StuI/PstI-digested pHBT-5’6xMyc or NotI/PstI-digested pHBT-5’GFP plasmids using the Gibson assembly system (New England Biolabs). Rice protoplasts were isolated from 10-day-old rice seedlings (O. sativa cv. Donjin), and then co-transformed with pHBT-6xMyc-OsJAZ9 and pHBT-GFP-SLR1, pHBT-6xMyc-OsJAZ9 and pHBT-GFP-OsbHLH148 or pHBT-GFP-SLR1, and pHBT-6xMyc-OsPIL14 as previously described (Yang et al., 2014) with slight modification. Around 2.5 × 106 protoplasts were co-transformed with 3 μg of pHBT-6xMyc-OsJAZ9 and pHBT-GFP-SLR1, pHBT-6xMyc-OsJAZ9 and pHBT-GFP-OsbHLH148 or pHBT-GFP-SLR1 and pHBT-6xMyc-OsPIL14. The co-transformed protoplasts were incubated in the dark at 28°C for 10 h, and then harvested by centrifugation at 300 g for 1 min. To extract total proteins, the protoplasts were homogenized with IP buffer 25 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5 % Triton X-100, 1 mM EDTA, 1 mM DTT and Protease inhibitor cocktail (Roche) and then sonicated. For Co-IP, GFP antibody (Nacalai tesque) or Myc antibody (Santa Cruz Biotechnology) was added to the protein extracts and immunocomplexes were precipitated using 40 μl of Protein G Agarose Beads (Sigma). The immunocomplexes were isolated from the beads by boiling in sample buffer for 5 min and were separated using SDS-PAGE.
Bimolecular Fluorescence Complementation (BiFC) Assay
The BiFC assay was used to test for the interaction between OsJAZ9 and SLR1 as described previously (Citovsky et al., 2006). Full-length cDNAs of OsJAZ9 and SLR1 were amplified by RT-PCR from total RNA extracted from 14-day-old rice (Dongjin). The amplified cDNA of SLR1 was inserted into the pSATN vector and the cDNA of OsJAZ9 was inserted into the pSATC vector using the EcoRI and SmaI sites (Tzfira et al., 2005). The nYFP-SLR1 and OsJAZ9-cYFP constructs, empty nYFP and OsJAZ9-cYFP or nYFP-SLR1 and empty cYFP were introduced into onion epidermal cells by tungsten particle bombardment as described by Sanford et al. (1987) with slight modifications. The nYFP-SLR1 and OsJAZ9-cYFP, empty nYFP and OsJAZ9-cYFP or nYFP-SLR1 and empty cYFP plasmids were mixed at a 1:1 (w/w) ratio and 50 μg of DNA was adsorbed onto 10 μg of 1-μm tungsten particles according to the manufacturer’s instructions (Bio-Rad). The particles were bombarded into onion epidermal cells at a pressure of 1000 kPa using a portable Helios gene gun system (model PDS-1000/He, Bio-Rad).
Accession Numbers
Sequence data from this article can be found in the Rice Annotation Project (RAP) or the Arabidopsis Information Resource (TAIR) databases under the following accession numbers:OsJAZ1 (Os10g0392400), OsJAZ2 (Os03g0180900), OsJAZ3 (Os03g0180800), OsJAZ4 (Os03g0181100), OsJAZ5 (Os03g0402800), OsJAZ6 (Os07g0615200), OsJAZ7 (Os09g0439200), OsJAZ8 (Os09g0401300), OsJAZ9 (Os08g0428400), OsJAZ10 (Os04g0653000), OsJAZ11 (Os04g0395800), OsJAZ12 (Os02g0732400), SLR1 (Os03g0707600), SLRL1 (Os01g0646300), SLRL2 (Os05g0574900), OsPIL14 (Os07g0143200), OsbHLH148 (Os03g0741100), OsDREB1A (Os09g0522200), OsDREB1B (Os09g0522000), OsDREB1G (Os02g0677300), and RGA (At2G01570).
Results
Identification of the Interactions Between OsJAZ8, OsJAZ9, and SLR1
To explore the interaction between JAZs and DELLA proteins in rice, we performed yeast two-hybrid (Y2H) assays using the 12 OsJAZs as bait and SLR1 as prey (Figure 1A). The yeast cells (Saccharomyces cerevisiae Y2H GOLD) co-transformed with the OsJAZ8 or OsJAZ9 bait plasmid together with the SLR1 prey plasmid survived in media containing aureobasidin A (Abs A), which selects for yeast lines with a direct interaction between the bait and prey proteins (Chang et al., 2017). Yeast lines transformed with other OsJAZ bait plasmids did not survive on Abs A. These results indicated that OsJAZ8 and OsJAZ9 interact with rice SLR1. In Arabidopsis, six JAZs, AtJAZ1, 3, 4, 9, 10, and 11, interact with the Arabidopsis DELLA protein RGA (Hou et al., 2010; Yang et al., 2012). Examination of the interaction between RGA and the 12 OsJAZs showed that RGA interacted only with OsJAZ8 and OsJAZ9, as did SLR1 (Supplementary Figure 1).
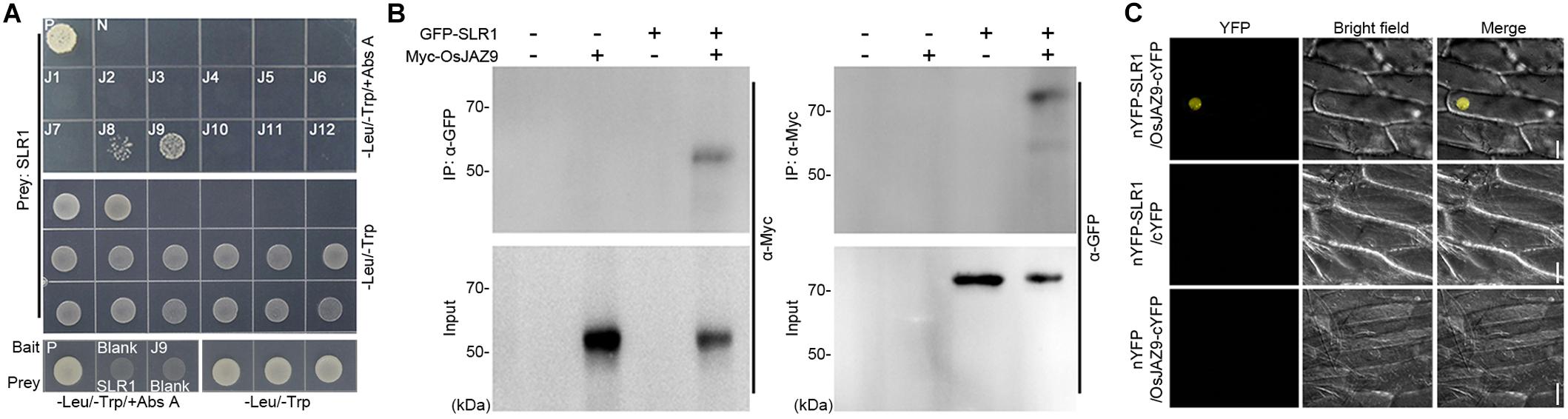
Figure 1. Analysis of the interaction between JAZ and DELLA proteins in rice. (A) A Y2H assay showing the interaction between OsJAZs and SLR1. Co-transformation with the p53 bait and T prey plasmid was used for a positive control (P) and co-transformation with the LAM bait and T prey plasmid was used for a negative control (N). -Leu/-Trp/+Abs A indicates aureobasidin A-containing DDO media to test for the JAZs-DELLA interaction. -Leu/-Trp means DDO media to verify yeast transformation of the indicated bait and prey plasmid. J1–J12 indicates the OsJAZ1–OsJAZ12 bait plasmids used for yeast transformation. Blank indicates empty bait or prey plasmid. (B) Co-IP results showing the interaction between OsJAZ9 and SLR1. Total proteins were extracted from rice protoplasts co-transformed with 6xMyc-OsJAZ9 and GFP-OsSLR1. IP indicates immunoprecipitation, and α-GFP (left) and α-Myc (right) indicate the antibodies used for the immunoprecipitation. α-Myc (left) and α-GFP (right) are the antibodies used for western blotting to detect the interaction between OsJAZ9 and SLR1. (C) A bimolecular fluorescence complementation (BiFC) assay showing the interaction between OsJAZ9 and SLR1 protein. Onion epidermal cells were co-transfected with nYFP-SLR1 and OsJAZ9-cYFP (top), nYFP-SLR1 and empty cYFP (middle), or empty nYFP and OsJAZ9-cYFP (bottom) plasmids by tungsten particle bombardment. The yellow channel image is shown in the YFP panel and the bright-field panel shows the differential interference contrast image. Scale bars = 50 μm.
To verify the interaction between OsJAZ9 and SLR1 in vivo, we performed Co-IP assays using rice protoplasts co-transformed with 6xMyc-OsJAZ9 and GFP-SLR1 (Figure 1B). When protein extracts were immunoprecipitated with anti-GFP antibody, 6xMyc-OsJAZ9 was co-immunoprecipated with GFP-SLR1. Similar results were obtained by immunoprecipitating 6xMyc-OsJAZ9 with anti-Myc antibody, indicating that OsJAZ9 interacts with SLR1. Bimolecular fluorescence complementation (BiFC) assays using nYFP-SLR1 and OsJAZ9-cYFP plasmids further supported the interaction between OsJAZ9 and SLR1 (Figure 1C). When these plasmids were introduced into onion epidermal cells by tungsten particle bombardment, fluorescent signals were observed in the nucleus. However, the onion epidermal cells co-transformed with nYFP-SLR1 and empty cYFP, or empty nYFP and OsJAZ9-cYFP did not show YFP signals. These observations showed that the JA signaling repressor OsJAZ9 and the GA signaling repressor SLR1 interact.
We also tested the interaction between OsJAZs and other rice DELLA proteins such as SLRL1 and SLRL2. SLRL1 did not interact with any of the OsJAZs in the Y2H assay and SLRL2 interacted with OsJAZ9 but not with the other OsJAZs (Supplementary Figure 2). This showed that OsJAZ9 has a broader interaction with SLR proteins compared with OsJAZ8 proteins.
The N-Terminal Region of OsJAZ9 Interacts With the C-Terminal Region of SLR1
To understand how OsJAZ9 interacts with SLR1, we generated a series of OsJAZ9 bait plasmids encoding truncated OsJAZ9 proteins and carried out Y2H assays (Figures 2A,B). The yeast line co-transformed with the SLR1 prey plasmid and the truncated JAZ9N bait, which encodes the N-terminal region upstream of the ZIM domain, survived on the Abs A media. The yeast line co-transformed with the SLR1 prey plasmid and the JAZ9NZ bait plasmid, which encodes the N-terminal region and the ZIM domain, also survived on the Abs A media. However, the yeast lines carrying JAZ9Z and JAZ9J, without the N-terminal region, as baits did not survive in this condition. The N-terminal region of OsJAZ9 also interacted with RGA proteins (Figure 2C). These observations indicated that the N-terminal region of OsJAZ9 is responsible for its interaction with DELLA proteins.
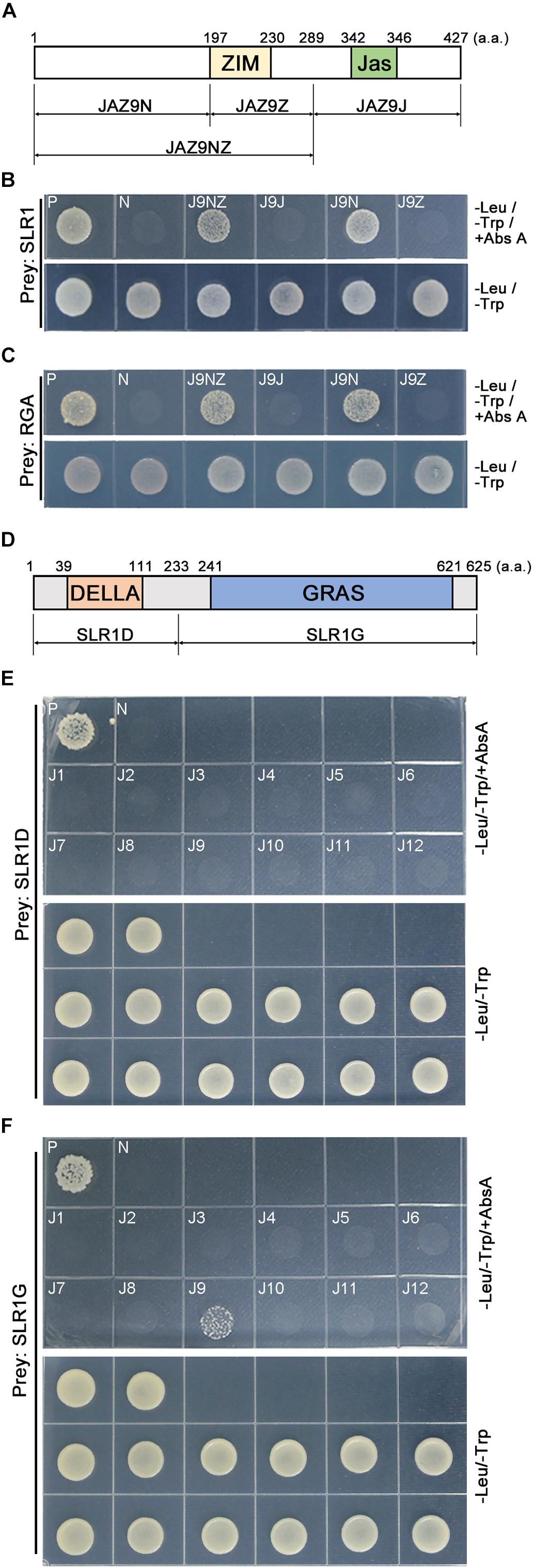
Figure 2. Identification of the domains in OsJAZ9 and SLR1 proteins that mediate their interaction. (A) Schematic of the truncated OsJAZ9 proteins used to analyze the interaction with DELLA proteins. The colored boxes indicate the ZIM (yellow) and Jas (green) domains (Seo et al., 2011). (B,C) Truncated OsJAZ9 bait constructs were co-transformed with SLR1 (B) or Arabidopsis RGA (At2G01570) (C) prey constructs and plated out on DDO media containing Abs A. -Leu/-Trp/+Abs A and -Leu/-Trp indicate DDO media with and without aureobasidin A, respectively. (D) Schematic of the truncated SLR1 proteins used to test the interaction with JAZ proteins. The colored boxes indicate the DELLA (orange) and GRAS (blue) domains (Ueguchi-Tanaka et al., 2007; Hirano et al., 2010). (E,F) Yeast lines were co-transformed with OsJAZ bait plasmids (J1–J12) together with truncated SLR1D (E) or SLR1G (F) prey plasmids and plated on Abs-A containing media.
To identify the SLR1 domain responsible for its interaction with OsJAZ9, we generated two types of truncated SLR1 prey encoding SLR1D and SLR1G, which contain the DELLA domain and the GRAS domain, respectively (Figure 2D). In the Y2H assay, we found that the yeast transformed with the truncated SLR1G prey plasmid together with the OsJAZ9 bait plasmid survived on the Abs A media. However, the yeast line carrying the SLR1D prey plasmid did not survive on Abs A (Figures 2E,F). These results showed that the interaction between OsJAZ9 and SLR1 is mediated through the N-terminal region of OsJAZ9 and the GRAS domain of SLR1 in rice.
OsJAZ9 and SLR1 Show Similar Expression Patterns
To investigate the spatial expression patterns of OsJAZ9 and SLR1 in rice, we extracted total RNAs from the root, shoot base, leaf sheath, leaf blade, and flower and measured the transcript levels of these genes by RT-qPCR (Figure 3). In 2-week-old plants, the expression of OsJAZ9 was higher in the leaf sheath and leaf blade than in the other tissues (Figure 3A). However, in 14-week-old plants, the roots exhibited higher expression of OsJAZ9 than the leaves and flowers (Figure 3B), indicating that tissue-specific expression of OsJAZ9 changes as the plant develops.
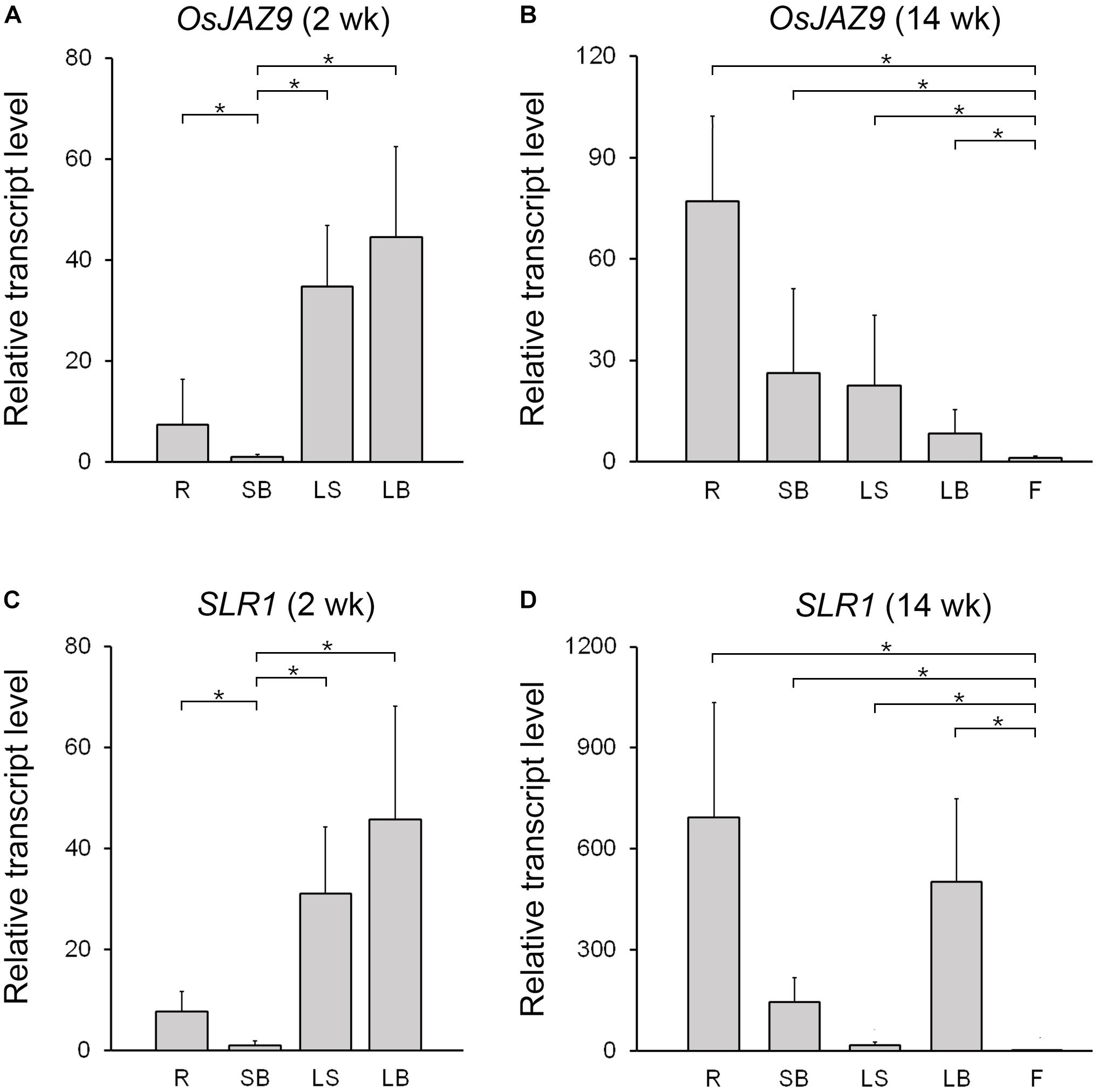
Figure 3. Expression patterns of OsJAZ9 and SLR1 in rice. (A,B) Expression of OsJAZ9 in 2- (A) and 14-week-old rice (B). (C,D) Expression patterns of SLR1 in 2- (C) and 14-week-old rice (D). Tissues tested were: root (R), shoot base (SB), leaf sheath (LS), leaf blade (LB), and flower (F). Total RNAs were extracted from the indicated samples, and the expression level was analyzed by RT-qPCR. Data represent mean values of three biological replicates, and error bars indicate SD. Asterisks show statistically significant differences between the indicated samples (p-value < 0.01, Student’s t-test). OsTubA2 (Os11g0247300) was used as an internal control and relative expression levels are shown as fold values.
If the interaction between OsJAZ9 and SLR1 is involved in crosstalk between JA and GA and the coordination of plant defense and growth, it would be expected that SLR1 and OsJAZ9 would have similar patterns of expression during development. To test this idea, we analyzed the expression pattern of SLR1 in the root, shoot base, leaf sheath, leaf blade, and flower of 2- and 14-week-old plants. The expression of SLR1 was higher in the leaf sheath and leaf blade than in the other tissues in 2-week-old plants (Figure 3C). In 14-week-old plants, the level of SLR1 mRNA was higher in the roots than in the leaves, as was OsJAZ9 (Figure 3D). These results show that the spatial and temporal expression patterns of OsJAZ9 and SLR1 are similar during rice development, consistent with the hypothesis that these two genes may function in the same tissues.
Expression of OsbHLH148, OsPIL14, and OsDREB1s Is Antagonistically Regulated by JA and GA
Previous studies in Arabidopsis showed that the interaction between JAZs and DELLAs regulates the antagonistic interplay between JA and GA (Hou et al., 2010; Yang et al., 2012). To understand the function of OsJAZ9 in JA and GA responses, we attempted to identify genes that are antagonistically regulated by JA and GA. Similar to the previous results with Arabidopsis JAZs and RGA, we found that the transcript levels of OsJAZ9 and SLR1 were not antagonistically regulated by JA and GA (Supplementary Figure 3).
The JA-responsive gene OsbHLH148 and the GA-responsive gene OsPIL14 encode key transcription factors controlling JA and GA responses in rice, and their expression is deeply involved in plant growth and stress responses (Seo et al., 2011; Du et al., 2013; Cordeiro et al., 2016). Because OsbHLH148 promotes the expression of drought-induced genes, such as OsDREB1A, OsDREB1B, and OsDREB1G, but OsPIL14 represses the expression of OsDREB1B (Seo et al., 2011; Cordeiro et al., 2016), we expected that expression of OsbHLH148 and OsPIL14 would be antagonistically regulated by JA and GA. To test this, we analyzed changes in their transcript levels in response to JA and GA treatments (Figures 4A,B; Supplementary Figure 4). The transcript level of OsbHLH148 was strongly increased by the JA treatment, but slightly reduced by the GA treatment. The transcript level of OsHLH148 in the plants co-treated with JA and GA was much lower than that in the plants treated with JA alone. This indicated that GA nullifies the effect of JA on OsbHLH148 expression (Figure 4A). For OsPIL14, the GA treatment increased OsPIL14 expression and the JA treatment reduced OsPIL14 expression. However, JA+GA co-treatment negated the effects of both JA and GA on the expression of OsPIL14 (Figure 4B). These results showed that the expression of OsbHLH148 and OsPIL14 is antagonistically regulated by JA and GA.
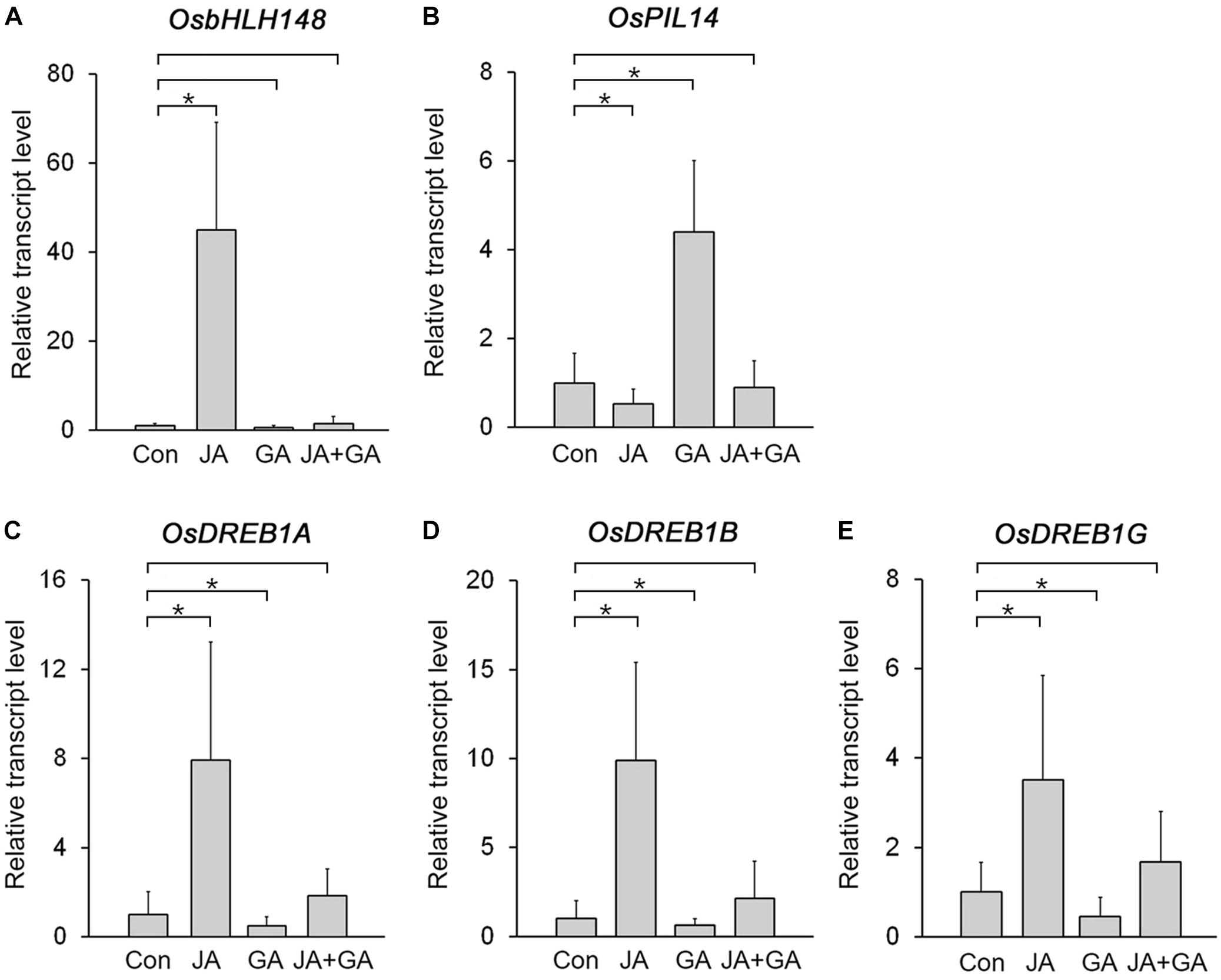
Figure 4. Identification of rice genes antagonistically regulated by JA and GA. (A,B) RT-qPCR analysis showing changes in transcript levels of OsbHLH148 (A) and OsPIL14 (B) in the leaf blade of 8-week-old WT plants (Dongjin) treated with 10 μM MeJA (JA), 10 μM GA3 (GA), or 10 μM MeJA + 10 μM GA3 (JA+GA) for 3 h. (C–E) RT-qPCR analysis showing changes in transcript levels of OsDREB1A (C), OsDREB1G (D), and OsDERB1G (E) in the leaf blades of 8-week-old WT plants (Dongjin) treated with 10 μM MeJA (JA), 10 μM GA3 (GA), or 10 μM MeJA + 10 μM GA3 (JA+GA) for 3 h. Control plants (Con) were not treated with JA, GA, or JA+GA. Data represent mean values of three biological replicates, and error bars indicate SD. Asterisks indicate statistically significant differences between the corresponding samples and their control (p-value < 0.01, Student’s t-test). OsTubA2 was used as an internal control and relative expression levels are shown as fold values.
If JA and GA antagonistically regulate the expression of OsbHLH148 and OsPIL14, then expression of OsDREB1s should also be antagonistically regulated by JA and GA, because OsbHLH148 activates OsDREB1s and OsPIL14 suppresses OsDREB1s (Seo et al., 2011; Cordeiro et al., 2016). As expected, the expression of OsDREB1s was upregulated in response to JA, but downregulated by GA. The JA+GA co-treatment diminished the effects of JA and GA on the expression of OsDREB1s, as was observed with OsbHLH148 and OsPIL14 (Figures 4C–E), supporting the idea that expression of OsbHLH148 and OsPIL14 was antagonistically regulated by JA and GA. In addition, analysis of protein–protein interactions using Co-IP showed that OsJAZ9 and SLR1 interact with OsbHLH148 and OsPIL14, respectively (Supplementary Figure 5). This finding indicates that OsbHLH148 and OsPIL14 are involved in the OsJAZ9 and SLR1-mediated antagonistic interplay between JA and GA in rice.
Overexpression of OsJAZ9 Promotes the GA Response
To understand the function of OsJAZ9 in the antagonistic interaction between JA and GA, we analyzed the effect of OsJAZ9 overexpression on the expression of OsbHLH148 and OsPIL14. To do this, we generated transgenic rice lines overexpressing OsJAZ9 under the control of the constitutively overexpressing PHOSPHOGLUCONATE DEHYDROGENASE1 (PGD1) promoter (Supplementary Figure 6; Park et al., 2010). We identified two independent lines (lines 10 and 15) of transgenic plants with a single-copy insertion of the PGD1::OsJAZ9 transgene (Supplementary Figure 6A). Expression levels of OsJAZ9 in these transgenic plants were approximately 6–11 fold higher than that in wild-type plants (Supplementary Figure 6B). In these OsJAZ9-overexpressing transgenic plants (OsJAZ9-Ox), the transcript levels of OsbHLH148 were downregulated (Figure 5A), indicating that overexpression of OsJAZ9 reduces the JA response in rice.
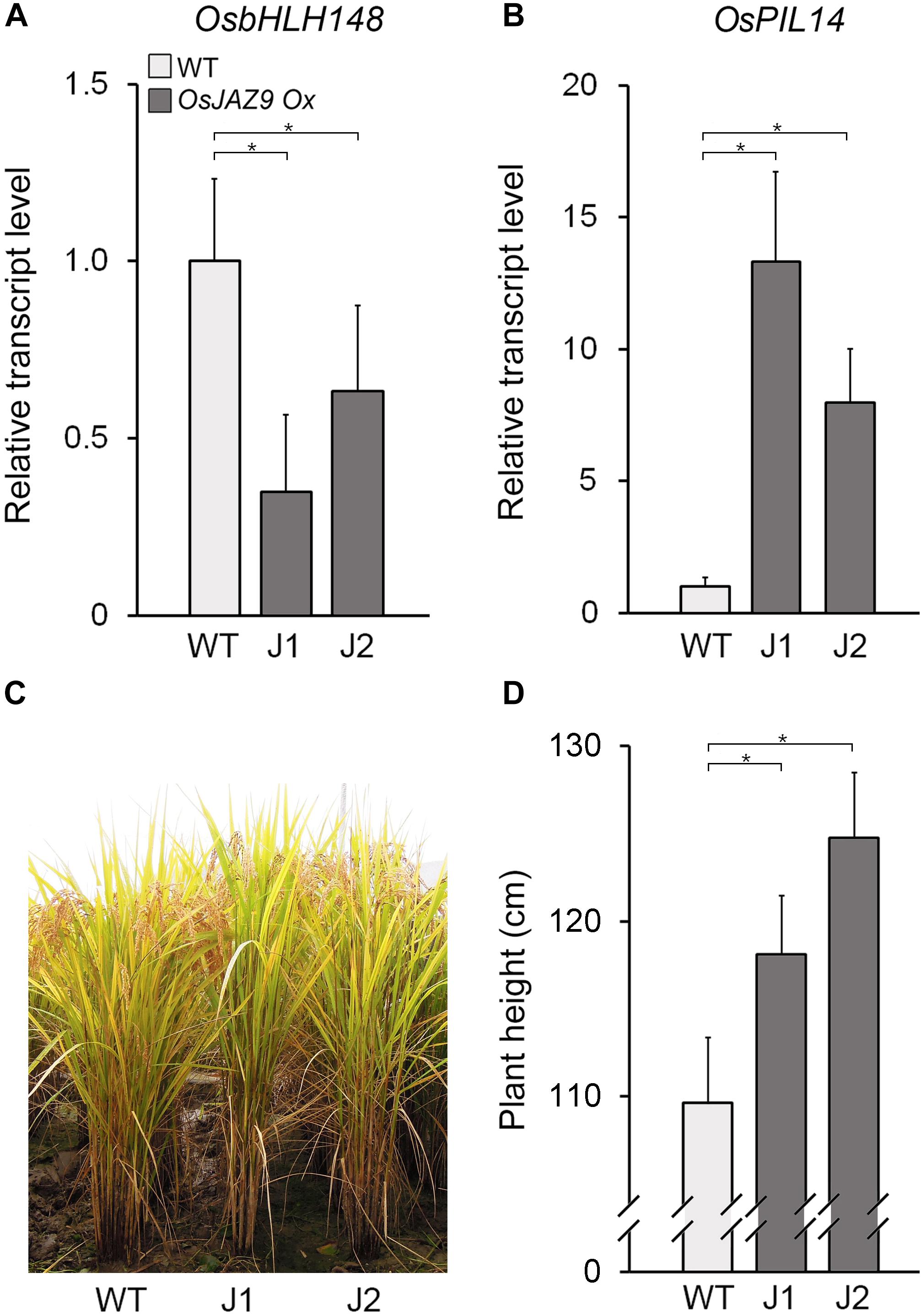
Figure 5. Overexpression of OsJAZ9 promotes the GA response in rice. (A,B) RT-qPCR analysis showing the expression levels of the JA-responsive gene OsbHLH148 (A) and the GA-responsive gene OsPIL14 (B) in the leaf blades of 8-week-old OsJAZ9-overexpressing transgenic rice (PGD1::OsJAZ9). J1 and J2 indicate two independent lines of PGD1::OsJAZ9 plants. Data represent mean values of three biological replicates, and error bars indicate SD. OsTubA2 was used as an internal control and relative expression levels are shown as fold values. (C,D) Growth of PGD1::OsJAZ9 plants grown in field conditions for 25 weeks (C), and quantification of plant height (D) (n > 30). Error bars indicate SD. Asterisks indicate statistically significant differences between the corresponding samples and their control (p-value < 0.01, Student’s t-test).
These findings were partially supported by the phenotype of the OsJAZ9-Ox plants, which had longer roots compared with wild-type plants (Supplementary Figure 7). Furthermore, the length of the OsJAZ9-Ox plants relative to wild type increased in response to JA. This indicated that OsJAZ9-Ox plants are less sensitive to JA than wild-type plants. In contrast to the expression of JA-responsive OsbHLH148, the expression of the GA-responsive gene OsPIL14 was higher in the OsJAZ9-Ox plants than in wild-type plants (Figures 5B), and the OsJAZ9-Ox plants were taller than wild type (Figures 5C,D). Because the GA response promotes rice growth (Huang et al., 1998; Matsukura et al., 1998), these results indicate that the taller phenotype in the OsJAZ9-overexpressing transgenic plants resulted from an enhanced GA response.
The GA Response Is Reduced in the osjaz9 Knockout Mutants
If OsJAZ9 regulates both JA and GA responses, the mutant rice line lacking OsJAZ9 activity would be expected to exhibit some changes in the JA and GA responses. To address this, we analyzed the JA and GA responses in two independent osjaz9 knockout mutants (osjaz9-1 and osjaz9-2), generated by CRISPR/Cas9 (Jang et al., 2016). In both of the independent osjaz9 knockout mutants, the expression of the JA-responsive gene OsbHLH148 tended to be higher than that in the wild-type plants, but not significantly so (Figure 6A). The root growth of osjaz9 mutants was similar to that of wild-type plants in JA-untreated conditions (Supplementary Figure 8). However, in JA-treated conditions, the root length of osjaz9 knockout mutants was significantly shorter than that of wild-type plants, indicating that osjaz9 mutants are more sensitive to JA than wild-type plants. In contrast to the expression of OsbHLH148, the expression of the GA-responsive gene OsPIL14 was lower in the osjaz9 knockout mutants than in the wild-type plants (Figures 6B). Furthermore, the osjaz9 knockout mutants were shorter than the wild-type plants (Figures 6C,D). These findings indicate that the osjaz9 plants exhibit reduced GA responses compared to the wild-type plants. Together with the results of the OsJAZ9-overexpressing plants, these observations supported the idea that OsJAZ9 antagonistically regulates the JA and GA responses.
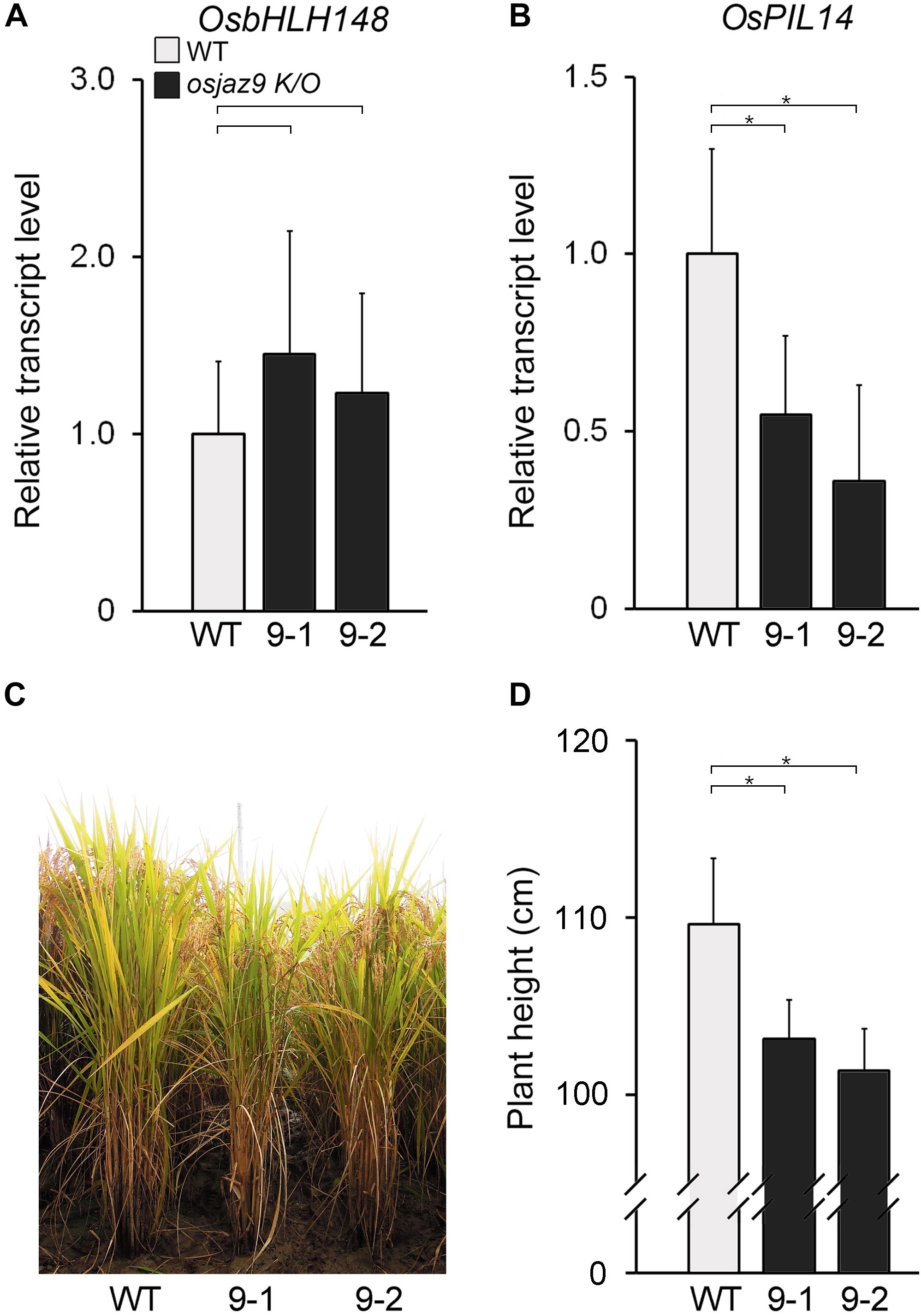
Figure 6. The reduced GA response in the osjaz9 mutants. (A,B) RT-qPCR analysis showing the expression levels of OsbHLH148 (A) and OsPIL14 (B) in the leaf blades of 8-week-old osjaz9 mutant plants. 9-1 and 9-2 indicate two independent mutants, osjaz9-1, and osjaz9-2. Data represent mean values of three biological replicates, and error bars indicate SD. OsTubA2 was used as an internal control and relative expression levels are shown as fold values. (C,D) Images of osjaz9 plants grown in field conditions for 25 weeks (C), and quantification of plant height (D) (n > 30). Error bars indicate SD. Asterisks indicate statistically significant differences between the corresponding samples and their control (p-value < 0.01, Student’s t-test).
To verify this, we further analyzed the expression of SLR1 and OsDREB1s in OsJAZ9-Ox and osjaz9 knockout mutant plants (Supplementary Figure 9). Unlike SLR1, whose expression was not antagonistically regulated by JA and GA, the OsDREB1s showed opposite expression patterns in the OsJAZ9-Ox plants and the osjaz9 mutants: the OsJAZ9-Ox plants exhibited reduced expression of OsDREB1s and the osjaz9 mutants showed increased expression of OsDREB1s. These findings supported the hypothesis that OsJAZ9 modulates the antagonistic interplay between JA and GA signaling in rice, and the interaction between OsJAZ9 and OsSLR1 is deeply involved in this process.
Discussion
In this study, we found that OsJAZ8 and OsJAZ9, 2 of the 12 OsJAZs, interact with SLRs. By contrast, 6 of the 12 Arabidopsis JAZs interact with RGA (Hou et al., 2010; Yang et al., 2012). Therefore, compared with Arabidopsis, the interaction between OsJAZs and SLRs is simpler and more specific, because fewer OsJAZs interact with each SLR. The ability of OsJAZ8 and OsJAZ9 to interact with DELLAs was supported by phylogenetic analysis (Supplementary Figure 10). OsJAZ8 and OsJAZ9 are the most similar to each other of the OsJAZs, and are closely related to AtJAZ1, 3, 4, 9, 10, and 11, which interact with RGA. Although OsJAZ8 and OsJAZ9 interact with rice SLR1 and Arabidopsis RGA, only OsJAZ9 interacted with SLRL2. The broader interaction of OsJAZ9 proteins with SLRs indicates that OsJAZ9 might be largely responsible for the interplay between JA and GA through the interaction with DELLA proteins in rice, and the Co-IP and BiFC results showing the interaction between OsJAZ9 and SLR1 protein support this idea.
In this study, we examined the transcript levels of the JA-responsive gene OsbHLH148 and the GA-responsive gene OsPIL14 in OsJAZ9-Ox plants and osjaz9 knockout mutants to understand the function of OsJAZ9 in the regulation of JA and GA responses. In rice, OsbHLH148 and OsPIL14 play essential roles in the regulation of JA and GA responses (Seo et al., 2011; Cordeiro et al., 2016). We found that the transcript levels of OsbHLH148 and OsPIL14 are antagonistically regulated by JA and GA. This indicated that regulation of the expression of OsbHLH148 and OsPIL14 is involved in the antagonistic interplay between JA and GA in rice, and the finding that OsbHLH148 and OsPIL14 interact with OsJAZ9 and SLR1, respectively, supported this. When the transcript levels of OsbHLH148 and OsPIL14 were measured in OsJAZ9-Ox and osjaz9 knockout mutants, we found that OsJAZ9-Ox plants exhibited decreased expression of OsbHLH148, but increased expression of OsPIL14 while osjaz9 plants showed increased expression of OsbHLH148, but decreased expression of OsPIL14. These findings indicate that OsJAZ9 antagonistically regulates JA and GA responses. These findings were supported by characterization of SLR1-overexpressing plants (SLR1-Ox) (Supplementary Figure 11). Similar to overexpression of OsJAZ9, overexpression of SLR1 affected the expression of OsbHLH148 and OsPIL14. However, the expression pattern was opposite between OsJAZ9-Ox and SLR1-Ox plants: in SLR1-overexpressing plants, the expression of OsbHLH148 was upregulated while the expression of OsPIL14 was downregulated. Taking these results together with the finding that OsJAZ9 interacts with SLR1, we suggest that OsJAZ9 antagonistically regulates the JA and GA responses in rice, and the interaction between OsJAZ9 and SLR1 is deeply involved in this process.
Author Contributions
YC and GJ conceived the original screening and research plans. TU performed most of the experiments. HL, SL, SC, and PC provided technical assistance to TU. YC, K-BO, and GJ analyzed the data. GJ, J-KK, and YC wrote the article with contributions of all the authors.
Funding
This work was carried out with the support of the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ0112152017 to YC and PJ01364301 to GJ) Rural Development Administration, Republic of Korea, and the National Research Foundation of Korea Grant funded by the Korean Government (MOE) (NRF-2014R1A1A2054261 to YC).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
A graduate research assistantship to TU and SL from the Brain Korea 21 Plus project of the MOE is acknowledged.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01866/full#supplementary-material
References
Achard, P., Gusti, A., Cheminant, S., Alioua, M., Dhondt, S., Coppens, F., et al. (2009). Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19, 1188–1193. doi: 10.1016/j.cub.2009.05.059
Chang, S. H., Lee, S., Um, T. Y., Kim, J. K., Choi, Y. D., and Jang, G. (2017). pTAC10, a key subunit of plastid-encoded RNA polymerase, promotes chloroplast development. Plant Physiol. 174, 435–449. doi: 10.1104/pp.17.00248
Cheng, H., Qin, L., Lee, S., Fu, X., Richards, D. E., Cao, D., et al. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131, 1055–1064. doi: 10.1242/dev.00992
Cheng, H., Song, S., Xiao, L., Soo, H. M., Cheng, Z., Xie, D., et al. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genetics 5:e1000440. doi: 10.1371/journal.pgen.1000440
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Citovsky, V., Lee, L. Y., Vyas, S., Glick, E., Chen, M. H., Vainstein, A., et al. (2006). Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 362, 1120–1131. doi: 10.1016/j.jmb.2006.08.017
Cordeiro, A. M., Figueiredo, D. D., Tepperman, J., Borba, A. R., Lourenço, T., Abreu, I. A., et al. (2016). Rice phytochrome-interacting factor protein OsPIF14 represses OsDREB1B gene expression through an extended N-box and interacts preferentially with the active form of phytochrome B. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms 1859, 393–404. doi: 10.1016/j.bbagrm.2015.12.008
Creelman, R. A., and Mullet, J. E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Biol. 48, 355–381. doi: 10.1146/annurev.arplant.48.1.355
De Lucas, M., Daviere, J. M., Rodriguez-Falcon, M., Pontin, M., Iglesias-Pedraz, J. M., Lorrain, S., et al. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484. doi: 10.1038/nature06520
Dill, A., and Sun, T. P. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159, 777–785.
Du, H., Liu, H., and Xiong, L. (2013). Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Frontiers in Plant Science 4:397. doi: 10.3389/fpls.2013.00397
Farmer, E. E., Alméras, E., and Krishnamurthy, V. (2003). Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr. Opin. Plant Biol 6, 372–378. doi: 10.1016/S1369-5266(03)00045-1
Feng, S., Martinez, C., Gusmaroli, G., Wang, Y., Zhou, J., Wang, F., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479. doi: 10.1038/nature06448
Fonseca, S., Chini, A., Hamberg, M., Adie, B., Porzel, A., Kramell, R., et al. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350. doi: 10.1038/nchembio.161
Fu, X., and Harberd, N. P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421, 740–743. doi: 10.1038/nature01387
Fukao, T., and Bailey-Serres, J. (2008). Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. U.S.A. 105, 16814–16819. doi: 10.1073/pnas.0807821105
Grunewald, W., Vanholme, B., Pauwels, L., Plovie, E., Inze, D., Gheysen, G., et al. (2009). Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep 10, 923–928. doi: 10.1038/embor.2009.103
Heinrich, M., Hettenhausen, C., Lange, T., Wünsche, H., Fang, J., Baldwin, I. T., et al. (2013). High levels of jasmonic acid antagonize the biosynthesis of gibberellins and inhibit the growth of Nicotiana attenuata stems. Plant J. 73, 591–606. doi: 10.1111/tpj.12058
Hirano, K., Asano, K., Tsuji, H., Kawamura, M., Mori, H., Kitano, H., et al. (2010). Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 22, 2680–2696. doi: 10.1105/tpc.110.075549
Hou, X., Ding, L., and Yu, H. (2013). Crosstalk between GA and JA signaling mediates plant growth and defense. Plant Cell Rep. 32, 1067–1074. doi: 10.1007/s00299-013-1423-4
Hou, X., Lee, L. Y., Xia, K., Yan, Y., and Yu, H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19, 884–894. doi: 10.1016/j.devcel.2010.10.024
Huang, S., Raman, A. S., Ream, J. E., Fujiwara, H., Cerny, R. E., and Brown, S. M. (1998). Overexpression of 20-oxidase confers a gibberellin-overproduction phenotype in Arabidopsis. Plant Physiol. 118, 773–781. doi: 10.1104/pp.118.3.773
Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., et al. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13, 999–1010. doi: 10.1105/tpc.13.5.999
Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14, 57–70. doi: 10.1105/tpc.010319
Jang, G., Chang, S. H., Um, T. Y., Lee, S., Kim, J. K., and Choi, Y. D. (2017). Antagonistic interaction between jasmonic acid and cytokinin in xylem development. Scientific Reports 7, 10212. doi: 10.1038/s41598-017-10634-1
Jang, G., Lee, S., Um, T. Y., Chang, S. H., Lee, H. Y., Chung, P. J., et al. (2016). Genetic chimerism of CRISPR/Cas9-mediated rice mutants. Plant Biotechnology Reports 10, 425–435. doi: 10.1007/s11816-016-0414-7
King, K. E., Moritz, T., and Harberd, N. P. (2001). Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159, 767–776.
Lee, S., Cheng, H., King, K. E., Wang, W., He, Y., Hussain, A., et al. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes & Development 16, 646–658. doi: 10.1101/gad.969002
Lorenzo, O., and Solano, R. (2005). Molecular players regulating the jasmonate signalling network. Curr. Opin. Plant Biol 8, 532–540. doi: 10.1016/j.pbi.2005.07.003
Matsukura, C., Itoh, S. I., Nemoto, K., Tanimoto, E., and Yamaguchi, J. (1998). Promotion of leaf sheath growth by gibberellic acid in a dwarf mutant of rice. Planta 205, 145–152. doi: 10.1007/s004250050306
Nakamura, Y., Kato, T., Yamashino, T., Murakami, M., and Mizuno, T. (2007). Characterization of a set of phytochrome-interacting factor-like bHLH proteins in Oryza sativa. Biosci. Biotechnol. Biochem. 71, 1183–1191. doi: 10.1271/bbb.60643
Navarro, L., Bari, R., Achard, P., Lisón, P., Nemri, A., Harberd, N. P., et al. (2008). DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 18, 650–655. doi: 10.1016/j.cub.2008.03.060
Park, S. H., Yi, N., Kim, Y. S., Jeong, M. H., Bang, S. W., Choi, Y. D., et al. (2010). Analysis of five novel putative constitutive gene promoters in transgenic rice plants. J. Exp. Bot. 61, 2459–2467. doi: 10.1093/jxb/erq076
Piskurewicz, U., Jikumaru, Y., Kinoshita, N., Nambara, E., Kamiya, Y., and Lopez-Molina, L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20, 2729–2745. doi: 10.1105/tpc.108.061515
Qi, T., Huang, H., Wu, D., Yan, J., Qi, Y., Song, S., et al. (2014). Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 26, 1118–1133. doi: 10.1105/tpc.113.121731
Sanford, J. C., Klein, T. M., Wolf, E. D., and Allen, N. (1987). Delivery of substances into cells and tissues using a particle bombardment process. Particulate Science and Technology 5, 27–37. doi: 10.1080/02726358708904533
Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., et al. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299, 1896–1898. doi: 10.1126/science.1081077
Seo, J. S., Joo, J., Kim, M. J., Kim, Y. K., Nahm, B. H., Song, S. I., et al. (2011). OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant Journal 65, 907–921. doi: 10.1111/j.1365-313X.2010.04477.x
Shyu, C., Figueroa, P., Depew, C. L., Cooke, T. F., Sheard, L. B., Moreno, J. E., et al. (2012). JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 24, 536–550. doi: 10.1105/tpc.111.093005
Silverstone, A. L., Ciampaglio, C. N., and Sun, T.-P. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10, 155–169. doi: 10.1105/tpc.10.2.155
Staswick, P. E., and Tiryaki, I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16, 2117–2127. doi: 10.1105/tpc.104.023549
Stintzi, A. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. U.S.A. 97, 10625–10630. doi: 10.1073/pnas.190264497
Sun, T. P. (2011). The molecular mechanism and evolution of the GA–GID1–DELLA signaling module in plants. Curr. Biol. 21, R338–R345. doi: 10.1016/j.cub.2011.02.036
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Tian, C., Wan, P., Sun, S., Li, J., and Chen, M. (2004). Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 54, 519–532. doi: 10.1023/B:PLAN.0000038256.89809.57
Todaka, D., Nakashima, K., Maruyama, K., Kidokoro, S., Osakabe, Y., Ito, Y., et al. (2012). Rice phytochrome-interacting factor-like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress. Proc. Natl. Acad. Sci. U.S.A. 109, 15947–15952. doi: 10.1073/pnas.1207324109
Tzfira, T., Tian, G. W., Vyas, S., Li, J., Leitner-Dagan, Y., Krichevsky, A., et al. (2005). pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 57, 503–516. doi: 10.1007/s11103-005-0340-5
Ubeda-Tomás, S., Swarup, R., Coates, J., Swarup, K., Laplaze, L., Beemster, G. T., et al. (2008). Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat. Cell Biol. 10, 625–628. doi: 10.1038/ncb1726
Ueguchi-Tanaka, M., Nakajima, M., Katoh, E., Ohmiya, H., Asano, K., Saji, S., et al. (2007). Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19, 2140–2155. doi: 10.1105/tpc.106.043729
Wasternack, C., and Hause, B. (2002). Jasmonates and octadecanoids: signals in plant stress responses and development. Progress in Nucleic Acid Research and Molecular Biology 72, 165–221. doi: 10.1016/S0079-6603(02)72070-9
Wild, M., and Achard, P. (2013). The DELLA protein RGL3 positively contributes to jasmonate/ethylene defense responses. Plant Signal. Behav. 8, e23891. doi: 10.4161/psb.23891
Wild, M., Davière, J. M., Cheminant, S., Regnault, T., Baumberger, N., Heintz, D., et al. (2012). The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 24, 3307–3319. doi: 10.1105/tpc.112.101428
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M., and Turner, J. G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. doi: 10.1126/science.280.5366.1091
Yan, Y., Stolz, S., Chételat, A., Reymond, P., Pagni, M., Dubugnon, L., et al. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19, 2470–2483. doi: 10.1105/tpc.107.050708
Yang, D. L., Yao, J., Mei, C. S., Tong, X. H., Zeng, L. J., Li, Q., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. U.S.A. 109, E1192–E1200. doi: 10.1073/pnas.1201616109
Keywords: jasmonic acid, gibberellic acid, antagonistic interaction, OsJAZ9, SLR1, Oryza sativa
Citation: Um TY, Lee HY, Lee S, Chang SH, Chung PJ, Oh K-B, Kim J-K, Jang G and Choi YD (2018) Jasmonate Zim-Domain Protein 9 Interacts With Slender Rice 1 to Mediate the Antagonistic Interaction Between Jasmonic and Gibberellic Acid Signals in Rice. Front. Plant Sci. 9:1866. doi: 10.3389/fpls.2018.01866
Received: 23 May 2018; Accepted: 04 December 2018;
Published: 18 December 2018.
Edited by:
Randy D. Allen, Oklahoma State University, United StatesReviewed by:
Seonghee Lee, University of Florida, United StatesJohan Memelink, Leiden University, Netherlands
Copyright © 2018 Um, Lee, Lee, Chang, Chung, Oh, Kim, Jang and Choi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Geupil Jang, yk3@jnu.ac.kr; geupil.jang@gmail.com Yang Do Choi, choiyngd@snu.ac.kr
 Tae Young Um
Tae Young Um Han Yong Lee1
Han Yong Lee1 Pil Joong Chung
Pil Joong Chung Ju-Kon Kim
Ju-Kon Kim Geupil Jang
Geupil Jang Yang Do Choi
Yang Do Choi