- 1Department of Bioenergy Science and Technology, Chonnam National University, Gwangju, South Korea
- 2Department of Plant Biology, Carnegie Institution for Science, Stanford, CA, United States
- 3Department of Molecular Biosciences, The Institute for Cellular and Molecular Biology, University of Texas, Austin, TX, United States
Anthocyanins are flavonoid compounds that protect plant tissues from many environmental stresses including high light irradiance, freezing temperatures, and pathogen infection. Regulation of anthocyanin biosynthesis is intimately associated with environmental changes to enhance plant survival under stressful environmental conditions. Various factors, such as UV, visible light, cold, osmotic stress, and pathogen infection, can induce anthocyanin biosynthesis. In contrast, high temperatures are known to reduce anthocyanin accumulation in many plant species, even drastically in the skin of fruits such as grape berries and apples. However, the mechanisms by which high temperatures regulate anthocyanin biosynthesis in Arabidopsis thaliana remain largely unknown. Here, we show that high ambient temperatures repress anthocyanin biosynthesis through the E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) and the positive regulator of anthocyanin biosynthesis ELONGATED HYPOCOTYL5 (HY5). We show that an increase in ambient temperature decreases expression of genes required in both the early and late steps of the anthocyanin biosynthesis pathway in Arabidopsis seedlings. As a result, seedlings grown at a high temperature (28∘C) accumulate less anthocyanin pigment than those grown at a low temperature (17∘C). We further show that high temperature induces the degradation of the HY5 protein in a COP1 activity-dependent manner. In agreement with this finding, anthocyanin biosynthesis and accumulation do not respond to ambient temperature changes in cop1 and hy5 mutant plants. The degradation of HY5 derepresses the expression of MYBL2, which partially mediates the high temperature repression of anthocyanin biosynthesis. Overall, our study demonstrates that high ambient temperatures repress anthocyanin biosynthesis through a COP1-HY5 signaling module.
Introduction
Anthocyanins are water-soluble pigments found throughout the plant kingdom and are derived from the flavonoid branch of the phenylpropanoid pathway. Anthocyanins play key roles in diverse physiological processes, such as protecting plant tissues from damage after exposure to UV, freezing temperature, and pathogens; assist in the attraction of pollinators and seed dispersers, scavenge free radicals produced under stress conditions, and modulate auxin transport (Falcone Ferreyra et al., 2012). The anthocyanin biosynthesis pathway has been studied in numerous plant species and most of the genes involved in this process have been identified. In the model plant Arabidopsis thaliana, the genes encoding anthocyanin biosynthesis enzymes are grouped into two classes: early biosynthetic genes [chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), and flavonoid 3′-hydroxylase (F3′H)], all of which are common to other flavonoids; and late biosynthetic genes [dihydroflavonol 4-reductase (DFR), leucoanthocyanidin oxygenase (LDOX), UDP-glucose flavonoid 3-O-glucosyltransferase (UF3GT), and anthocyanin acyltransferase (AAT)] that are specific to the anthocyanin pathway (Figure 1) (Tanaka et al., 2008).
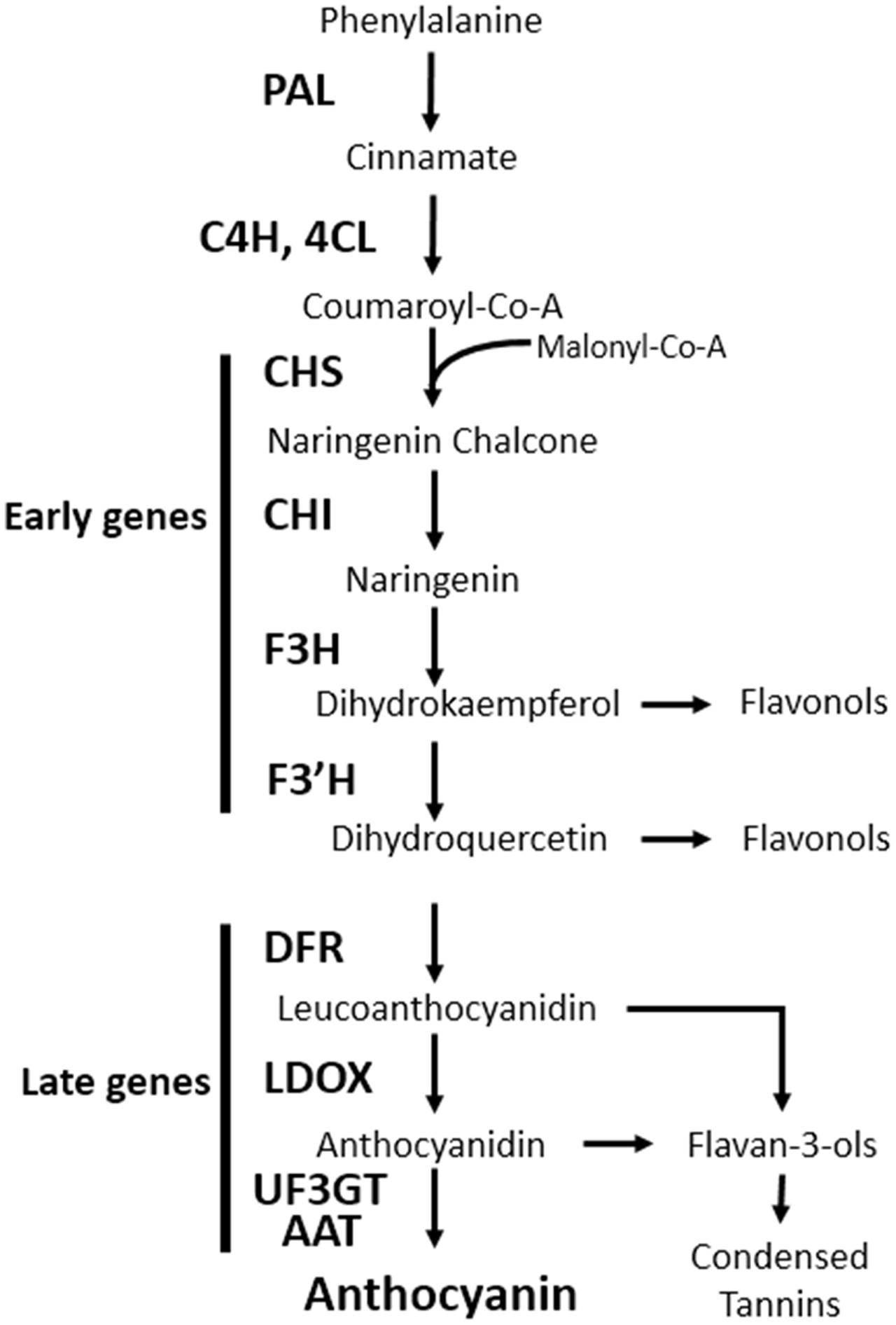
FIGURE 1. Schematic diagram of the anthocyanin biosynthetic pathway in Arabidopsis thaliana. PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumaroyl: CoA-ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; DFR, dihydroflavonol reductase; LDOX, leucoanthocyanidin dioxygenase; UF3GT, UDP-glucose flavonoid 3-O-glucosyltransferase; AAT, anthocyanin acyltransferase.
Anthocyanin pigment accumulation is mainly regulated via the transcriptional regulation of anthocyanin biosynthetic genes (Tanaka et al., 2008). Several transcription factors that regulate the expression of anthocyanin biosynthetic genes have been identified. The R2R3-type MYB transcription factors PAP1/MYB75, PAP2/MYB90, MYB113, and MYB114 positively regulate anthocyanin biosynthesis in vegetative tissues (Borevitz et al., 2000; Teng et al., 2005; Gonzalez et al., 2008). Three members of the bHLH transcription factor subgroup IIIf, GLABRA3 (GL3), ENHANCER OF GLABRA3 (EGL3), and TRANSPARENT TESTA8 (TT8), are positive regulators of anthocyanin biosynthesis (Zhang et al., 2003). The bHLH and MYB transcription factors form a ternary complex with the WD40 protein TTG1 to activate anthocyanin biosynthesis, particularly the late biosynthetic steps (Zhang et al., 2003; Gonzalez et al., 2008). In contrast, the small MYB protein MYBL2 negatively regulates anthocyanin biosynthesis by repressing the expression of the late biosynthetic genes DFR and LDOX (Dubos et al., 2008; Matsui et al., 2008).
Since anthocyanin biosynthesis is a metabolically expensive process, it is closely regulated with respect to surrounding environmental conditions. Thus, the regulation of anthocyanin biosynthetic genes is associated with many environmentally responsive signaling pathways. For example, UV, visible, and far-red light irradiation can induce anthocyanin biosynthesis. Photoreceptors including UV-B RESISTANCE8 (UVR8), phytochromes (PHYs), and cryptochromes (CRYs) mediate light induction of anthocyanin biosynthesis (Sponga et al., 1986; Mancinelli et al., 1991; Heijde et al., 2013). The ring-finger E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) regulates anthocyanin biosynthesis downstream of the photoreceptors. COP1 has been shown to induce the degradation of PAP1 and PAP2 to repress anthocyanin biosynthesis in the absence of light (Maier et al., 2013). COP1 also interacts with a bZIP transcription factor, LONG HYPOCOTYL5 (HY5), that directly binds to the promoters of both early and late anthocyanin biosynthetic genes and activates their expression (Chattopadhyay et al., 1998; Shin et al., 2007). In addition to light, anthocyanin accumulation is also induced by cold stress, a response partly mediated by HY5 (Catala et al., 2011). Osmotic stress, nitrogen deficiency, and pathogen infection have also been shown to induce anthocyanin biosynthesis (Lea et al., 2007; Tanaka et al., 2008). In addition, it has been shown that anthocyanin accumulation is affected by ambient temperature in various plant species. For example, anthocyanin accumulation in the skins of ripening fruit (grapevine and apple) is reduced in response to high temperatures (Mori et al., 2007; Lin-Wang et al., 2011; Movahed et al., 2016; Pastore et al., 2017). However, the effects of ambient temperature changes on anthocyanin biosynthesis in Arabidopsis, are poorly understood.
Temperature-related changes in plant development and growth are critical for survival in a variable environment. For example, young seedlings respond to increases in ambient temperature by elongation of hypocotyls and petioles, hyponastic leaf growth, and leaf thinning; these responses are collectively called thermomorphogenesis (Quint et al., 2016). The morphological changes are considered to increase plant survival under heat stress, in part by enhancing leaf transpiration efficiency (Crawford et al., 2012; Zhu et al., 2016). The bHLH transcription factor PHYTOCHROME INTERACTING FACTOR4 (PIF4) is a key regulator of thermomorphogenesis (Koini et al., 2009; Proveniers and van Zanten, 2013; Choi and Oh, 2016). HY5 has also been reported to negatively regulate thermomorphogenic growth (Delker et al., 2014). Although thermomorphogenesis, especially hypocotyl elongation, has been extensively studied, it is still not clear how high ambient temperatures impinge on other physiological processes including anthocyanin biosynthesis.
In this study, we show that a high temperature represses anthocyanin biosynthesis through HY5 in Arabidopsis. Elevation of ambient temperature decreases the expression of several anthocyanin biosynthetic genes and results in a lower level of anthocyanin accumulation in Arabidopsis seedlings. HY5 and COP1 both appear to be required for the thermo-sensitivity of anthocyanin accumulation and expression of anthocyanin biosynthetic genes. We also show that the level of the HY5 protein, but not the level of HY5 mRNA, is reduced by high temperature in a COP1 activity-dependent manner. Taken together, our study demonstrates that the control of anthocyanin biosynthesis in response to high ambient temperature changes is largely exerted through degradation of HY5.
Results
High Temperature Represses the Accumulation of Anthocyanin Pigments
In order to identify developmental responses to an elevation in ambient temperature, we performed gene ontology (GO) analyses using high temperature-regulated genes identified in a previous RNA-sequencing experiment (Jung et al., 2016). These high temperature-regulated genes were identified by comparing the transcriptomes of wild type (Ler) seedlings grown at 22°C with those of wild type seedlings grown at 27°C at Zeitgeber Time (ZT) 0, 1, 4, 8, 12, 16, 20, and 22 h. The GO analyses revealed that genes for the biosynthesis of anthocyanin-containing compounds were highly enriched in the high temperature-repressed genes at ZT0 (Figure 2A and Supplementary Table 1). Most anthocyanin biosynthetic genes, including both early and late genes, were transcriptionally repressed in seedlings grown at 27°C compared to those grown at 20°C (Figure 2B). These results suggest that high ambient temperatures may directly repress anthocyanin pigment accumulation. To test this hypothesis, we grew Arabidopsis seedlings on MS medium containing 3% sucrose at different temperatures (17, 20, and 28°C) under continuous light (Figure 2C). As reported previously, the increase in ambient temperature to 28°C dramatically induced hypocotyl elongation (Figure 2D) (Koini et al., 2009). In addition to this known morphological change, we also observed that anthocyanin pigment accumulation was lower in these seedlings than in seedlings grown at 17 or 20°C (Figures 2D,E). To determine whether anthocyanin pigment accumulation was affected by short and transient ambient temperature changes, seedlings were grown for 4 days at 17°C and then transferred to 28°C for 24 h (Figure 2F). Anthocyanin pigment levels in seedlings exposed to 28°C for 24 h were significantly lower than in seedlings kept at 17°C (Figures 2G,H). These results indicate that high ambient temperature represses anthocyanin pigment accumulation in young seedlings.
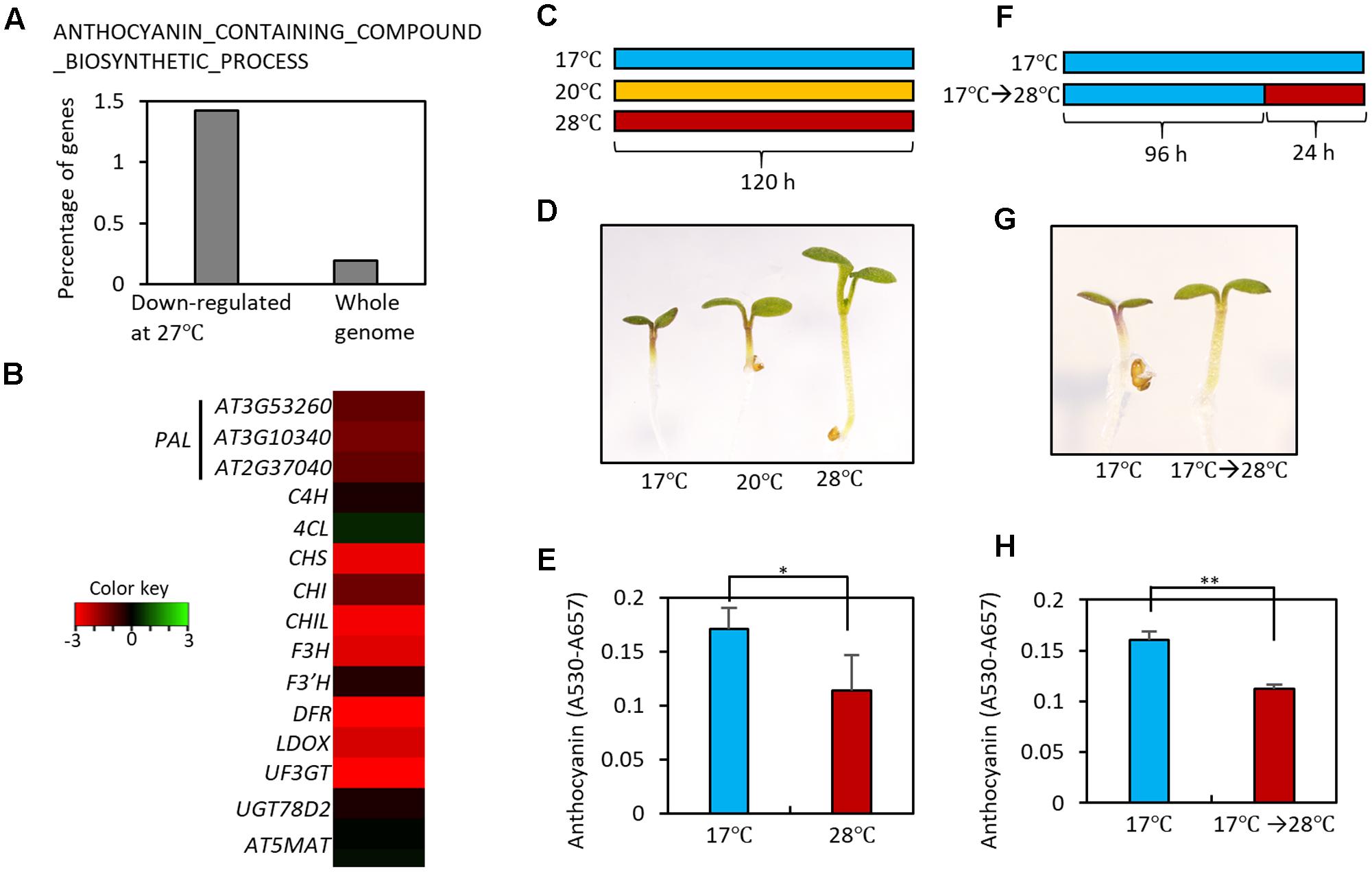
FIGURE 2. High temperature represses the accumulation of anthocyanin pigments. (A) Gene Ontology (GO) analyses showed that genes in the ANTHOCYANIN_CONTAINING_COMPOUND_BIOSYNTHETIC_PROCESS are highly enriched among high temperature (27°C)-repressed genes identified in a previous RNA-seq experiment (Jung et al., 2016). Other molecular processes enriched in the high temperature-repressed genes are shown in Supplementary Table 1. (B) Heat map of the anthocyanin biosynthetic genes at Zeitgeber Time 0 (ZT0) in previous RNA-seq experiment (Jung et al., 2016). Red color indicates genes that were down-regulated by high temperatures and green color indicates genes that were up-regulated by high temperatures. (C) Growth conditions in experiments (D,E). Seedlings were grown on MS medium containing 3% sucrose at different temperatures (17, 20, or 28°C) for 5 days under continuous white light, and then harvested for anthocyanin extraction. (D,E) A lower level of anthocyanin accumulation was found in seedlings grown at the higher temperatures. Representative seedlings are shown in (D) and quantification of anthocyanin content is shown in (E). Error bars in (E) indicate s.d. (n = 3). ∗P < 0.05 (Student’s t-test). (F) Growth conditions in experiments (G,H). Seedlings were either grown at 17°C for 4 days and then at 28°C for 24 h before harvesting, or were kept at 17°C as a control. (G,H) Anthocyanin accumulation was reduced by the high temperature treatment. Representative seedlings are shown in (G) and quantification of anthocyanin content is shown in (H). Error bars in (H) indicate s.d. (n = 3). ∗∗P < 0.01 (Student’s t-test).
High Temperature Transcriptionally Represses Anthocyanin Biosynthetic Genes
To elucidate the molecular mechanisms underlying the high temperature repression of anthocyanin accumulation, we performed a qRT-PCR analysis to examine the expression levels of anthocyanin biosynthesis genes in seedlings grown at 28°C for 24 h or kept at 17°C. The qRT-PCR analysis revealed that expression of the early anthocyanin biosynthetic genes CHS and CHI was lower by about 20 to 40% in seedlings exposed to 28°C than in those kept at 17°C (Figure 3). The late anthocyanin biosynthesis genes DFR and LDOX also showed a twofold reduction in expression after the high temperature treatment (Figure 3). The expression patterns of the anthocyanin biosynthetic genes were consistent with the observation of lower levels of anthocyanin pigments in seedlings grown at 28°C than in those grown at 17°C (Figure 2), indicating that the decreased anthocyanin accumulation induced by the elevated ambient temperature was caused by transcriptional reduction of both early and late anthocyanin biosynthetic genes.
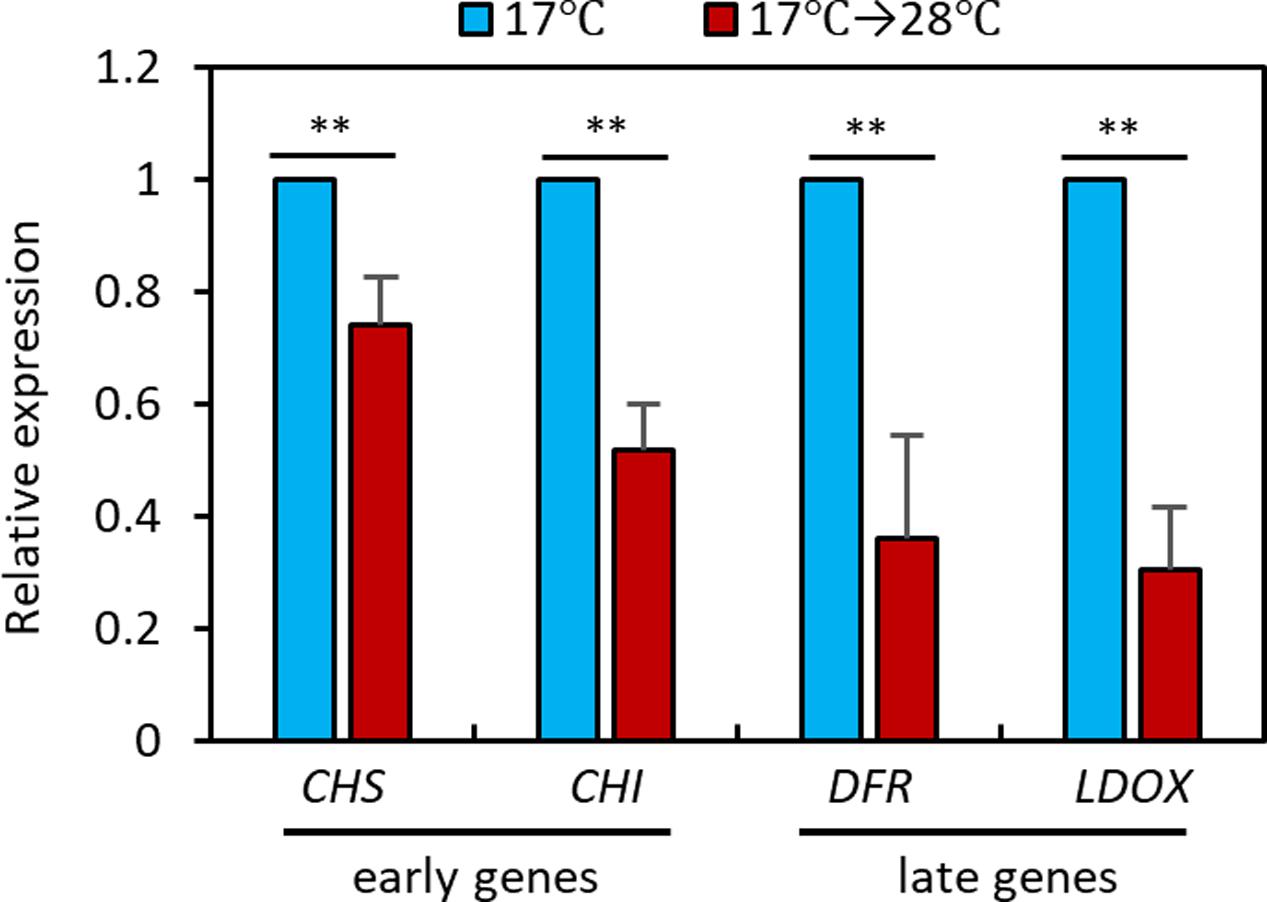
FIGURE 3. High temperature represses expression of anthocyanin biosynthetic genes. The qRT-PCR analyses showed that expression of anthocyanin biosynthetic genes was decreased at 28°C. Wild type seedlings were grown at 17°C under continuous white light for 4 days and then either transferred to 28°C for 24 h (17°C → 28°C) or kept at 17°C before harvesting for total RNA extraction. Gene expression levels were normalized to PP2A (AT1G13320) and are presented as values relative to those of seedlings kept at 17°C. Error bars indicate s.d. (n = 3). ∗∗P < 0.01 (Student’s t-test).
HY5 Mediates the High Temperature Regulation of Anthocyanin Biosynthesis
PIF4 is a key transcription factor in high temperature acclimation processes including thermomorphogenesis and acceleration of flowering (Choi and Oh, 2016; Quint et al., 2016). Moreover, it was recently reported that PIF4, and its closest homolog PIF5, negatively regulate anthocyanin biosynthesis under red light (Liu et al., 2015). Thus, it was likely that PIF4 mediated the high temperature regulation of anthocyanin biosynthesis. To test whether this was the case, we measured anthocyanin pigment levels in pif4;pif5 double mutant seedlings grown at 17°C or at 17°C for 4 days followed by 28°C for 24 h. As shown in Figure 4A, anthocyanin levels in pif4;pif5 seedlings were significantly reduced by the high temperature treatment in a similar manner to that in the wild type, suggesting that PIF4 and PIF5 are not major factors that mediate the high temperature repression of anthocyanin biosynthesis.
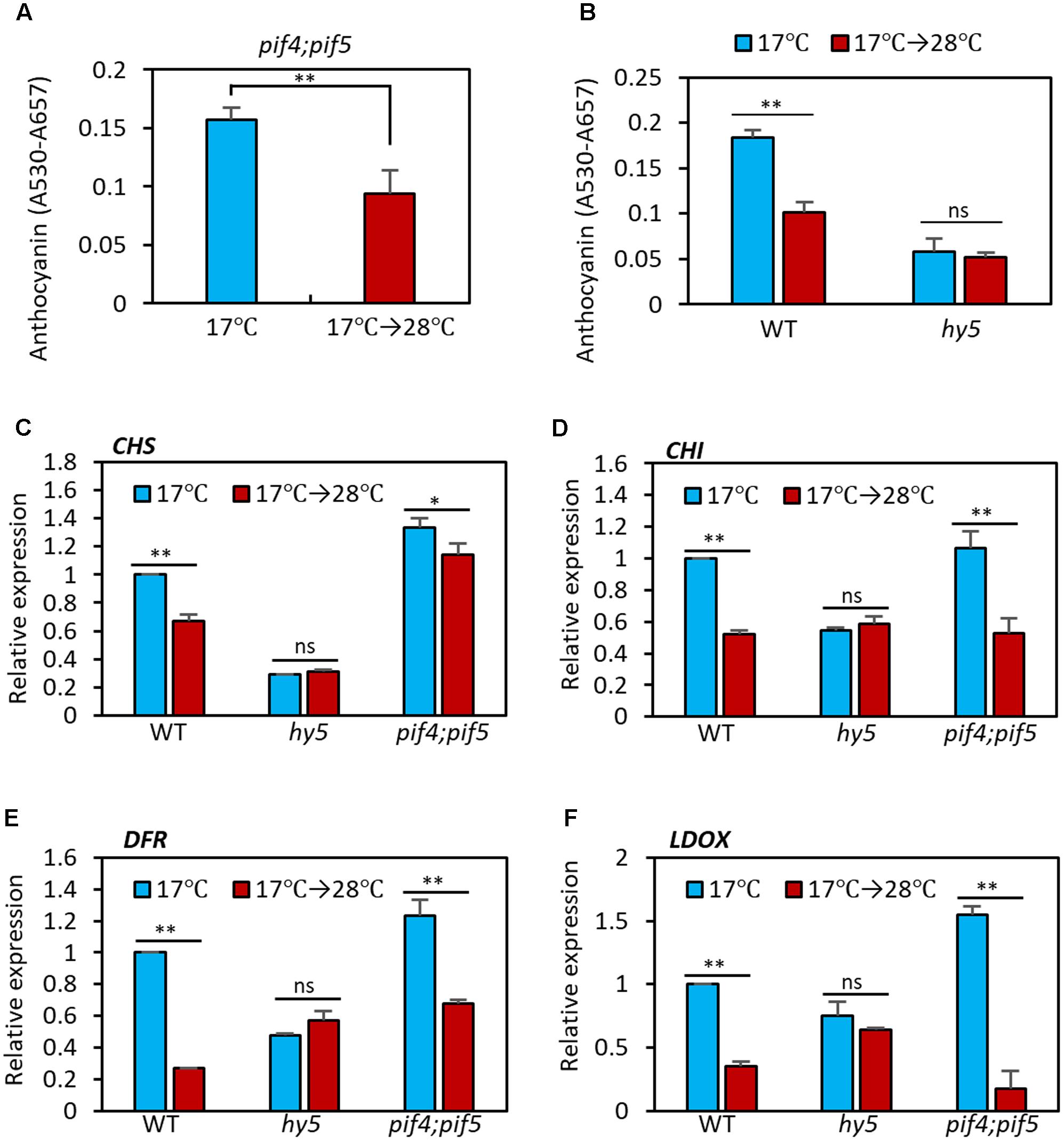
FIGURE 4. Anthocyanin biosynthesis is insensitive to high temperature in hy5 mutant. (A) Anthocyanin accumulation was repressed by high temperature treatment in pif4;pif5 double mutant seedlings. The pif4;pif5 seedlings were grown at 17°C under continuous white light for 4 days and then either transferred to 28°C for 24 h (17°C → 28°C) or kept at 17°C before harvesting for anthocyanin extraction. (B) Anthocyanin accumulation in the hy5 mutant was not significantly affected by an increase in ambient temperature. Seedlings of wild type and hy5 mutant were grown under the same conditions as (A). Error bars in (A,B) indicate s.d. (n = 3). ∗∗P < 0.01 (Student’s t-test); ns, not significant (P ≥ 0.05). (C–F) The qRT-PCR analyses showed that expression of both early and late anthocyanin biosynthetic genes were insensitive to high temperature in the hy5 mutant. Seedlings were grown at 17°C for 4 days and then transferred to 28°C for 24 h (17°C → 28°C) or kept at 17°C before harvesting for total RNA extraction. Gene expression levels were normalized to PP2A (AT1G13320) and are presented as values relative to those of wild type seedlings kept at 17°C. Error bars indicate s.d. (n = 3). ∗∗P < 0.01 (Student’s t-test) and ns, not significant (Student’s t-test P ≥ 0.05).
The bZIP transcription factor HY5 is a positive regulator of anthocyanin biosynthesis and directly regulates expression of several anthocyanin biosynthetic genes, including both early and late genes (Shin et al., 2007, 2013). HY5 also regulates the expression of anthocyanin-promoting transcription factors such as PAP1 (Shin et al., 2013). Moreover, HY5 has previously been reported to positively regulate cold-induced gene expression and cold acclimation and negatively regulate high temperature-induced hypocotyl elongation (Catala et al., 2011; Delker et al., 2014). We therefore examined whether HY5 mediates the high temperature repression of anthocyanin biosynthesis. As previously reported (Shin et al., 2007), anthocyanin pigment levels were lower in hy5 mutant seedlings at 17°C than in the wild type (Figure 4B). However, in contrast to the wild type, anthocyanin levels were not significantly affected by high temperature exposure in the hy5 mutant (Figure 4B), supporting the idea that HY5 plays a dominant role in ambient temperature regulation of anthocyanin biosynthesis.
Next, we examined the transcriptional responses of anthocyanin biosynthetic genes to an increase in ambient temperature in wild type, hy5, and pif4;pif5 seedlings. In the wild type and pif4;pif5 seedlings, expression of CHS and CHI (early genes) was reduced about 20–40% by the high temperature treatment and expression of DFR and LDOX (late genes) was reduced by about 60–80% (Figures 4C–F). The expression of UF3GT was also dramatically decreased by the high temperature in wild type (Supplementary Figure 1). In contrast, in the hy5 mutant, none of these genes showed any significant difference in level of expression after the high temperature treatment (Figures 4C–F and Supplementary Figure 1). The expression patterns of these four anthocyanin biosynthetic genes were consistent with the significant reduction in anthocyanin pigment accumulation after high temperature in the wild type and pif4;pif5 seedlings, and also with the thermo-insensitivity of the hy5 mutant (Figure 4B). Thus, HY5 appears to mediate the transcriptional regulation of anthocyanin biosynthesis genes in response to an ambient temperature change.
High Temperature Induces HY5 Protein Degradation
Given that HY5 is a positive regulator of anthocyanin biosynthesis, the observation that high temperature represses anthocyanin biosynthesis suggests that HY5 activity is decreased as temperature increases. To identify the molecular mechanism(s) involved in the high temperature reduction of HY5 activity, we first determined whether high temperature controls HY5 at the transcriptional level. Although a previous study reported that the level of HY5 mRNA was decreased at high temperature (Delker et al., 2014), we did not find any significant change in HY5 expression in response to 6 or 24 h at 28°C under our growth conditions (Figures 5A–C), implicating that high temperature represses HY5 protein activity post-transcriptionally. Consistent with our observation, HY5 expression was not discernibly affected by high temperatures in a recent study (Park et al., 2017). Next, we examined the effect of high temperature on HY5 protein stability. To investigate protein stability, we measured HY5-GFP levels (driven by a 35S promoter) by western blot analysis. This analysis showed that the HY5 protein level was slightly decreased after 6 h of high-temperature treatment and was further decreased after 24 h (Figure 5D). Since HY5 mRNA levels were not significantly affected by high temperature (Figures 5B,C), these results suggest that high ambient temperature represses the anthocyanin biosynthesis mainly through destabilization of HY5 protein.
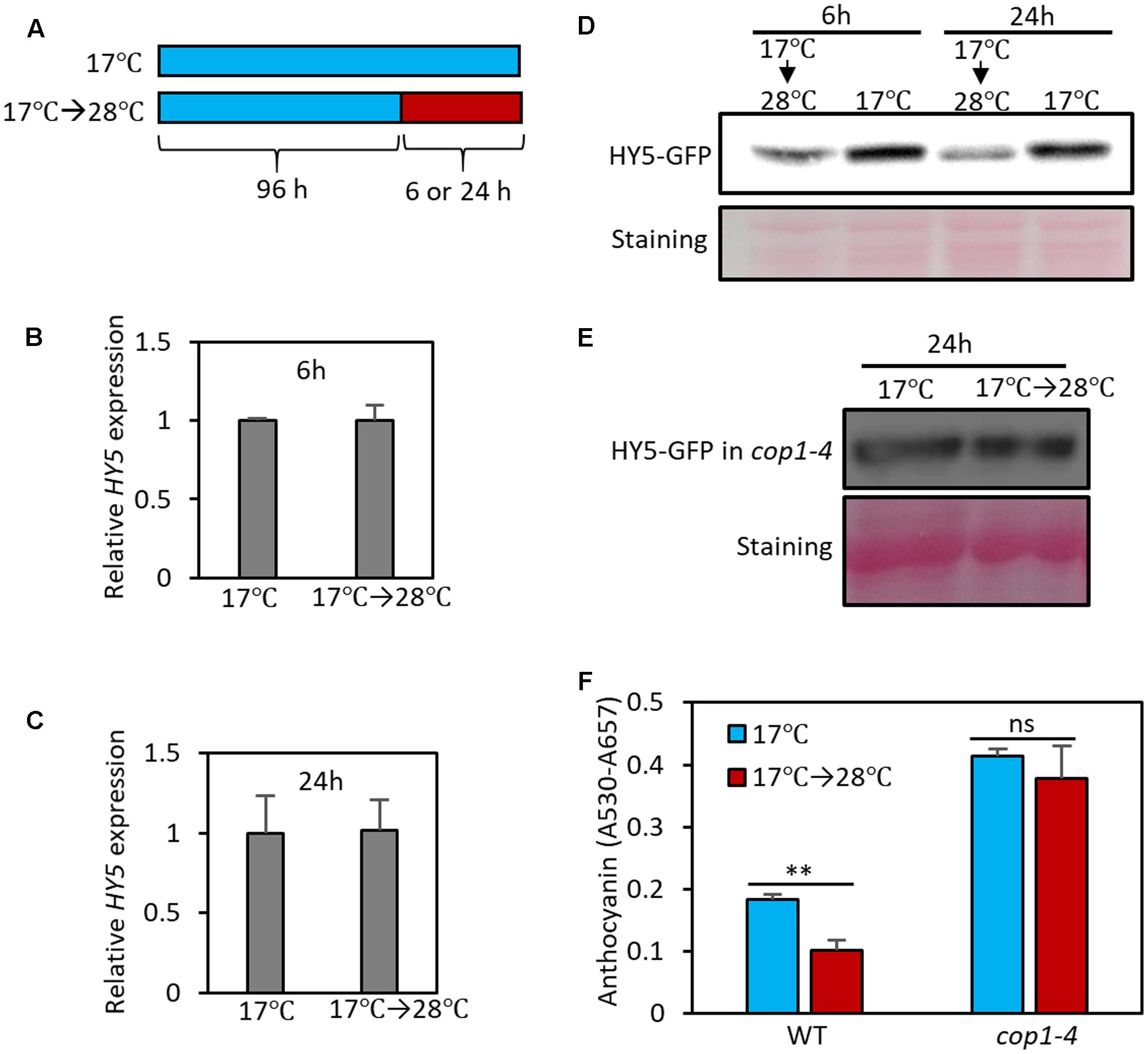
FIGURE 5. High temperature induces the degradation of HY5 protein. (A) Growth conditions in experiments (B–F). Seedlings were grown on MS medium containing 3% sucrose at 17°C for 4 days and then transferred to 28°C for 6 or 24 h (17°C → 28°C) or kept at 17°C. The seedlings were then harvested for extraction of total RNA, protein, and anthocyanin. (B,C) The expression of HY5 was not significantly affected by the 6- or 24-h high temperature treatment. Gene expression levels were normalized to PP2A (AT1G13320) and are presented as values relative to that of seedlings kept at 17°C. Error bars indicate s.d. (n = 3). (D,E) Western blotting with anti-GFP antibody shows that the stability of HY5 protein is reduced at high temperature in wild type, but not in cop1-4 mutant. Total protein was extracted from 35S::HY5-GFP (D) or 35S::HY5-GFP;cop1-4 (E) mutant plants. Equal loading of samples is shown by Ponceau S staining. (F) Anthocyanin pigment accumulation was not affected by high temperature (24 h) in cop1-4 mutant seedlings. Error bars indicate s.d. (n = 3). ∗∗P < 0.01 (Student’s t-test) and ns, not significant (Student’s t-test P ≥ 0.05).
HY5 protein stability is known to be regulated by light through E3 ligase COP1-mediated degradation (Osterlund et al., 2000). In the dark, nuclear-localized COP1 directly interacts with HY5 and induces ubiquitination and degradation of the protein; as a result, HY5 protein levels are low in the dark. In contrast, in the light, COP1 is translocated to the cytosol by light-activated signals, resulting in HY5 protein accumulation in the nucleus (Osterlund et al., 2000). To examine whether COP1 mediates high temperature-induced HY5 degradation, we determined HY5-GFP levels in the cop1 loss-of-function mutant (cop1-4) background. In contrast to wild type plants, the HY5 protein level was not significantly decreased by exposure to high temperature in the cop1-4 mutant (Figure 5E), indicating that COP1 activity is required for high temperature-induced HY5 degradation. Consistent with the patterns of HY5 protein, the anthocyanin levels of cop1-4 mutant seedlings were higher than those of wild type and were thermo-insensitive (Figure 5F). These results suggest that COP1 is involved in the high temperature repression of anthocyanin by destabilizing the HY5 protein.
MYBL2 Is Transcriptionally Induced at High Temperatures
MYBL2 is a negative regulator of late anthocyanin biosynthetic genes such as DFR and LDOX (Dubos et al., 2008; Matsui et al., 2008). It has been shown that HY5 directly binds to the promoter of MYBL2 and reduces its expression, thereby indirectly activating DFR and LDOX transcription (Wang et al., 2016). Since the level of the HY5 protein is reduced at high temperatures (Figure 5D), it is highly likely that MYBL2 expression is also temperature-regulated. To examine this possibility, we compared MYBL2 expression in seedlings grown at 17°C to that of seedlings grown at 17°C for 4 days followed by 28°C for 24 h. We found that MYBL2 expression was increased about twofold by the high temperature treatment in wild type seedlings (Figure 6A). Interestingly, MYBL2 expression was constantly high irrespective of ambient temperature in the hy5 mutant, suggesting that the high temperature induced MYBL2 expression through inactivation of HY5. This interpretation is consistent with a recent report showing that HY5 negatively regulates MYBL2 expression, both directly and indirectly through miRNA858a activity (Wang et al., 2016). Given that the expression of DFR and LDOX is reduced at high temperatures, the expression patterns of MYBL2 were consistent with its function as a transcriptional repressor of DFR and LDOX and further suggests that MYBL2 contributes to the temperature regulation of anthocyanin biosynthesis. Therefore, we determined the anthocyanin accumulation of mybl2 loss-of-function mutant (mybl2-1) seedlings grown at two different ambient temperatures (17 and 17°C followed by 28°C). Anthocyanin accumulation was less affected by high temperature treatment (28°C) in mybl2-1 mutant than in wild type seedlings and, as a result, the anthocyanin levels of the high temperature-treated mybl2 seedlings were significantly higher than those of wild type treated with 28°C (Figure 6B). These results indicate that MYBL2 partially mediates the high temperature repression of anthocyanin biosynthesis. Therefore, it appears that high temperature represses anthocyanin biosynthesis through two distinct hierarchical, connected, and complementary mechanisms: (1) inactivation of the positive anthocyanin regulator HY5, and (2) transcriptional activation of MYBL2, which negatively regulates DFR and LDOX expression.
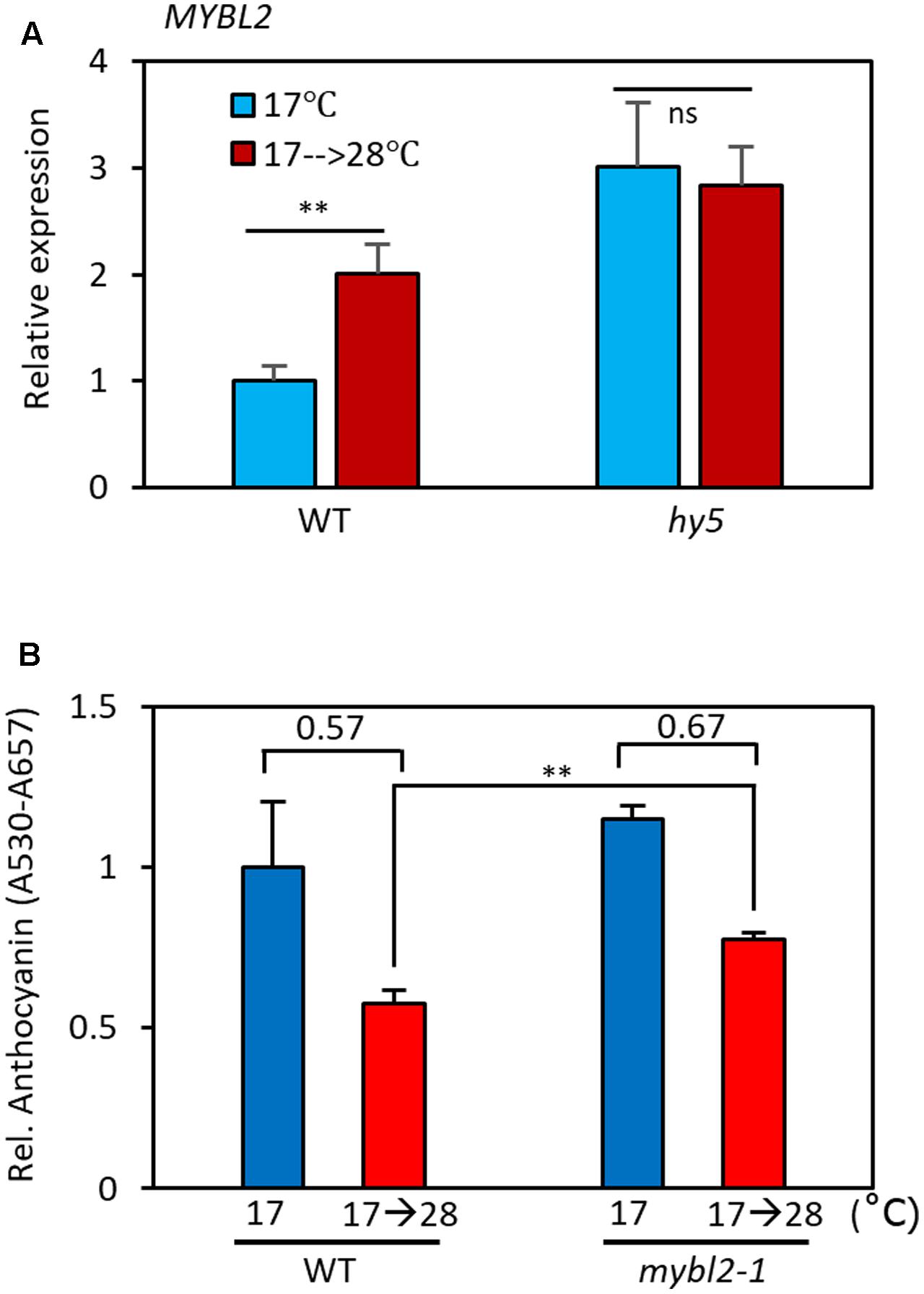
FIGURE 6. MYBL2 partially mediates the high temperature repression of anthocyanin biosynthesis. (A) Expression of MYBL2 in wild type and hy5 mutant seedlings grown at 17°C for 4 days and then transferred to 28°C for 24 h (17°C → 28°C) or kept at 17°C. MYBL2 expression levels were normalized to PP2A (AT1G13320) and are presented as values relative to that of wild type seedlings kept at 17°C. Error bars indicate s.d. (n = 3) and ns, not significant (Student’s t-test P ≥ 0.05). (B) Anthocyanin levels of wild type and mybl2 mutant seedlings grown at two different temperatures. The wild type and mybl2-1 mutant seedlings were grown at 17°C under continuous white light for 4 days and then either transferred to 28°C for 24 h (17°C → 28°C) or kept at 17°C before harvesting for anthocyanin extraction. ∗∗P < 0.01 (Student’s t-test) and numbers indicate the ratio of anthocyanin levels (17°C → 28°C/17°C).
Discussion
In the present study, we demonstrated that anthocyanin biosynthesis is repressed by an elevation in ambient temperature and that a COP1-HY5 signaling module mediates the high-temperature regulation of anthocyanin biosynthesis. We propose that high temperature induces HY5 protein degradation through COP1, as recently suggested by Park et al. (2017), which directly leads to an induced expression of the negative regulator of anthocyanin biosynthesis (MYBL2), and reduced expression of both early (CHS and CHI) and late (DFR, LDOX, and UF3GT) anthocyanin biosynthetic genes. As a consequence, anthocyanin pigment levels are lower in seedlings grown at a high temperature (Figure 7).
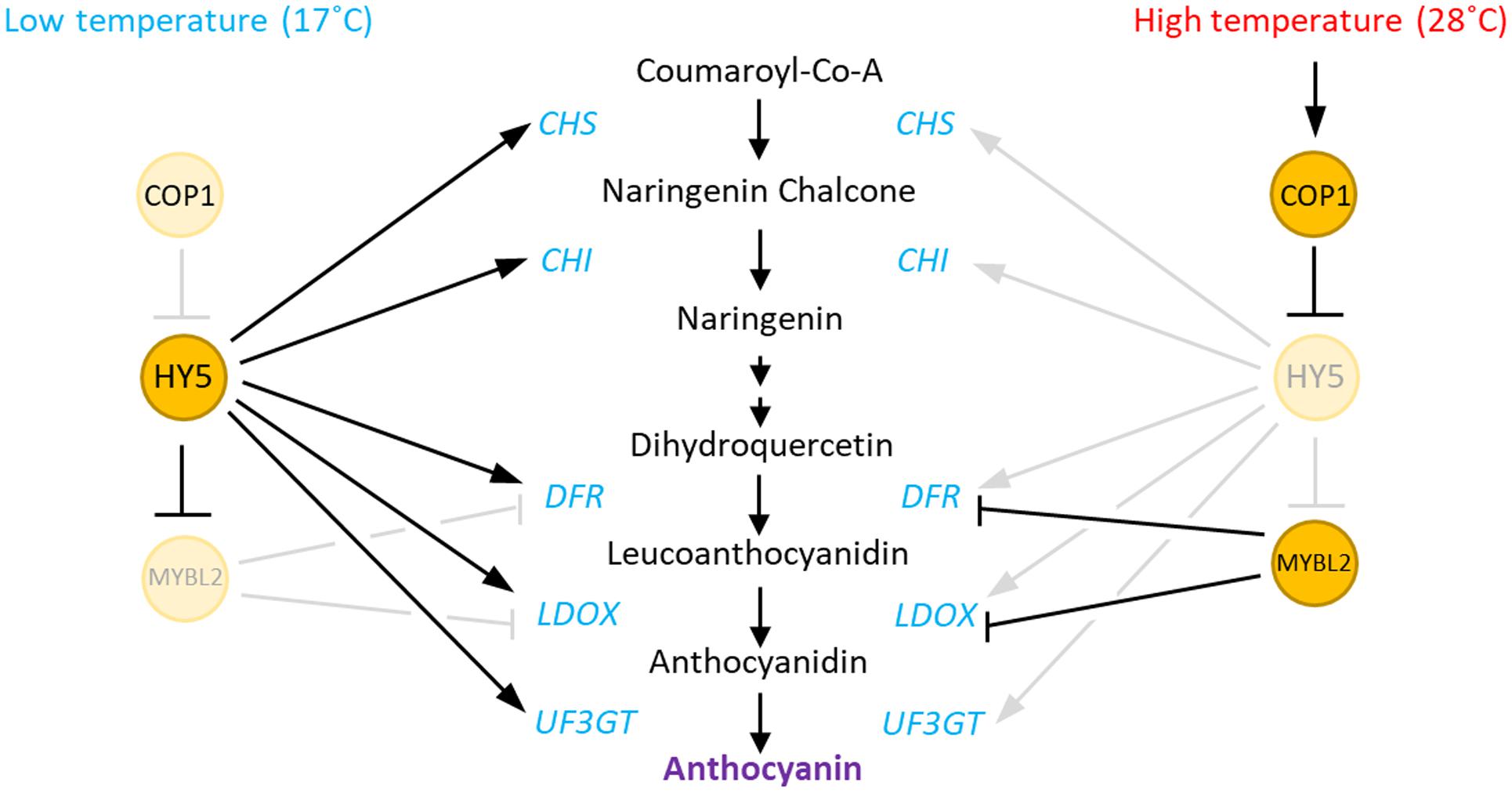
FIGURE 7. Hypothetical model of the regulation of anthocyanin biosynthesis by high temperature in Arabidopsis. At low ambient temperatures, HY5 directly activates the expression of both early (CHS and CHI) and late (DFR, LDOX, and UF3GT) anthocyanin biosynthetic genes. HY5 also indirectly represses the expression of the late genes by repression of MYBL2, which encodes a repressor of DFR and LDOX. In contrast, at high ambient temperatures, increased COP1 activity induces the degradation of HY5 protein, which results in low expression of both the early and late anthocyanin biosynthetic genes. The HY5 degradation also transcriptionally activates MYBL2, which also contributes the repression of the late genes at high temperatures.
High ambient temperature induces dramatic morphological changes in Arabidopsis seedlings, including the promotion of hypocotyl elongation and hyponastic leaf growth. PIF4 is a key regulator of the thermo-induced morphological changes and thus, hypocotyl elongation, in pif4 mutant seedlings (and also in the pif4;pif5 double mutant) is insensitive to high temperatures (Koini et al., 2009). In contrast to its thermo-insensitive hypocotyl elongation phenotype, anthocyanin accumulation and expression of anthocyanin biosynthetic genes in the pif4;pif5 double mutant were significantly decreased in response to high temperatures. These results suggest that the anthocyanin biosynthesis is regulated by the high temperatures, independent of the PIF4-mediated morphogenic responses. Although the anthocyanin biosynthetic genes were down-regulated by high temperatures, basal levels of CHS, DFR, and LDOX were slightly higher in the pif4;pif5 double mutant than in the wild type, indicating the negative functions of PIF4 and PIF5 in anthocyanin biosynthesis. In agreement with these observations, a previous study showed that these factors negatively regulate anthocyanin biosynthesis under red light (Liu et al., 2015).
Our results show that anthocyanin accumulation is thermo-insensitive in the cop1-4 mutant. COP1 activity is known to be regulated by light receptor phytochromes (Yi and Deng, 2005). The light-activated phytochromes induce translocation of COP1 protein from the nucleus to the cytoplasm, thereby reducing the nuclear COP1 level; as a result, nuclear HY5 protein levels increase. Given that COP1 mediates the high temperature-induced degradation of HY5, it is likely that COP1 activity is increased at high temperatures. However, the expression of COP1 was not significantly affected by high temperatures (Supplementary Figure 2), suggesting that COP1 is post-transcriptionally regulated by ambient temperature. In support of this hypothesis, a recent study reported that the nuclear COP1 protein level is increased at elevated temperatures, which is responsible for the high temperature-induced degradation of HY5 (Park et al., 2017). It was shown that the activities of phytochromes are regulated by ambient temperature as well as light (Jung et al., 2016; Legris et al., 2016). Phytochromes switch between an inactive Pr state and an active Pfr state depending on light conditions. Increased temperature promotes the reversion of active Pfr to the Pr state and the activities of phytochromes are decreased at high temperature (Jung et al., 2016; Legris et al., 2016). Therefore, it is likely that the high activity of COP1 at high temperature may be due to reduced phytochrome activities. Further experiments are required to determine if HY5 protein stability is regulated by temperature through the phytochrome-COP1 interacting module.
The accumulation of anthocyanin pigments in the ripening fruit of grapevine or apple has been reported to be reduced at high temperatures (Mori et al., 2007; Lin-Wang et al., 2011). In grapevine (Vitis vinifera L.), the expression of the HY5 homologous gene VviHY5 is highly correlated with flavonol levels, and ectopic expression of VviHY5 transcriptionally activates flavonol-related genes (FLS4 and GT5) (Loyola et al., 2016), indicating a positive role for the HY5 ortholog in flavonol biosynthesis. In addition, a recent study proposed that VviHY5 may regulate anthocyanin biosynthesis by controlling MYBA genes (Matus et al., 2017). The expression of VviHY5 increases in response to high temperatures under light conditions (Loyola et al., 2016). Even if VviHY5 expression increases with high temperature, this may be irrelevant if the VviHY5 protein is also degraded as in Arabidopsis. Therefore, further studies are required to determine if the COP1-HY5 module mediates the high temperature repression of anthocyanin biosynthesis in other species.
Anthocyanin biosynthesis is increased at cold temperatures, and this increase has been suggested to enhance the survival of plants under cold stress (Catala et al., 2011). By contrast, the benefit of suppressing anthocyanin biosynthesis at high temperatures is unknown. Given that both morphological changes and anthocyanin biosynthesis are energy-demanding processes, the energy balance between these two processes might be optimized in response to the environmental conditions. In this respect, the suppression of anthocyanin biosynthesis might be more beneficial for plants grown at high temperature because thermomorphogenic growth at a cost of reduced anthocyanin biosynthesis might enhance survival under high temperature stress.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana plants were grown in a greenhouse with 16 h light/8 h dark cycles at 20 to 24°C for general growth and seed harvesting. All the A. thaliana plants (hy5, pif4;pif5, mybl2-1, and cop1-4) used in this study were of the Col-0 ecotype background. The hy5 mutants were obtained from the Salk Institute (Salk_ 056405C). The pif4;pif5 seeds were previously described (Lorrain et al., 2008). The cop1-4 mutant was previously described (McNellis et al., 1994). The mybl2-1 mutant was previously described (Dubos et al., 2008). The 35S:HY5-GFP (in Col-0 and cop1-4 background) transgenic plants were described previously (Park et al., 2017).
Gene Ontology (GO) Analysis
The list of high temperature-repressed genes for GO analysis was obtained from a previous RNA-seq experiment (Jung et al., 2016). In that experiment, genes with expression levels that were reduced more than four-fold at ZT0 in seedlings grown at 27°C compared to seedlings at 22°C were defined as high temperature-repressed genes. Low-expression genes [sum of FPKM values (22°C + 27°C) < 0.2] were excluded from the analysis. Enriched GO terms of the high temperature-repressed genes (a total of 915 genes) were identified using the Plant GeneSet Enrichment Analysis Toolkit (Yi et al., 2013) and heat maps for the expression of the anthocyanin biosynthetic genes were generated using Heatmapper web-based tools (Babicki et al., 2016).
Anthocyanin Extraction
Seeds were sterilized using 70% (v/v) ethanol and 0.01% (v/v) Triton X-100 and were then plated on MS medium (Duchefa) containing 3% sucrose. After 3 days of stratification at 4°C, the plates were placed under white light for 6 h to promote seed germination. Seedlings were incubated at 17°C under continuous light conditions (light intensity: 30 μmol m-2 s-1) for 5 days or incubated at 17°C for 4 days followed by incubation at 28°C for 24 h. For anthocyanin extraction, 30 seedlings from each condition were placed in 600 μL of 1% HCl in methanol (v/v) (1% methanolic HCL = 45 mL methanol + 450 mL HCl). The samples were then incubated overnight in the dark at 4°C with shaking. After overnight incubation, 400 μL water and 400 μL chloroform were added to each sample and mixed. After centrifugation, the absorbance of the supernatant was measured at 530 and 657 nm, and the concentration of anthocyanin was calculated by A530 - 0.25∗A657 (Rabino and Mancinelli, 1986).
qRT-PCR Gene Expressions Analysis
Seedlings were grown at 17°C for 5 days or grown at 17°C for 4 days followed by incubation at 28°C for 24 h. Seedlings (about 50 for each biological replicate) were then harvested and ground in liquid nitrogen for total RNA extraction. Total RNA was extracted from the seedlings using a MiniBEST Plant RNA extraction kit (TaKaRa). M-MLV reverse transcriptase (Fermentas) was used to synthesize cDNA from the RNA. Quantitative real-time PCR (qRT-PCR) was performed using a CFX96 Real-Time PCR detection system (Bio-Rad) and the EvaGreen master mix (Solgent). Gene expression levels were normalized to that of PP2A (PROTEIN PHOSPHATASE 2A SUBUNIT, AT1G13320) and are shown relative to the expression levels in wild type (Czechowski et al., 2005). Three biological replicates were analyzed for each qRT-PCR experiment. The gene specific primers used here are listed in Supplementary Table 2.
Protein Extraction and Western Blot Analysis
Five-day-old seedlings (about 50 seedlings in total) were harvested and ground in liquid nitrogen. Proteins were extracted with 2× protein extraction buffer (100 mM Tris-HCl, pH 6.8, 25% glycerol, 2% SDS, 0.01% bromophenol blue, with β-mercaptoethanol added to 10% before use). Western blot analysis was performed to determine HY5-GFP protein levels using an anti-GFP antibody (Santa Cruz, sc-9996). The original images for the western blot and Ponceau S staining are shown in Supplementary Figure 3.
Author Contributions
SK, EO, and GH conceived and designed the research, performed the experiments, and wrote the manuscript, SL, J-YZ, TN, and IP performed the experiments and analyzed the data, JK wrote the manuscript.
Funding
Research was supported by a grant from the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (grant no. NRF-2016R1C1B2008821), the Basic Research Laboratory Program of the National Research Foundation funded by the Korean government (grant no. 2017R1A4A1015620), Chonnam National University (grant no. 2016-2514), and BK21 Plus project of the National Research Foundation of Korea Grant.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer NA and handling Editor declared their shared affiliation.
Acknowledgments
The pif4;pif5 seeds were kindly supplied by Christian Fankhauser (University of Lausanne). The mybl2-1 seeds were kindly provided by Loïc Lepiniec (INRA, AgroParisTech, ERL CNRS). The 35S::HY5-GFP;cop1-4 seeds were kindly provided by Chung-Mo Park (Seoul National University).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01787/full#supplementary-material
References
Babicki, S., Arndt, D., Marcu, A., Liang, Y., Grant, J. R., Maciejewski, A., et al. (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 44, W147–W153. doi: 10.1093/nar/gkw419
Borevitz, J. O., Xia, Y., Blount, J., Dixon, R. A., and Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2394. doi: 10.1105/tpc.12.12.2383
Catala, R., Medina, J., and Salinas, J. (2011). Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 16475–16480. doi: 10.1073/pnas.1107161108
Chattopadhyay, S., Ang, L. H., Puente, P., Deng, X. W., and Wei, N. (1998). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10, 673–683. doi: 10.1105/tpc.10.5.673
Choi, H., and Oh, E. (2016). PIF4 integrates multiple environmental and hormonal signals for plant growth regulation in Arabidopsis. Mol. Cells 39, 587–593. doi: 10.14348/molcells.2016.0126
Crawford, A. J., Mclachlan, D. H., Hetherington, A. M., and Franklin, K. A. (2012). High temperature exposure increases plant cooling capacity. Curr. Biol. 22, R396–R397. doi: 10.1016/j.cub.2012.03.044
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K., and Scheible, W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. doi: 10.1104/pp.105.063743
Delker, C., Sonntag, L., James, G. V., Janitza, P., Ibanez, C., Ziermann, H., et al. (2014). The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep. 9, 1983–1989. doi: 10.1016/j.celrep.2014.11.043
Dubos, C., Le Gourrierec, J., Baudry, A., Huep, G., Lanet, E., Debeaujon, I., et al. (2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55, 940–953. doi: 10.1111/j.1365-313X.2008.03564.x
Falcone Ferreyra, M. L., Rius, S. P., and Casati, P. (2012). Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3:222. doi: 10.3389/fpls.2012.00222
Gonzalez, A., Zhao, M., Leavitt, J. M., and Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Heijde, M., Binkert, M., Yin, R., Ares-Orpel, F., Rizzini, L., Van De Slijke, E., et al. (2013). Constitutively active UVR8 photoreceptor variant in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 20326–20331. doi: 10.1073/pnas.1314336110
Jung, J. H., Domijan, M., Klose, C., Biswas, S., Ezer, D., Gao, M., et al. (2016). Phytochromes function as thermosensors in Arabidopsis. Science 354, 886–889. doi: 10.1126/science.aaf6005
Koini, M. A., Alvey, L., Allen, T., Tilley, C. A., Harberd, N. P., Whitelam, G. C., et al. (2009). High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19, 408–413. doi: 10.1016/j.cub.2009.01.046
Lea, U. S., Slimestad, R., Smedvig, P., and Lillo, C. (2007). Nitrogen deficiency enhances expression of specific MYB and bHLH transcription factors and accumulation of end products in the flavonoid pathway. Planta 225, 1245–1253. doi: 10.1007/s00425-006-0414-x
Legris, M., Klose, C., Burgie, E. S., Rojas, C. C., Neme, M., Hiltbrunner, A., et al. (2016). Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354, 897–900. doi: 10.1126/science.aaf5656
Lin-Wang, K., Micheletti, D., Palmer, J., Volz, R., Lozano, L., Espley, R., et al. (2011). High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 34, 1176–1190. doi: 10.1111/j.1365-3040.2011.02316.x
Liu, Z., Zhang, Y., Wang, J., Li, P., Zhao, C., Chen, Y., et al. (2015). Phytochrome-interacting factors PIF4 and PIF5 negatively regulate anthocyanin biosynthesis under red light in Arabidopsis seedlings. Plant Sci. 238, 64–72. doi: 10.1016/j.plantsci.2015.06.001
Lorrain, S., Allen, T., Duek, P. D., Whitelam, G. C., and Fankhauser, C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323. doi: 10.1111/j.1365-313X.2007.03341.x
Loyola, R., Herrera, D., Mas, A., Wong, D. C., Holl, J., Cavallini, E., et al. (2016). The photomorphogenic factors UV-B RECEPTOR 1, ELONGATED HYPOCOTYL 5, and HY5 HOMOLOGUE are part of the UV-B signalling pathway in grapevine and mediate flavonol accumulation in response to the environment. J. Exp. Bot. 67, 5429–5445. doi: 10.1093/jxb/erw307
Maier, A., Schrader, A., Kokkelink, L., Falke, C., Welter, B., Iniesto, E., et al. (2013). Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 74, 638–651. doi: 10.1111/tpj.12153
Mancinelli, A. L., Rossi, F., and Moroni, A. (1991). Cryptochrome, phytochrome, and anthocyanin production. Plant Physiol. 96, 1079–1085. doi: 10.1104/pp.96.4.1079
Matsui, K., Umemura, Y., and Ohme-Takagi, M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55, 954–967. doi: 10.1111/j.1365-313X.2008.03565.x
Matus, J. T., Cavallini, E., Loyola, R., Holl, J., Finezzo, L., Dal Santo, S., et al. (2017). A group of grapevine MYBA transcription factors located in chromosome 14 control anthocyanin synthesis in vegetative organs with different specificities compared with the berry color locus. Plant J. 91, 220–236. doi: 10.1111/tpj.13558
McNellis, T. W., Von Arnim, A. G., Araki, T., Komeda, Y., Misera, S., and Deng, X. W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6, 487–500. doi: 10.1105/tpc.6.4.487
Mori, K., Goto-Yamamoto, N., Kitayama, M., and Hashizume, K. (2007). Loss of anthocyanins in red-wine grape under high temperature. J. Exp. Bot. 58, 1935–1945. doi: 10.1093/jxb/erm055
Movahed, N., Pastore, C., Cellini, A., Allegro, G., Valentini, G., Zenoni, S., et al. (2016). The grapevine VviPrx31 peroxidase as a candidate gene involved in anthocyanin degradation in ripening berries under high temperature. J. Plant Res. 129, 513–526. doi: 10.1007/s10265-016-0786-3
Osterlund, M. T., Hardtke, C. S., Wei, N., and Deng, X. W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. doi: 10.1038/35013076
Park, Y. J., Lee, H. J., Ha, J. H., Kim, J. Y., and Park, C. M. (2017). COP1 conveys warm temperature information to hypocotyl thermomorphogenesis. New Phytol. 215, 269–280. doi: 10.1111/nph.14581
Pastore, C., Dal Santo, S., Zenoni, S., Movahed, N., Allegro, G., Valentini, G., et al. (2017). Whole plant temperature manipulation affects flavonoid metabolism and the transcriptome of grapevine berries. Front. Plant Sci. 8:929. doi: 10.3389/fpls.2017.00929
Proveniers, M. C., and van Zanten, M. (2013). High temperature acclimation through PIF4 signaling. Trends Plant Sci. 18, 59–64. doi: 10.1016/j.tplants.2012.09.002
Quint, M., Delker, C., Franklin, K. A., Wigge, P. A., Halliday, K. J., and Zanten, M. (2016). Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2:15190. doi: 10.1038/nplants.2015.190
Rabino, I., and Mancinelli, A. L. (1986). Light, temperature, and anthocyanin production. Plant Physiol. 81, 922–924. doi: 10.1104/pp.81.3.922
Shin, D. H., Choi, M., Kim, K., Bang, G., Cho, M., Choi, S. B., et al. (2013). HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 587, 1543–1547. doi: 10.1016/j.febslet.2013.03.037
Shin, J., Park, E., and Choi, G. (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 49, 981–994. doi: 10.1111/j.1365-313X.2006.03021.x
Sponga, F., Deitzer, G. F., and Mancinelli, A. L. (1986). Cryptochrome, phytochrome, and the photoregulation of anthocyanin production under blue light. Plant Physiol. 82, 952–955. doi: 10.1104/pp.82.4.952
Tanaka, Y., Sasaki, N., and Ohmiya, A. (2008). Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54, 733–749. doi: 10.1111/j.1365-313X.2008.03447.x
Teng, S., Keurentjes, J., Bentsink, L., Koornneef, M., and Smeekens, S. (2005). Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139, 1840–1852. doi: 10.1104/pp.105.066688
Wang, Y., Wang, Y., Song, Z., and Zhang, H. (2016). Repression of MYBL2 by Both microRNA858a and HY5 leads to the activation of anthocyanin biosynthetic pathway in Arabidopsis. Mol. Plant 9, 1395–1405. doi: 10.1016/j.molp.2016.07.003
Yi, C., and Deng, X. W. (2005). COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15, 618–625. doi: 10.1016/j.tcb.2005.09.007
Yi, X., Du, Z., and Su, Z. (2013). PlantGSEA: a gene set enrichment analysis toolkit for plant community. Nucleic Acids Res. 41, W98–W103. doi: 10.1093/nar/gkt281
Zhang, F., Gonzalez, A., Zhao, M., Payne, C. T., and Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869. doi: 10.1242/dev.00681
Keywords: Arabidopsis, high temperature stress, anthocyanin, HY5, flavonoid, gene expression
Citation: Kim S, Hwang G, Lee S, Zhu J-Y, Paik I, Nguyen TT, Kim J and Oh E (2017) High Ambient Temperature Represses Anthocyanin Biosynthesis through Degradation of HY5. Front. Plant Sci. 8:1787. doi: 10.3389/fpls.2017.01787
Received: 13 July 2017; Accepted: 02 October 2017;
Published: 20 October 2017.
Edited by:
Kimberley Cathryn Snowden, Plant & Food Research, New ZealandReviewed by:
José Tomás Matus, Centre for Research in Agricultural Genomics (CRAG), SpainNick Albert, Plant & Food Research, New Zealand
Copyright © 2017 Kim, Hwang, Lee, Zhu, Paik, Nguyen, Kim and Oh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eunkyoo Oh, eoh@jnu.ac.kr; matins81@gmail.com
†These authors have contributed equally to this work.
 Sara Kim
Sara Kim Geonhee Hwang1†
Geonhee Hwang1† Inyup Paik
Inyup Paik Thom Thi Nguyen
Thom Thi Nguyen Jungmook Kim
Jungmook Kim Eunkyoo Oh
Eunkyoo Oh