- 1Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, Institute of Vegetables and Flowers – Chinese Academy of Agricultural Sciences, Beijing, China
- 2College of Horticulture, China Agricultural University, Beijing, China
Cuticular waxes covering the outer plant surface impart a whitish appearance. Wax-less cabbage mutant shows glossy in leaf surface and plays important roles in riching cabbage germplasm resources and breeding brilliant green cabbage. This is the first report describing the characterization and fine-mapping of a wax biosynthesis gene using a novel glossy Brassica oleracea mutant. In the present paper, we identified a glossy cabbage mutant (line10Q-961) with a brilliant green phenotype. Genetic analyses indicated that the glossy trait was controlled by a single recessive gene. Preliminary mapping results using an F2 population containing 189 recessive individuals revealed that the Cgl1 gene was located at the end of chromosome C08. Several new markers closely linked to the target gene were designed according to the cabbage reference genome sequence. Another population of 1,172 recessive F2 individuals was used to fine-map the Cgl1 gene to a 188.7-kb interval between the C08SSR61 simple sequence repeat marker and the end of chromosome C08. There were 33 genes located in this region. According to gene annotation and homology analyses, the Bol018504 gene, which is a homolog of CER1 in Arabidopsis thaliana, was the most likely candidate for the Cgl1 gene. Its coding and promoter regions were sequenced, which indicated that the RNA splice site was altered because of a 2,722-bp insertion in the first intron of Bol018504 in the glossy mutant. Based on the FGENESH 2.6 prediction and sequence alignments, the PLN02869 domain, which controls fatty aldehyde decarbonylase activity, was absent from the Bol018504 gene of the 10Q-961 glossy mutant. We inferred that the inserted sequence in Bol018504 may result in the glossy cabbage mutant. This study represents the first step toward the characterization of cuticular wax biosynthesis in B. oleracea, and may contribute to the breeding of new cabbage varieties exhibiting a brilliant green phenotype.
Introduction
Cuticular waxes, which are composed of a range of lipid compounds, act as a hydrophobic layer and cover the outer surface of aerial plant tissues (Millar et al., 1999). To adapt to environmental changes, plants secrete waxes onto the surface or into the interior of cuticles to form the first barrier against ultraviolet radiation, plant pathogens, and insects (Mariani and Wolters-Arts, 2000; Kunst and Samuels, 2003). Cuticular waxes can reduce non-stomatal water evapotranspiration and increase water retention, as well as prevent pollen, dust, and air pollutants from falling onto plant surfaces (Kerstiens, 1996; Barthlott et al., 1998). Moreover, cuticular waxes affect various physiological functions, such as preventing fruit cracking and influencing morphological development and pigmentation of leaves and fruits. Additionally, cuticular waxes regulate plant fertility by affecting pollen development (Koch and Ensikat, 2008; Koch et al., 2009).
Since Dellaert (Dellaert et al., 1979) discovered the first waxy cuticle eceriferum mutant (cer) in Arabidopsis thaliana, numerous genes involving in wax biosynthesis, export and regulation have hitherto been identified. In A. thaliana, ABCG11, CER1, CER3, CER4, CER5, CER6, CER7, CER10, CFL1, FATB, HDG1, KCR1, LACS1, LTPG1, MAH1, MYB30/41/96, PAS2, W1N1/SHN1, WBC11, and WSD1 (Samuels et al., 2008; Bernard and Joubès, 2013; Lee and Suh, 2013) were mapped and cloned. Among them, CER4, CER6, CER10, FATB, WSD1, and MAH1 were related to wax biosynthesis, while CER5, CER7, WBC11, W1N1/SHN1, MYB30/41/96, CFL1, and HDG1 were responsible for the transportation and regulation (Samuels et al., 2008; Lee and Suh, 2013). Besides, OsGL1-1 to OsGL1-11, OsGL1, OsWSL2 and OsWSL3 in rice (Oryza sativa) (Islam et al., 2009; Qin et al., 2011; Mao et al., 2012; Zhou et al., 2013; Gan et al., 2016), ZmGL1, ZmGL2, and ZmGL8 in maize (Zea mays) (Tacke et al., 1995; Xu et al., 2002; Sturaro et al., 2005), HvCUT1.1, HvCUT1.2, HvCUT1.3, HvFDH1.1, HvCER1.1, and HvCER1.2 in barley (Hordeum vulgare) (Richardson et al., 2007), and W1, W2, Iw1, Iw2, Iw3, Wx-7A, Wx-4A, Wx-7D, and Wax1 in wheat (Triticum aestivum Linn.) (Dubcovsky et al., 1997; Murai et al., 1999; Lu et al., 2015) were mapped. Within Brassica species, BnCER1 in canola (Brassica napus) (Pu et al., 2013) and BrWax1 in Chinese cabbage (Brassica rapa L. ssp. pekinensis) (Zhang et al., 2013) were also mapped and characterized.
Among Brassica species, most studies focused on genetic analyses of waxy mutants. The glossy trait in broccoli (Brassica oleracea L. var. italica Plenck) (Anstey and Moore, 1954; Farnham, 2010), Brussels sprouts (B. oleracea L. var. gemmifera Zenk) (North and Priestley, 1962), and B. napus ‘Nilla glossy’ (Thompson, 1972; Jianguo et al., 1995) is controlled by recessive genes respectively, but dominant genes in B. napus wl mutant (Jianguo et al., 1992) and collard (B. oleracea L. var. sabauda DC.) (Priestley and Wills, 1966). However, no genes related to glossy trait in cabbage have been identified, and the molecular mechanism of cuticular wax biosynthesis and secretion in B. oleracea has yet to be fully characterized.
In this study, using simple sequence repeat (SSR) markers and two F2 populations, Cgl1, a gene controlling glossy trait in cabbage, was fine mapped in a 188.7 kb region at the end of chromosome C08. According to gene annotation and homology analyses, Bol018504 was identified as the candidate gene of Cgl1 from 33 genes in target region. Moreover, further analysis on coding and promoter sequences of Bol018504 were made to validate the fine-mapping result. Findings in the present research will contribute to a more comprehensive understanding of plant cuticular wax metabolic networks and accelerate the breeding of cabbage cultivars exhibiting the brilliant green trait.
Materials and Methods
Plant Materials and Growth Conditions
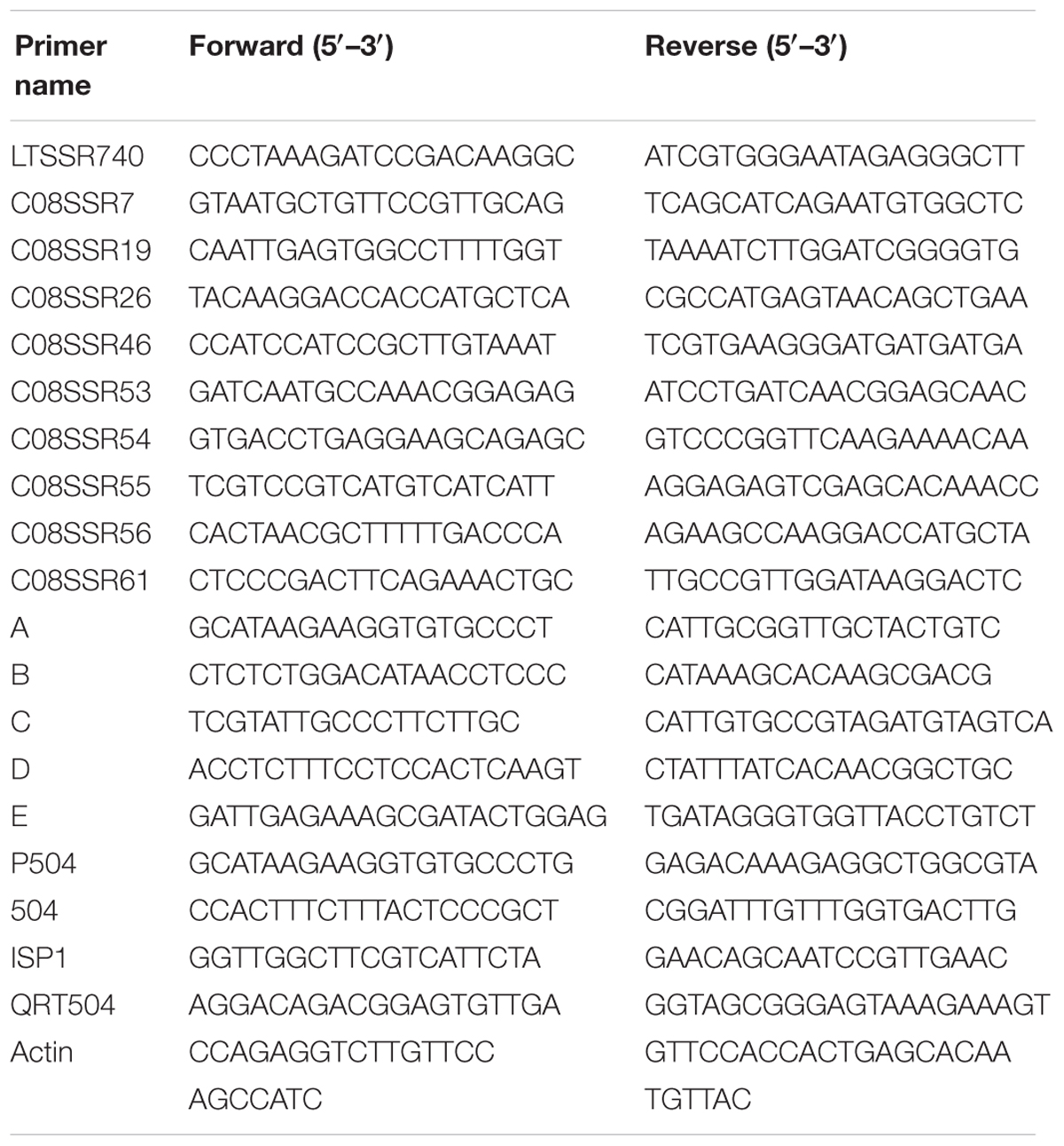
Line 10Q-961 (a glossy mutant of inbred cabbage line10Q-962) (Figure 1), and the waxy inbred cabbage line 10Q-206 were used as parents to generate an F1 hybrids. The F2 population was generated through self-pollination of F1 plants. To acquire more polymorphism markers and accelerate the mapping process, another F2 population was produced from a cross between line 10Q-961 and a waxy Chinese kale doubled haploid line, M-36.
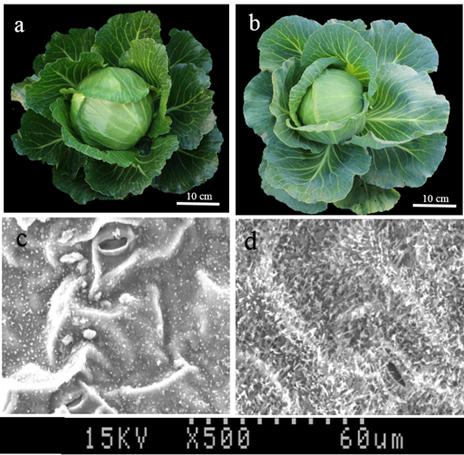
FIGURE 1. Plant appearance of mutant 10Q-961, wild-type 10Q-962, and the micro-structure observation of leaf blade. (a) Mutant 10Q-961, (b) wild-type 10Q-962, (c) microstructure of 10Q-961 blade ventral, (d) microstructure of 10Q-962 blade ventral.
Plant phenotype was investigated visually at three-real-leaf stage. All plants were grown in spring and autumn each year in a solar greenhouse at the experimental station in Changping (39°54′ N, Beijing, China). All plant materials were provided by the Cabbage and Broccoli Research Group, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences.
Map-Based Cloning of Cgl1
Genomic DNA from young leaves was extracted using CTAB method with modifications (Tel-Zur et al., 1999). For preliminary mapping, bulked segregant analysis (BSA) (Chantret et al., 2000) was performed with DNA pools from 10 waxy F2 individuals and 10 glossy F2 individuals. 189 glossy individuals of the F2 population which produced from a cross between line 10Q-961 and 10Q-206 were used in preliminary mapping. About 5, 000 F2 seeds (10Q-961 × M-36, self-pollination) were planted, and 1,172 individuals with a glossy phenotype were selected for fine mapping purposes.
Simple Sequence Repeat Markers, Polymerase Chain Reaction, and Polyacrylamide Gel Electrophoresis
A total of 866 SSR markers evenly distributed on nine chromosomes were designed according to the whole genome sequence of B. oleracea1 (Liu et al., 2014). Primers were designed to produce 100–460-bp amplicons with 40–50% GC content and a melting temperature of 57–61°C.
A polymerase chain reaction (PCR) was completed in a 10 μl reaction volume containing 2 μl DNA template (50 ng μl-1), 1 μl 10× PCR buffer [200 mM Tris-HCl pH 8.3, 200 mM KCl, 100 mM (NH4)2SO4, 20 mM MgSO4], 0.8 μl dNTP (2.5 mM), 0.5 μl forward primer (5 μM), 0.5 μl reverse primer (5 μM), 0.2 μl Taq DNA polymerase (2.5 U μl-1), and 5 μl double-distilled H2O. The PCR conditions were as follows: initial denaturation at 96°C for 8 min, followed by 33 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 40 s. The PCR program was completed with a final extension step at 72°C for 7 min. The PCR was conducted using a Veriti 96-Well Thermal Cycler (Applied Biosystems, Carlsbad, CA, USA). Amplicons were analyzed using 8% polyacrylamide gel electrophoresis at 160 V for 70 min, and visualized by silver staining (Bassam et al., 1991).
To analyze the Bol018504 candidate gene, five primer pairs were designed using DNAMAN 7.0 based on the ORF and putative promoter sequence of Bol018504. A PCR amplification was completed using the Q5 Ultra High Fidelity DNA polymerase (New England Biolabs, Inc.) following its manufacturer’s instructions. Genomic DNA of line 10Q-961 and wild-type line 10Q-962 were used as PCR templates, and the resulting amplicons were analyzed by agarose gel electrophoresis.
Quantitative Reverse-Transcription PCR (qRT-PCR) Analysis of the Candidate Gene
Total RNA was prepared from young leaves of 10Q-961 and 10Q-962 using a RNAprep pure Plant Kit (TIANGEN, Beijing, China), according to the manufacturer’s instructions. The extracted RNA was treated with RNase-free DNase I (Fermentas, Harrington, QC, Canada) to eliminate genomic DNA contamination. 1 μg of RNA was used to synthesize oligo (dT)-primed first-strand cDNA using a PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa, Kyoto, Japan) following the protocol provided by the manufacturer. Primer QRT504 was designed for quantitative analysis of Bol018504. Primer sequences of both actin and Bol018504 used in qRT-PCR analysis are listed in Table 1. qRT-PCR was carried out using the SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) kit (Takara) to analyze gene expression. Amplification was performed on a CFX96 TouchTM real-time PCR detection system (Bio-Rad, Hercules, CA, USA). Three technical replicates were used for each cDNA sample and three samples (biological replicates) were tested. The relative mRNA expression was calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
Data Analysis
For each marker, individuals with the 10Q-961 mutant allele were categorized as “b.” Individuals with the 10Q-206 or M-36 alleles were categorized as “a.” Those with the F1 allele were categorized as “h.” The Kosambi mapping function was used to calculate genetic distances between markers (Kosambi, 1943) and the genetic map was constructed using MapDraw (Liu and Meng, 2003).
Results
Preliminary Mapping of the Cgl1 Locus
In this study, F1 hybrids of 10Q-961 (P1) × M-36 (P2) were all exhibit waxy phenotype as M-36. In F2 population, the ratio of waxy to glossy plants was 3.085:1 (398:129), which was confirmed to be 3:1 by the Chi-square test (χ2 = 0.076 < χ2 0.05 = 3.841). The ratio of waxy to glossy plants was 0.933:1 (236:253), which was confirmed to be 1:1 by the Chi-square test (χ2 = 0.591 < χ20.05 = 3.841) in BC1P1 population, and all the plants in BC1P2 backcross population exhibited waxy phenotype. These genetic analysis indicated that the glossy traits in line 10Q-961 was controlled by a single recessive gene. 886 SSR primers were used to screen the polymorphisms between the parent lines 10Q-961 and 10Q-206. A total of 61 of the 866 primer pairs showed polymorphisms. Markers that were polymorphic between parents were screened in two bulks. Of all the primers, only marker LTSSR740 was identified polymorphisms between the two pools (Figure 2). The F2 population of the 10Q-961 × 10Q-206 cross consisted of 189 recessive plants and was used to validate the LTSSR740 marker. Six recombinants (Figure 3) were detected, and the calculated genetic distance between LTSSR740 and Cgl1 was 3.17 cM. According to the location of LTSSR740 in the ‘02-12’ cabbage reference genome, Cgl1 was localized to the end of chromosome C08 (40,596,166–41,516,064).
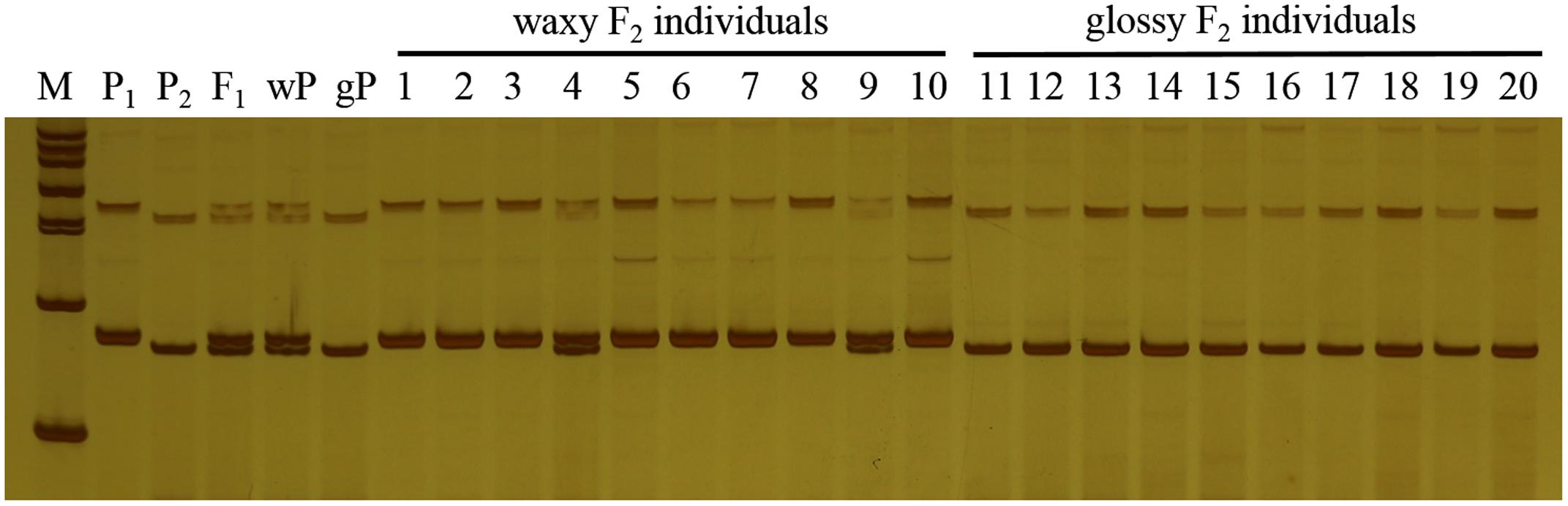
FIGURE 2. Polymorphisms of LTSSR740 in parents, F1, gene pools, and partial individuals of the F2 population. M, DNA ladder; P1, line 10Q-206 with waxy phenotype; P2, line 10Q-961 with glossy phenotype; F1, hybrid; wP, waxy pool; and gP, glossy pool. Lanes 1–10, waxy F2 individuals; lanes 11–20, glossy F2 individuals.
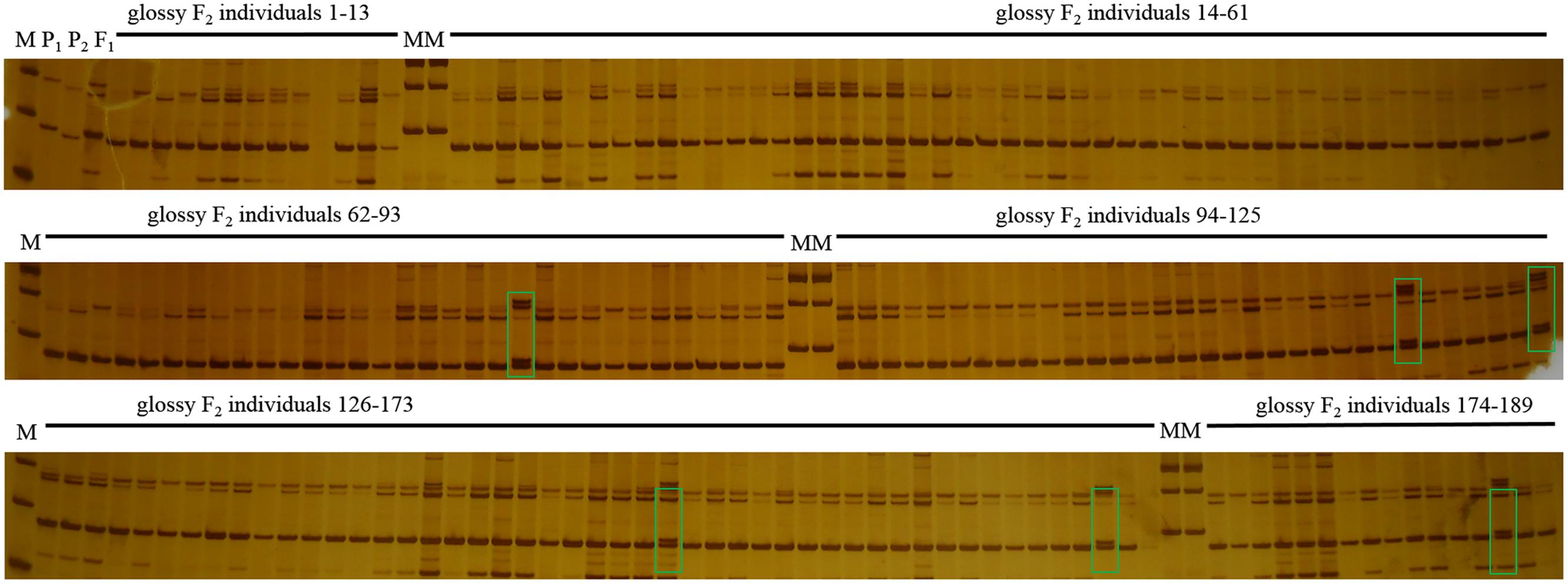
FIGURE 3. Polymorphisms of LTSSR740 in parents, F1, and 189 individuals of the F2 population. M, DNA ladder; P1, 10Q-206 with waxy phenotype; P2, 10Q-961 with glossy phenotype; F1, hybrid. Lanes 1–189, glossy F2 individuals. Green boxes represent recombinant plants.
Fine-Mapping of the Cgl1 Locus and Candidate Gene Analysis
To identify marker loci closely linked to Cgl1, 108 SSR markers were designed nearby marker LTSSR740. Nine of these markers, namely C08SSR7, C08SSR19, C08SSR26, C08SSR46, C08SSR53, C08SSR54, C08SSR55, C08SSR56, and C08SSR61 (Table 1), were polymorphic between lines 10Q-961 and M-36. These markers were used to screen 1,172 F2 individuals with the glossy phenotype, 15, 12, 6, 2, 1, 1, 1, 1, 1 recombinant plants were detected, respectively. This results indicated that Cgl1 was located between C08SSR61 and the end of chromosome C08 (41,327,369–41,516,064, genetic and physical map distances of 0.085 cM and 188.7 kb, respectively). The order of the nine SSR markers in the genetic map was consistent with that of the physical map (Figure 4). According to ‘02-12’ genome reference1, 33 genes were located in this 188.7-kb region. Based on gene annotations of cabbage and alignments with A. thaliana, Bol018504 was revealed to be highly homologous to AtCER1 which encodes an aldehyde decarbonylase catalyzing the process of conversion from C30 aldehydes to C29 alkanes of wax synthesis pathway. Thus, we tentatively designated Bol018504 as the candidate gene for the Cgl1 locus.
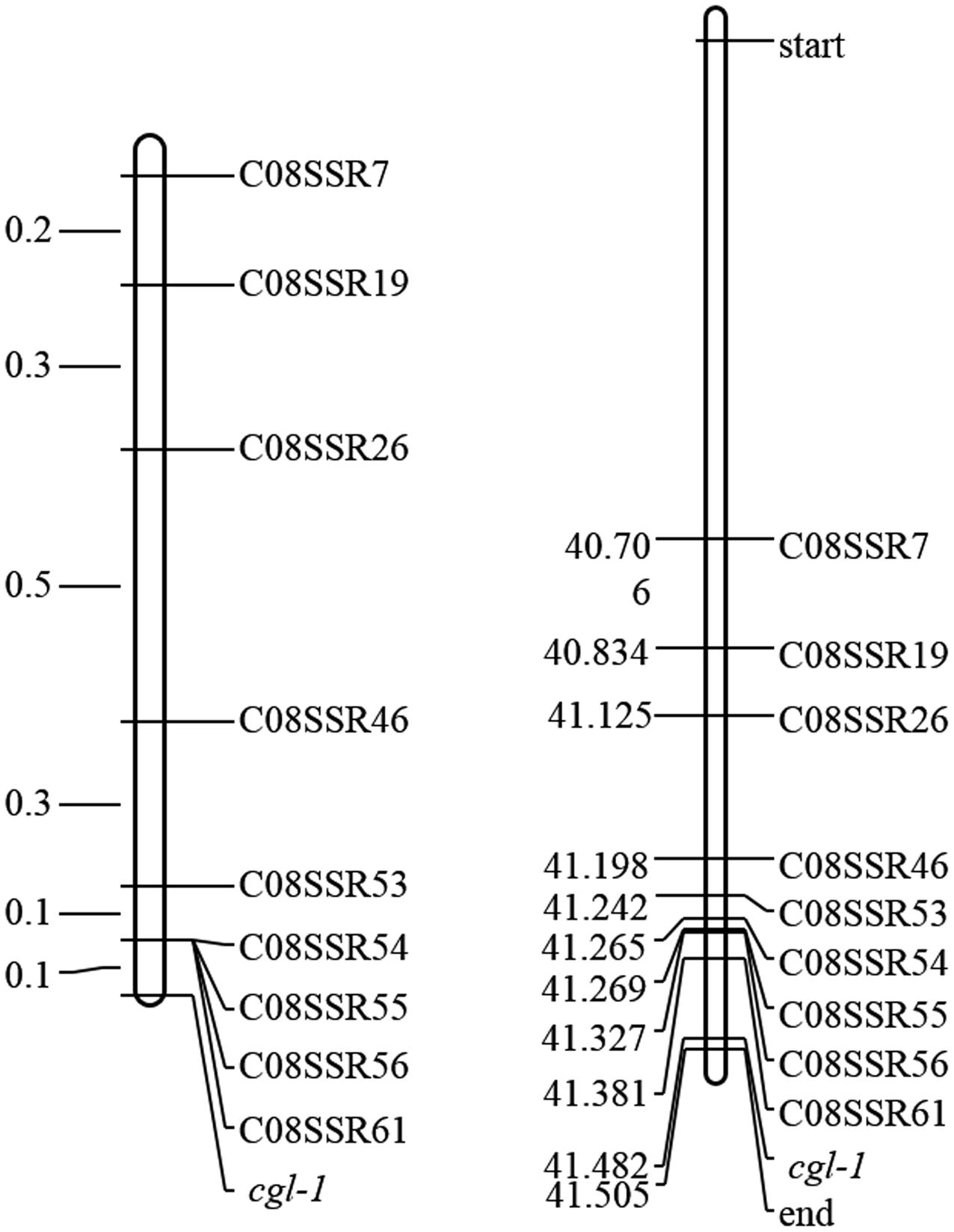
FIGURE 4. Genetic map of Cgl1 (left, unit: cM) and the corresponding physical map (right, unit: Mb).
Verification and Expression Level of the Bol018504 Candidate Gene
Five primer pairs (A–E) were designed spanning the full length of Bol018504 and its putative promoter region (Tables 1 and 2). There were no differences between the 10Q-961 and 10Q-962 amplification products generated from primer pairs A, C, D, and E. While primer pair B produced an approximately 1400-bp amplicon from the 10Q-962 template that was not amplified using the 10Q-961 template (Figure 5A). To avoid the possibility that primer pair B would not produce any amplicons from the 10Q-961 template, the full length Bol018504 gene and its promoter region were divided into two fragments (Table 2). Two primer pairs (P504 and 504) were used to amplify DNA templates from mutant and wild-type plants. Polymorphisms were detected only when using primer pair P504 (Figure 5B). The sequencing of the PCR products of the two primer pairs revealed that the primer pair 504 amplicons were identical between lines 10Q-961 and 10Q-962, while the primer pair P504 amplicon in the glossy mutant had a 2,722-bp insertion that was absent in the wild-type amplification product. To eliminate the possibility that the elongation time (1.0 min) was insufficient for primer pair B amplification of 10Q-961 DNA, it was increased to 2.5 min. The resulting approximately 4000-bp amplification product for line 10Q-961 and approximately 1400-bp product for line 10Q-962 (Figure 5C), and further confirmed the presence of a 2,722-bp insertion in line 10Q-961 by Sanger sequencing.
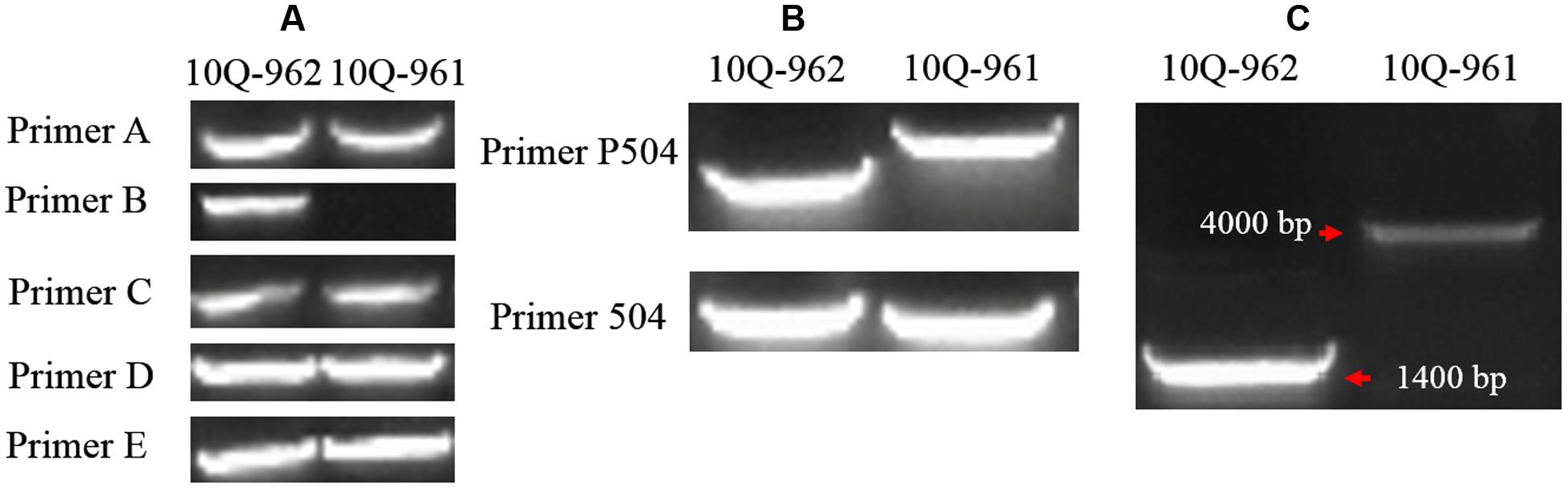
FIGURE 5. Amplification products of each primer pair using templates from lines 10Q-962 and 10Q-961. (A) Products of primer pairs A–E, (B) products of primer pairs P504 and 504, (C) products of primer pair B after the PCR extension time was increased.
According to the FGENESH 2.6 prediction, the full-length Bol018504 sequence in the wild-type line 10Q-962 (CGL1), comprising of 10 exons and nine introns (Figure 6A) was 3,351 bp, with coding sequence of 1,887 bp. While in the mutant line 10Q-961, Cgl1consisted of only nine exons and eight introns (Figure 6B), with ORF of 3,071 bp and coding sequence of 1,797 bp. The first exon of CGL1 was absent in Cgl1 and the positions of the second and third CGL1 exons differed in Cgl1, indicating the 2,722-bp insertion changed the RNA splice site. Using the National Center for Biotechnology Information BLAST tools, alignment analysis of the predicted Cgl1 amino acid sequence detected a missing PLN02869 domain, which controls fatty aldehyde decarbonylase activity. Thus, disruption of Cgl1 may result in inhibited conversion of aldehydes to alkanes and cause the glossy phenotype in mutant cabbage lines.
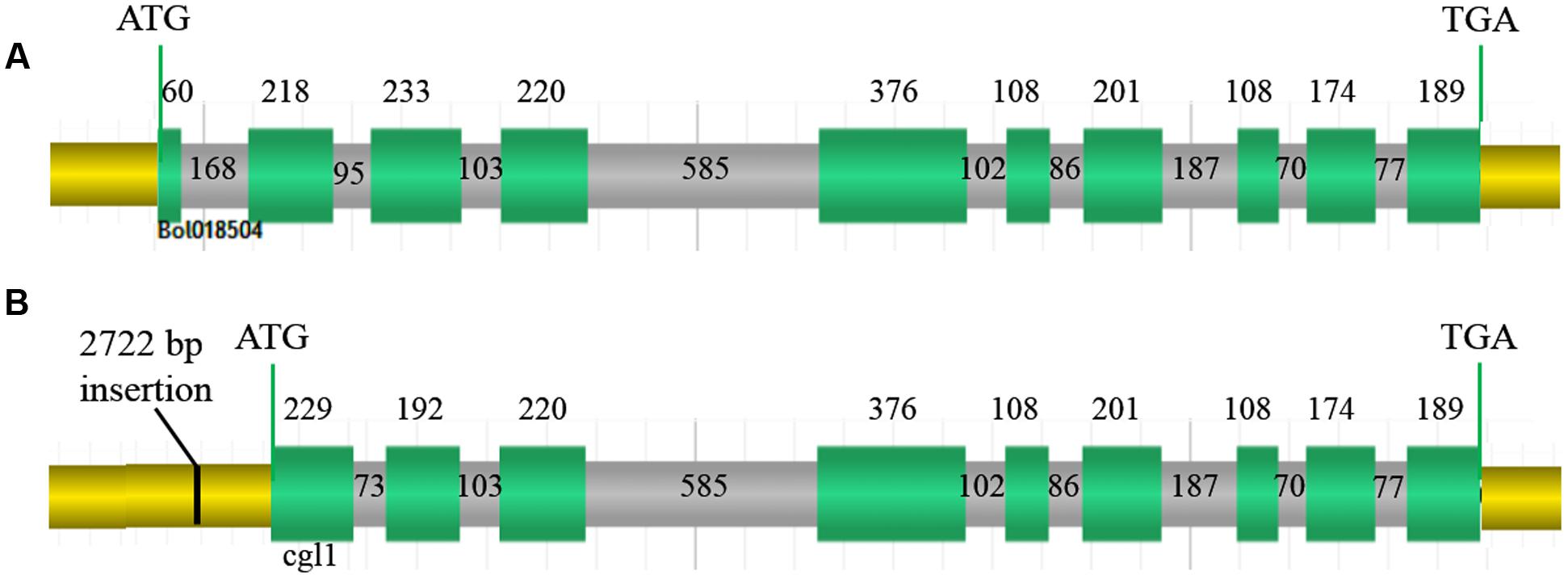
FIGURE 6. Genomic organization. (A) Genomic organization of Bol018504 in wild-type line 10Q-962, (B) genomic organization of Cgl1 in mutant line 10Q-961. Green sections, exons encoding amino acids; gray sections, introns; yellow sections, 5′- and 3′-untranslated regions; black line, 2,722-bp insertion. Numbers represent the lengths of each region in base pairs.
The expression level of Bol018504 between 10Q-961 and 10Q-962 were measured to determine whether the 2,722-bp insertion affected the expression of Bol018504. The expression levels of Bol018504 in leaf of the two materials at three-leaf stage seedlings were measured (Figure 7), and expression of the candidate gene was about 57.1 times higher in wild type 10Q-962 compared to mutant type10Q-961.
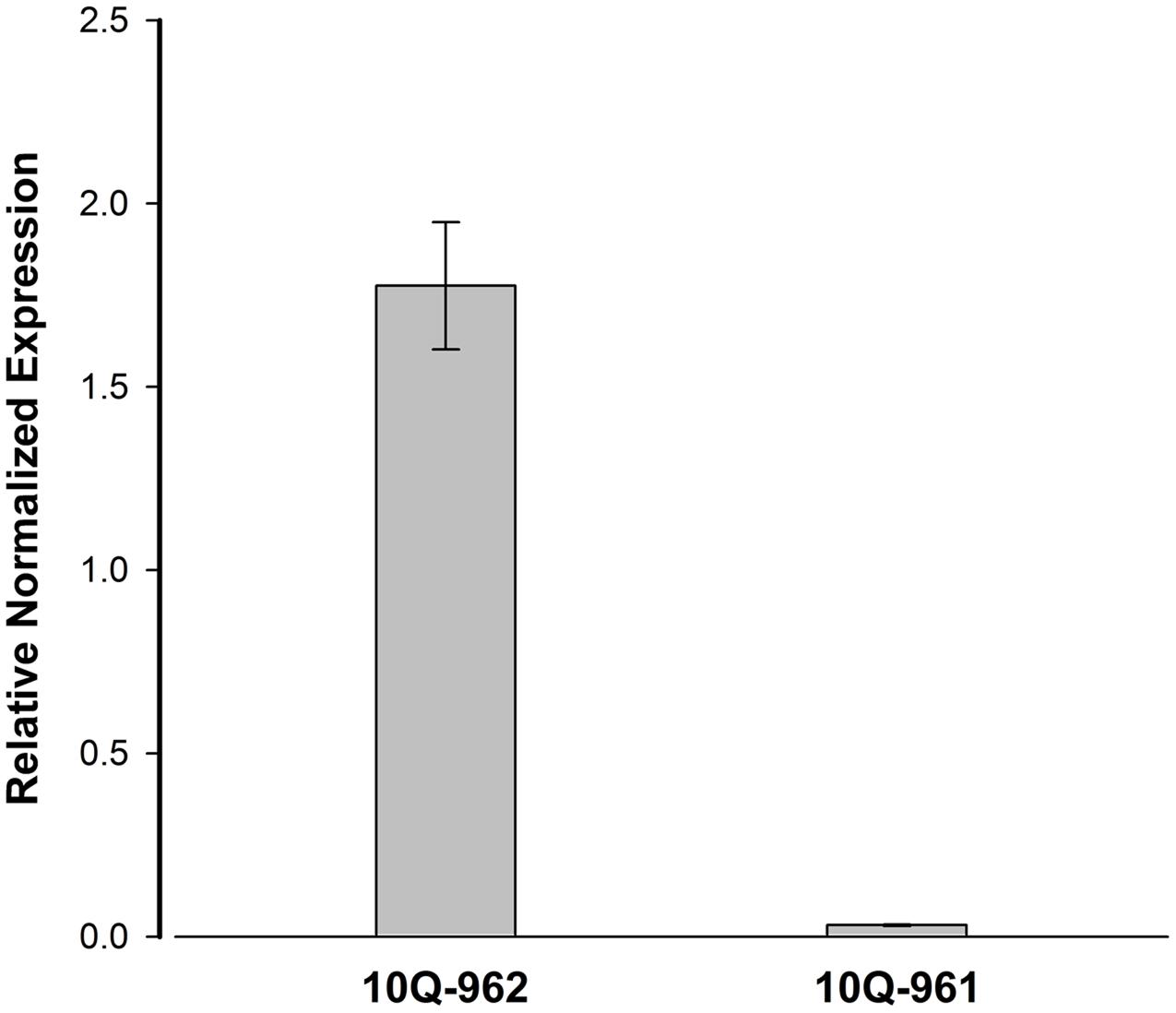
FIGURE 7. Real-time PCR analysis for the expression patterns of Bol018504 in mutant 10Q-961 and wild-type 10Q-962. Data represent means and standard errors of three replicates.
The 2,722-bp Insertion is Completely Linked with Mutant Phenotype in Recombinant Plants
Based on the insertion, a molecular marker, ISP1 (Table 1) which could amplified an approximately 3,300-bp and 600-bp amplification product from line 10Q-961 and 10Q-962 respectively was developed to analyze the recombinant plants. We found that the 2,722-bp insertion was completely linked with the mutant phenotype in the F2 mapping population (Figure 8). To determine whether this insertion was conserved in different varieties, a dominant glossy mutant and its wild type, another recessive glossy mutant and its wild type and six cabbage inbred lines available in our laboratory were tested. Agarose gel electrophoresis suggested that ISP1 marker produced approximately 600-bp amplicon in these 10 cabbage materials (Figure 8). This result indicated that the 2,722-bp insertion was conserved in glossy mutant 10Q-961, and that the ISP1 marker can distinguish the glossy mutant 10Q-961 from the other cabbage.
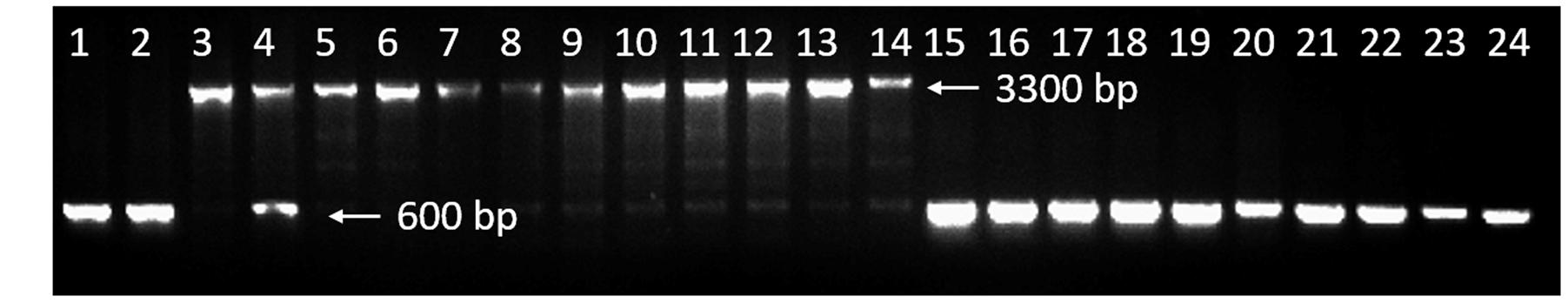
FIGURE 8. Validation of ISP1 marker. Lane 1, wild-type 10Q-962; lane 2, Chinese kale doubled haploid line, M-36; lane 3, mutant 10Q-961; lane 4, 10Q-961 × M-36 F1 hybrid; lanes 5–14, 10 recombinant plants in F2 mapping population; lanes 15–16, a dominant glossy mutant and its wild type; lanes 17–18, another recessive glossy mutant and its wild type; lanes 19–24, cabbage inbred lines 96–100, 87–534, xiafeng, mingde, Ikama and 8282.
Discussion
Our previous study revealed that the glossy trait in line 10Q-961 was controlled by a single recessive gene (Li et al., 2012). This result was also confirmed in this study. In the current report, 1,361 recessive glossy individuals from two different F2 populations were used to map the Cgl1 gene. Using a standard molecular genetic mapping strategy, Cgl1 was localized to a 188.7-kb region between the C08SSR61 SSR marker and the end of chromosome C08. Sequence and gene annotation analyses revealed that the Bol018504 candidate gene was highly homologous to AtCER1, which encodes an enzyme that catalyzes the presumed decarbonylation of aldehydes to alkanes (Aarts et al., 1995).
Our previous study involving scanning electron microscopy and gas chromatography-mass spectrometry analyses reported a decreased abundance of cuticular waxes in mutant line 10Q-961, with only 30.57% of the total wax content of the wild-type line 10Q-962; additional studies concluded that the reduced cuticular wax content in the mutant lines was mainly due to a decreased amount of C29 alkane; the lower C29 alkane levels were accompanied by a slightly increased C30 aldehyde content (Tang et al., 2015). These results were similar to those for the A. thaliana cer1-1 mutant (Bourdenx et al., 2011). In this study, the disruption of Bol018504 in mutant line 10Q-961 was the likely cause of its glossy phenotype. These results suggest that the cabbage CGL1 gene has the similar function as its A. thaliana homolog CER1.
According to comparative genome mapping, a triplication process occurred during cabbage evolution. Thus, an A. thaliana gene fragment may correspond to approximately three homologous copies in Brassica species (Cheng et al., 2014; Liu et al., 2014). There are two AtCER1 homologs in cabbage, namely Bol018504 and Bol035365. The Bol035365 fragment is an incomplete copy, with no sequence differences between mutant line 10Q-961 and wild-type line 10Q-962 (data not shown). Whether this gene is essential for CER1 protein function in cabbage is unclear. In this study, a 2,722-bp insertion in the first intron of Bol018504 may be responsible for the glossy mutant phenotype.
Brassica oleracea encompasses multiple cultivar groups that are classified based on the specialized morphology of their edible structures, namely kales (B. oleracea var. sabellica), cabbages (B. oleracea var. capitata f, alba), broccoli (B. oleracea var. italica), cauliflower (B. oleracea var. botrytis), brussels sprouts (B. oleracea var. gemmifera), kohlrabi (B. oleracea var. gongylodes), collard greens (B. oleracea var. viridis), savoy cabbage (B. oleracea var. capitata f, sabauda), romanesco broccoli (B. oleracea var. botrytis), broccolini (B. oleracea var. italica × alboglabra), and kai-lan (B. oleracea var. alboglabra). Brassica research has been enhanced through the publication of the B. rapa genome sequence and B. napus genome sequence (Wang et al., 2011; Chalhoub et al., 2014). Homolog sequences of this 2,722-bp insertion have been identified in Chinese cabbage (B. rapa; A09: 31,590,811–31,593,425) and canola (B. napus; C08: 38,256,925–38,259,552), but only the last 719 bp of the insertion is present in the cabbage genome (B. oleracea L. var. capitata; C02: 16,412,989–16,413,707). However, a 2,681-bp homolog sequences (C02: 7,222,204–7,224,885) of this 2,722-bp insertion could been identified in a doubled haploid B. oleracea kale-like type TO1000DH genome (Parkin et al., 2014). The current research has proved the existence of this 2,722-bp sequences in the wild-type 10Q-962, low sequence alignment against cabbage genome may be caused by sequencing or splicing error in cabbage genome assembly. Thus, further perfection of the cabbage genome sequences is supposed to be conducted.
A molecular marker, ISP1 which could distinguish the glossy mutant 10Q-961 from the other cabbage was developed. This molecular marker could be used for marker assistance selection in cabbage breeding and accelerate the breeding process of bright green cabbage. Additionally, few seeds were produced after self-pollination of 10Q-961 mutant line, which is consistent with observations for the cer1 A. thaliana mutant (Aarts et al., 1995). Future studies will be completed to determine whether Cgl1 influences fertility of cabbage.
To the best of our knowledge, CGL1 is the first wax synthesis gene mapped in B. oleracea. This achievement is an important advance for molecular research on wax synthesis in cabbage. Future functional studies on this gene will contribute to a more comprehensive understanding of plant cuticular wax metabolic networks.
Author Contributions
ZLiu developed the F2 populations, performed the experiments, analyzed the data, wrote and revised the manuscript. ZLiu, DL, and JT isolated the samples. LY, ZF, YL, MZ, YZ, ZZ, PS, HL, and ZLi conceived of the study and critically reviewed the manuscript. ZLiu and JT analyzed the sequencing data and designed the SSR primers. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was financially supported by grants from the National Natural Science Foundation of China (31672155), the Key Projects in the National Science & Technology Pillar Program during the Twelfth Five-Year Plan Period (2012BAD02B01), the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-IVFCAAS), and the Modern Agro-Industry Technology Research System (CARS-25-B-01), the National High Technology Research and Development Program of China (863 Program, 2012AA100101), and the Project of Science and Technology Commission of Beijing Municipality (Z141105002314020-1).
The work reported here was performed in the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, Beijing 100081, China.
Footnotes
References
Aarts, M. G., Keijzer, C. J., Stiekema, W. J., and Pereira, A. (1995). Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7, 2115–2127. doi: 10.1105/tpc.7.12.2115
Anstey, T. H., and Moore, J. F. (1954). Inheritance of glossy foliage and cream petals in green sprouting broccoli. J. Hered. 45, 39–41.
Barthlott, W., Neinhuis, C., Cutler, D., Ditsch, F., Meusel, I., Theisen, I., et al. (1998). Classification and terminology of plant epicuticular waxes. Bot. J. Linn. Soc. 126, 237–260. doi: 10.1111/j.1095-8339.1998.tb02529.x
Bassam, B. J., Caetano-Anollés, G., and Gresshoff, P. M. (1991). Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 196, 80–83. doi: 10.1016/0003-2697(91)90120-I
Bernard, A., and Joubès, J. (2013). Arabidopsis cuticular waxes: advances in synthesis, export and regulation. Prog. Lipid Res. 52, 110–129. doi: 10.1016/j.plipres.2012.10.002
Bourdenx, B., Bernard, A., Domergue, F., Pascal, S., Léger, A., Roby, D., et al. (2011). Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 156, 29–45. doi: 10.1104/pp.111.172320
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chantret, N., Sourdille, P., Röder, M., Tavaud, M., Bernard, M., and Doussinault, G. (2000). Location and mapping of the powdery mildew resistance gene MlRE and detection of a resistance QTL by bulked segregant analysis (BSA) with microsatellites in wheat. Theor. Appl. Genet. 100, 1217–1224. doi: 10.1007/s001220051427
Cheng, F., Wu, J., and Wang, X. (2014). Genome triplication drove the diversification of Brassica plants. Hortic. Res. 1:14024. doi: 10.1038/hortres.2014.24
Dellaert, L. M. W., Van Es, J. Y. P., and Koornneef, M. (1979). Eceriferum mutants in Arabidopsis thaliana (L.) Heynh. II. Phenotypic and genetic analysis. Arabid. Inf. Serv. 16, 10–26.
Dubcovsky, J., Echaide, M., Giancola, S., Rousset, M., Luo, M. C., Joppa, L. R., et al. (1997). Seed-storage-protein loci in RFLP maps of diploid, tetraploid, and hexaploid wheat. Theor. Appl. Genet. 95, 1169–1180. doi: 10.1007/s001220050678
Farnham, M. W. (2010). Glossy and nonglossy near-isogenic lines USVL115-GL, USVL115-NG, USVL188-GL, and USVL188-NG of broccoli. HortScience 45, 660–662.
Gan, L., Wang, X., Cheng, Z., Liu, L., Wang, J., Zhang, Z., et al. (2016). Wax crystal-sparse leaf 3 encoding a β-ketoacyl-CoA reductase is involved in cuticular wax biosynthesis in rice. Plant Cell Rep. 35, 1687–1698. doi: 10.1007/s00299-016-1983-1
Islam, M. A., Du, H., Ning, J., Ye, H., and Xiong, L. (2009). Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol. Biol. 70, 443–456. doi: 10.1007/s11103-009-9483-0
Jianguo, M., Wanqu, L., and Jisheng, W. (1992). Inheritance and agronomic performance of the waxless character in Brassica napus L. Plant Breed. 108, 256–259. doi: 10.1111/j.1439-0523.1992.tb00127.x
Jianguo, M., Wanqu, L., Qin, Y., and Bodnaryk, R. P. (1995). Inheritance of the waxless character of Brassica napus Nilla glossy. Can. J. Plant Sci. 75, 893–894. doi: 10.4141/cjps95-148
Kerstiens, G. (1996). Cuticular water permeability and its physiological significance. J. Exp. Bot. 47, 1813–1832. doi: 10.1093/jxb/47.12.1813
Koch, K., Bhushan, B., and Barthlott, W. (2009). Multifunctional surface structures of plants: an inspiration for biomimetics. Prog. Mater. Sci. 54, 137–178. doi: 10.1016/j.pmatsci.2008.07.003
Koch, K., and Ensikat, H. J. (2008). The hydrophobic coatings of plant surfaces: epicuticular wax crystals and their morphologies, crystallinity and molecular self-assembly. Micron 39, 759–772. doi: 10.1016/j.micron.2007.11.010
Kosambi, D. D. (1943). The estimation of map distances from recombination values. Ann. Eugen. 12, 172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x
Kunst, L., and Samuels, A. L. (2003). Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 42, 51–80. doi: 10.1016/S0163-7827(02)00045-0
Lee, S. B., and Suh, M. C. (2013). Recent advances in cuticular wax biosynthesis and its regulation in Arabidopsis. Mol. Plant 6, 246–249. doi: 10.1093/mp/sss159
Li, J. T., Yang, L. M., Fang, Z. Y., Liu, Y. M., Zhuang, M., Zhang, Y. Y., et al. (2012). First exploration on genetic law of glossy wax-less characteristics on cabbage (Brassica oleracea L. var. capitata L.) material 10Q-961. China Veg. 1, 37–41.
Liu, R. H., and Meng, J. L. (2003). [MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data]. Yi Chuan 25, 317–321.
Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I. A., et al. (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5:3930. doi: 10.1038/ncomms4930
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, P., Qin, J., Wang, G., Wang, L., Wang, Z., Wu, Q., et al. (2015). Comparative fine mapping of the Wax 1 (W1) locus in hexaploid wheat. Theor. Appl. Genet. 128, 1595–1603. doi: 10.1007/s00122-015-2534-9
Mao, B., Cheng, Z., Lei, C., Xu, F., Gao, S., Ren, Y., et al. (2012). Wax crystal-sparse leaf2, a rice homologue of WAX2/GL1, is involved in synthesis of leaf cuticular wax. Planta 235, 39–52. doi: 10.1007/s00425-011-1481-1
Mariani, M., and Wolters-Arts, M. (2000). Complex waxes. Plant Cell 12, 1795–1798. doi: 10.1105/tpc.12.10.1795
Millar, A. A., Clemens, S., Zachgo, S., Giblin, E. M., Taylor, D. C., and Kunst, L. (1999). CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11, 825–838. doi: 10.1105/tpc.11.5.825
Murai, J., Taira, T., and Ohta, D. (1999). Isolation and characterization of the three Waxy genes encoding the granule-bound starch synthase in hexaploid wheat. Gene 234, 71–79. doi: 10.1016/S0378-1119(99)00178-X
North, C., and Priestley, W. G. (1962). A glossy-leaved mutant of Brussels sprout. Hort. Res. 1, 95–99.
Parkin, I. A., Koh, C., Tang, H., Robinson, S. J., Kagale, S., Clarke, W. E., et al. (2014). Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol. 15:R77. doi: 10.1186/gb-2014-15-6-r77
Priestley, W. G., and Wills, A. B. (1966). Glossy-leaf in collard (Brassica oleracea L. var. Acephala DC): an incomplete dominant. Euphytica 15, 389–394. doi: 10.1007/BF00022185
Pu, Y., Gao, J., Guo, Y., Liu, T., Zhu, L., Xu, P., et al. (2013). A novel dominant glossy mutation causes suppression of wax biosynthesis pathway and deficiency of cuticular wax in Brassica napus. BMC Plant Biol. 13:215. doi: 10.1186/1471-2229-13-215
Qin, B. X., Tang, D., Huang, J., Li, M., Wu, X. R., Lu, L. L., et al. (2011). Rice OsGL1-1 is involved in leaf cuticular wax and cuticle membrane. Mol. Plant 4, 985–995. doi: 10.1093/mp/ssr028
Richardson, A., Boscari, A., Schreiber, L., Kerstiens, G., Jarvis, M., Herzyk, P., et al. (2007). Cloning and expression analysis of candidate genes involved in wax deposition along the growing barley (Hordeum vulgare) leaf. Planta 226, 1459–1473. doi: 10.1007/s00425-007-0585-0
Samuels, L., Kunst, L., and Jetter, R. (2008). Sealing plant surfaces: cuticular wax formation by epidermal cells. Plant Biol. 59, 683–707. doi: 10.1146/annurev.arplant.59.103006.093219
Sturaro, M., Hartings, H., Schmelzer, E., Velasco, R., Salamini, F., and Motto, M. (2005). Cloning and characterization of GLOSSY1, a maize gene involved in cuticle membrane and wax production. Plant Physiol. 138, 478–489. doi: 10.1104/pp.104.058164
Tacke, E., Korfhage, C., Michel, D., Maddaloni, M., Motto, M., Lanzini, S., et al. (1995). Transposon tagging of the maize Glossy2 locus with the transposable element En/Spm. Plant J. 8, 907–917. doi: 10.1046/j.1365-313X.1995.8060907.x
Tang, J., Liu, D.-M., Liu, Z.-Z., Yang, L.-M., Fang, Z.-Y., and Liu, Y.-M. (2015). Studies on characteristics of several glossy mutants in Cabbage. Acta Hortic. Sin. 42, 1093–1102.
Tel-Zur, N., Abbo, S., Myslabodski, D., and Mizrahi, Y. (1999). Modified CTAB procedure for DNA isolation from epiphytic cacti of the genera Hylocereus and Selenicereus (Cactaceae). Plant Mol. Biol. Rep. 17, 249–254. doi: 10.1023/A:1007656315275
Thompson, K. F. (1972). Cytoplasmic male-sterility in oil-seed rape. Heredity 29, 253–257. doi: 10.1038/hdy.1972.89
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Xu, X., Dietrich, C. R., Lessire, R., Nikolau, B. J., and Schnable, P. S. (2002). The endoplasmic reticulum-associated maize GL8 protein is a component of the acyl-coenzyme a elongase involved in the production of cuticular waxes. Plant Physiol. 128, 924–934. doi: 10.1104/pp.010621
Zhang, X., Liu, Z., Wang, P., Wang, Q., Yang, S., and Feng, H. (2013). Fine mapping of BrWax1, a gene controlling cuticular wax biosynthesis in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Mol. Breed. 32, 867–874. doi: 10.1007/s11032-013-9914-0
Keywords: cabbage, glossy mutant, fine-map, Cgl1, breeding
Citation: Liu Z, Fang Z, Zhuang M, Zhang Y, Lv H, Liu Y, Li Z, Sun P, Tang J, Liu D, Zhang Z and Yang L (2017) Fine-Mapping and Analysis of Cgl1, a Gene Conferring Glossy Trait in Cabbage (Brassica oleracea L. var. capitata). Front. Plant Sci. 8:239. doi: 10.3389/fpls.2017.00239
Received: 11 August 2016; Accepted: 08 February 2017;
Published: 20 February 2017.
Edited by:
Jun Yu, Beijing Institute of Genomics, ChinaReviewed by:
Chengdao Li, Murdoch University, AustraliaYong Xu, National Engineering Research Center for Vegetables, China
Copyright © 2017 Liu, Fang, Zhuang, Zhang, Lv, Liu, Li, Sun, Tang, Liu, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Limei Yang, yanglimei@caas.cn Zhenxian Zhang, zhangzx@cau.edu.cn
 Zezhou Liu
Zezhou Liu Zhiyuan Fang1
Zhiyuan Fang1 Honghao Lv
Honghao Lv Yumei Liu
Yumei Liu Zhansheng Li
Zhansheng Li Zhenxian Zhang
Zhenxian Zhang