- 1Key Laboratory of Agro-ecological Processes in Subtropical Region, Institute of Subtropical Agriculture, Chinese Academy of Sciences, Changsha, China
- 2Zhengzhou Tobacco Research Institute of China National Tobacco Corporation, Zhengzhou, China
- 3Postdoctoral Research Stations of Basic Medical Science, Zhengzhou University, Zhengzhou, China
- 4State Key Laboratory for Rice Biology, Biotechnology Institute, Zhejiang University, Hangzhou, China
Drought is a major environmental factor that limits plant growth and crop productivity. Genetic engineering is an effective approach to improve drought tolerance in various crops, including rice (Oryza sativa). Functional characterization of relevant genes is a prerequisite when identifying candidates for such improvements. We investigated OsSGL (Oryza sativa Stress tolerance and Grain Length), a novel DUF1645 domain-containing protein from rice. OsSGL was up-regulated by multiple stresses and localized to the nucleus. Transgenic plants over-expressing or hetero-expressing OsSGL conferred significantly improved drought tolerance in transgenic rice and Arabidopsis thaliana, respectively. The overexpressing plants accumulated higher levels of proline and soluble sugars but lower malondialdehyde (MDA) contents under osmotic stress. Our RNA-sequencing data demonstrated that several stress-responsive genes were significantly altered in transgenic rice plants. We unexpectedly observed that those overexpressing rice plants also had extensive root systems, perhaps due to the altered transcript levels of auxin- and cytokinin-associated genes. These results suggest that the mechanism by which OsSGL confers enhanced drought tolerance is due to the modulated expression of stress-responsive genes, higher accumulations of osmolytes, and enlarged root systems.
Introduction
Rice (Oryza sativa) is the most important food sources for more than half of the world’s population. However, water is already a scarce resource in many parts of the world, and increasingly frequent drought events present enormous challenges for sustainable rice production (Zhang, 2007). Drought is a major environmental factor that adversely affects plant growth and development, thus limiting agricultural productivity. Under stress conditions, plants can initiate a range of alterations at the molecular, cellular, and physiological levels, including stomata closure, reduced photosynthesis, higher accumulations of osmolytes, and the induction of many stress-responsive genes (Shinozaki and Yamaguchi-Shinozaki, 2007). Genetic engineering is considered an alternative for enhancing stress tolerance, and has greatly contributed to agronomic trait modifications in crop species. Many genes encoding functional proteins, transcription factors, and proteins involved in signaling pathways have been identified as abiotic stress-responsive genes (Shinozaki and Yamaguchi-Shinozaki, 2007; Masclaux-Daubresse et al., 2010; Turan et al., 2012). Tolerant plants have been engineered through heterologous expression of genes that encode functional or regulatory proteins (Golldack et al., 2011; Kim M. et al., 2013; Lu et al., 2013; Li et al., 2014).
Root morphology is one of the most promising traits for improving stress tolerance. An ideal root architecture and extensive root system can maximize water capture and allow access to moisture at the most suitable soil depth, thereby supporting shoot growth under drought conditions (Asch et al., 2005; Lynch, 2013). Genotypes with deeper roots generally have greater yields than shallow-rooted ones during drought periods (Ho et al., 2005; Hund et al., 2009; Lopes and Reynolds, 2010; Henry et al., 2011) and deeper rooting is more prevalent among species that grow in water-deficient environments (Schenk and Jackson, 2005). Some rice varieties with well-developed root systems have an advantage in grain yields under stress conditions (Price et al., 1997; Serraj et al., 2009). This improved yield potential can translate into better performance under stress scenarios but can also place a greater demand on water and other resources. However, root-specific expression of OsNAC5 and OsNAC9 in rice allows for the production of transgenic plants with an altered root architecture and enhanced drought tolerance without any yield penalty (Redillas et al., 2012; Jeong et al., 2013). Nelson et al. (2007) identified a transcription factor in Arabidopsis, AtNF-YB1, which confers drought tolerance. Transgenic maize (Zea mays) that constitutively expresses the orthologous gene ZmNF-YB2 is drought-tolerant and has improved yields under water-limited conditions. In addition, overexpression of DEEPER ROOTING 1 in rice confers the capacity for drought avoidance by altering root architecture (Uga et al., 2013). Overexpression of that gene results in a drought-related accumulation of lignin in those roots, which modifies cell wall architecture and enhances growth under stress conditions (Yoshimura et al., 2008).
Despite the tremendous progress made in determining gene functions in rice, many genes have not yet been characterized at the biochemistry and biological function levels. In eukaryotes, Domains of Unknown Function (DUF) proteins are part of several gene families that encode functionally uncharacterized proteins. A naming scheme for DUFs was established when DUF1 and DUF2 were added into the SMART database in the 1990s (Schultz et al., 1998). Those proteins have been categorized and enumerated as DUFX, where X represents the order of addition to the Protein-family (Pfam) database (Bateman et al., 2010). That database now contains over 3000 DUF families, and members within a particular family share conserved domains in their coding region (Bateman et al., 2010; Punta et al., 2012). Different DUF families play various roles in plant development and stress responses. Arabidopsis ESK1, a member of the DUF231 domain proteins, is a novel negative regulator of cold acclimation (Xin et al., 2007) while two other members, TBR and TBL3, are involved in cellulose synthesis and deposition of the secondary cell wall (Bischoff et al., 2010). The DUF640 domain-like gene TH1 is mainly expressed in young inflorescences of rice as well as in the spikelet lemmas and paleas, where it regulates their development (Li et al., 2012). Two Arabidopsis DUF1117 E3 ubiquitin ligase genes, AtRDUF1 and AtRDUF2, are induced by drought and ABA, and both single- and double-knockout mutants exhibit reduced tolerance to ABA-mediated drought stress (Kim et al., 2012). These studies indicate that DUF proteins play important roles in plant growth and stress adaptations. However, little is known about the molecular mechanisms by which those proteins confer drought tolerance.
To decipher the relevant genes in rice, we analyzed the genome expression profiles of rice Pei’ai 64S (Oryza sativa L.) under different stresses by rice microarray (unpublished data). One such gene, OsSGL, on Chromosome 2, encodes a putative DUF1645 domain-containing protein and lacks introns. Members of DUF1645 (PF07816) are derived from a number of hypothetical plant proteins. However, the gene family has not been characterized and no member has been functionally studied. Therefore, we investigated the role of OsSGL in the drought response. Transgenic rice plants over-expressing OsSGL showed enhanced tolerance during the seedling and vegetative stages, with altered transcript levels of stress-responsive genes, an enlarged root system, and higher osmolyte content. In addition, transgenic Arabidopsis that hetero-expressed this gene were more tolerant to osmotic stress.
Materials and Methods
Plant Materials and Growth Conditions
Rice seeds (O. sativa L. ssp. Indica cv 9311) were surface-sterilized with 75% ethanol for 1–2 min, followed by 50% sodium hypochlorite for 25 min and washed with distilled water at least three times. For rice root measurement, sterilized seeds were germinated on ½12-strength Murashige and Skoog (MS) (Murashige and Skoog, 1962) for 7 days at 28°C under the photoperiod of a 12 h light and 12 h dark photoperiod. Arabidopsis (A. thaliana ecotype Columbia (Col-0) was used as wild-type (WT) in this study. Seeds were sterilized with 75% ethanol and 10% bleach (v/v), followed by rinsing with sterile water for at least three times, and then vernalized for 2 days at 4°C. Plants were grown on ½12 MS medium containing 0.8% (w/v) sucrose and 0.75% (w/v) agar in a growth chamber maintained at 22°C and 60% relative humidity under the photoperiod of a 16 h light and 8 h darkness photoperiod.
Plasmid Construction and Transformation
The coding region of OsSGL was amplified from total RNA of rice Pei’ ai 64S, using an RT-PCR system (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The verified PCR fragment was digested and ligated into binary expression vector p1300-pJITl63, which was derived from pCAMBIA1300 and pJITl63, carrying the constitutive cauliflower mosaic virus (CaMV) 35S promoter. This yielded overexpression vector p1300-pJITl63-OsSGL. Thus, expression of OsSGL was controlled by the CaMV 35S promoter. All enzymes and reagents used for PCR or restriction digestion were purchased from Takara (Japan). Primer pairs used are listed in Supplementary Table S1.
For Arabidopsis transformation, binary construct p1300-pJITl63-OsSGL was introduced into Agrobacterium tumefaciens strain GV3101. Transformation was performed with the floral-dipping method (Clough and Bent, 1998). Transformants (T1) were selected on a ½12 MS medium supplemented with 40 mg L-1 hygromycin B. The T3 lines displaying 100% hygromycin resistance were considered homozygous and used for further experiments.
For rice transformation, plasmid p1300-pJITl63-OsSGL was introduced into A. tumefaciens strain EHA105, and embryogenic calli induced from rice cultivar 9311 (O. sativa L. ssp. Indica) which is widely used as a male parent in the production of high yield rice in china were used as the transformation receptor. Our method followed that described by Xiao et al. (2008). Hygromycin resistance was used to screen positive transgenic plants. Using primers specific for the hygromycin phosphotransferase gene, we performed PCR to confirm the presence of the T-DNA in the transformants. The expression level of OsSGL in positive transgenic plants was determined by semi-quantitative reverse-transcription PCR (RT-PCR) for Arabidopsis and quantitative real-time PCR (qPCR) for rice. Tubulin and Actin2 were used as the internal control in Arabidopsis and rice, respectively. All primers are listed in Supplementary Table S1. We used T3 homozygous plants in subsequent experiments.
Quantitative Real-Time PCR and Semi-quantitative RT-PCR
Total RNAs were extracted with TRIzol reagent (Invitrogen, Burlington, ON, Canada) according to the manufacturer’s instructions. Afterward, 1 μg of DNase-treated RNA was reverse-transcribed using a PrimeScriptTM 1st Strand cDNA synthesis kit (Takara) according to the manufacturer’s protocol. The qPCR analysis was conducted with FastStart Universal SYBR Green Master (Roche), and reactions were performed in an ABI 7900HT (Applied Biosystems) at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 58°C for 30 s. The internal controls were Tubulin and Actin2 for Arabidopsis and rice, respectively. Each sample was analyzed in triplicate and relative amounts of mRNA were calculated per the comparative threshold cycle method.
For semi-quantitative RT-PCR, two-step RT-PCR method was chosen. Total RNA extractions and cDNA synthesis were performed as described above. All PCR amplifications were conducted in a total volume of 20 μL under the following conditions: 22–27 cycles of denaturation (94°C, 30 s), annealing (58°C, 35 s), and extension (72°C, 30 s). The primer pairs for semi-quantitative RT-PCR and qPCR are listed in Supplementary Table S1.
Subcellular Localization
To determine the subcellular localization of OsSGL in rice protoplasts, we amplified the full-length open reading frame (ORF) of OsSGL without the termination codon. After verification by sequencing, the PCR fragment was cloned into vector pJITL63-GFP to produce an OsSGL-GFP fusion construct (CaMV35S-OsSGL-GFP), thus allowing the fusion gene to be driven by the CaMV 35S promoter. Construct CaMV35S-GFP was used as the control. Transformation mediated by polyethylene glycol (PEG) was performed as described previously (Zhang et al., 2011). After 6–18 h of incubation in the dark, the subcellular location of the recombinant proteins was observed with a confocal laser scanning microscope (Leica TCS 5 STED CW SP5).
Drought and Osmotic Treatment
For the analysis of osmotic tolerance in Arabidopsis, sterilized seeds of WT and OsSGL transgenic plants were sown in triplicate on ½12 MS medium containing different concentrations of mannitol (0 or 300 mM). After vernalization for 2 days at 4°C in the darkness, plates were transferred to a growth room (22°C, 16 h light/8 h dark) and kept for another 9 days. At the end of the treatment, survival rates were determined. Plants with green cotyledons were designated as survivors. For analyses of the effects of osmotic stress on root length, following vernalization, seeds of WT and transgenic plants were germinated on ½12 MS medium for 3 days. Plants with identical growth performance were transferred to ½12 MS medium containing 300 mM mannitol, and grown for another 12 days. Plates were placed vertically to facilitate comparison of root growth.
To determine drought tolerance in transgenic rice, T3 homozygous seeds were used for further analysis. Drought assays were performed in a controlled growth chamber PGC15.5 (Percival, Perry, IA, USA). For osmotic stress at the post-germination stage, sterilized seeds were sown on ½12 MS medium containing 0 or 400 mM mannitol for 11 d. Root and shoot were measured at the end of the treatments. For drought assays at seedling stage, WT and transgenic lines were planted in the same buckets. At the four-leaf-stage, watering was withheld and resumed 5 days later. After 3-week recovery, survival rates were determined.
Morphological Analysis of Roots
To determine the root morphological parameters at seedling stage, seeds were germinated on ½12 MS medium and kept growing for 7 days in a growth room. Three replicates were performed. Lengths of primary, adventitious and lateral roots were measured. For the determination of adventitious root, the five longest adventitious roots on each seedling were counted. Similar to measurement of adventitious root length, lateral root length was determined with the 15 longest lateral roots on each primary root. For root comparison at the vegetative and reproductive stages, plants of WT and transgenic lines were planted in the same big bucket. After 6 or 12 weeks, roots were rinsed clean by running water, root lengths and dry weights were measured.
Determining the Contents of Proline, Soluble Sugars, and Malondialdehyde (MDA)
Four-week-old rice seedlings were used for biochemical analysis. Both WT and transgenic plants were transferred from the basal nutrient solution to a nutrient solution supplemented with 20% PEG. After 2 days of treatment, proline and soluble sugar contents in harvested tissue samples were measured according to the sulphosalicylic acid method (Troll and Lindsley, 1955) and the anthrone method (Morris, 1948), respectively. The level of MDA was determined with thiobarbituric acid, as described by Kramer et al. (1991).
Gene Expression Analysis by RNA-sequencing
RNA samples were sent to the Beijing Genomics Institute (BGI) for further RNA-seq analysis. Briefly, total RNA was isolated with TRIzol reagent from the aerial parts of 15-day-old transgenic and WT 9311 rice plants that treated with 20% PEG (w/v) for 6 h. Material from 10 plants of each genotype was pooled for RNA extraction, and sequencing was performed in an Illumina HiSeq 2000 sequencing platform (Beijing, China). Clean reads were mapped to the Rice Annotation Project Database (RAP-DB) using TopHat Version 2.0.9 (Kim D. et al., 2013). The abundance of mapped reads was normalized to RPKM (reads per kilobase of exon model per million mapped reads) (Mortazavi et al., 2008). The DEGseq package was used for identifying genes differentially expressed between sample pairings (i.e., WT vs. transgenic), and P-values were adjusted according to the Benjamini and Hochberg method. RNA-seq read data in this article has been deposited in the National Center for Biotechnology Information (NCBI) SRA database under the accession number SRP094588.
Statistical Analysis
All statistical analyses were performed with Student’s t-tests using SPSS software version 13.0.
Results
Sequence and Phylogenetic Analyses of OsSGL
The cDNA of OsSGL is 1062 bp long and contains an ORF of 768 bp flanked by a 5′-untranslated region (105 bp) and a 3′-untranslated region (189 bp). This gene encodes a novel protein of 255 amino acid residues with a molecular mass of 26.73 kDa. Bioinformatics analysis showed that OsSGL has a conserved domain of DUF1645 (Pfam PF07816) from amino acid residues 69-2271. To investigate the possible evolutionary relationships between OsSGL and other DUF1645 proteins, we constructed a phylogenetic tree via the Neighbor-Joining method, using full-length amino acid sequences (Figure 1). This protein is relatively closely related to its homologs in maize, Triticum aestivum, Setaria italica, and Sorghum bicolor. However, the relationship between OsSGL and other rice DUF1645 proteins is more divergent, suggesting that they play different roles in rice.
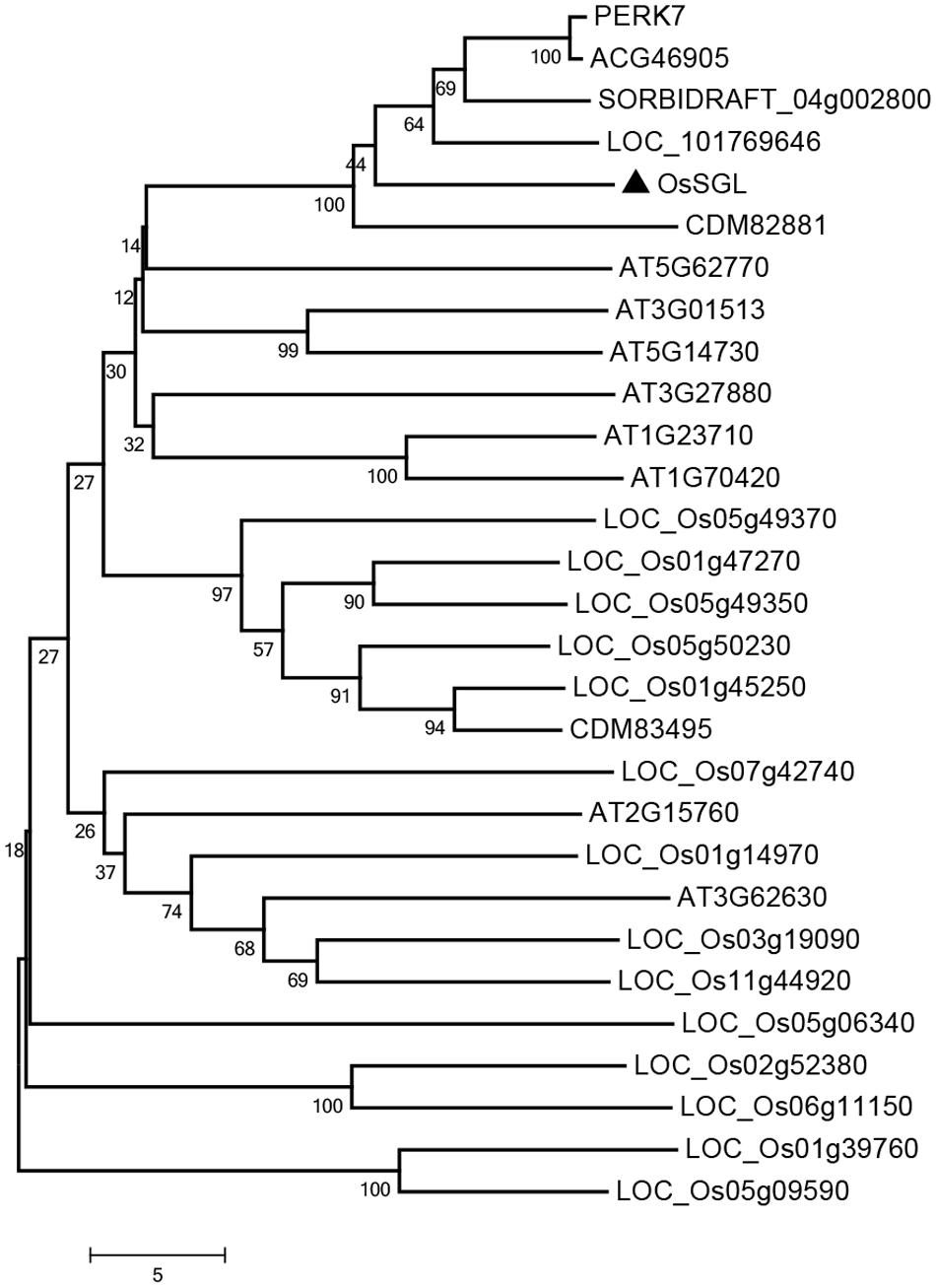
FIGURE 1. Phylogenetic relationship of OsSGL with other DUF1645 proteins. Multiple sequence alignment was performed via MEGA 6 and Neighbor-Joining method. Accession numbers include Oryza sativa: LOC_Os01g14970, LOC_Os01g39760, LOC_Os01g45250, LOC_Os01g47270, LOC_Os02g52380, LOC_Os03g19090, LOC_Os05g06340, LOC_Os05g09590, LOC_Os05g49350, LOC_Os05g49370, LOC_Os05g50230, LOC_Os06g11150, LOC_Os07g42740, and LOC_Os11g44920; Arabidopsis thaliana: AT1G23710, AT1G70420, AT2G15760, AT3G01513, AT3G27880, AT3G62630, AT5G14730, and AT5G62770; Triticum aestivum: CDM82881 and CDM83495; Zea mays: ACG46905 and PERK7; Setaria italica: LOC_101769646; and Sorghum bicolor: SORBIDRAFT_04g002800.
Expression Pattern of OsSGL and Its Subcellular Localization
To elucidate the physiological and functional relevance of OsSGL, we examined its expression profile in young roots, mature stems, panicles at the heading stage, mature leaf blades, and mature leaf sheaths. Our qPCR results revealed that this gene was constitutively expressed in almost all tissues examined, with expression being greatest in the roots and panicles (Figure 2A). We also performed qPCR analysis using 2-week-old rice seedlings exposed to osmotic, salt, or cold stress and found that expression was highly up-regulated by all three as well as by ABA treatment (Figure 2B).
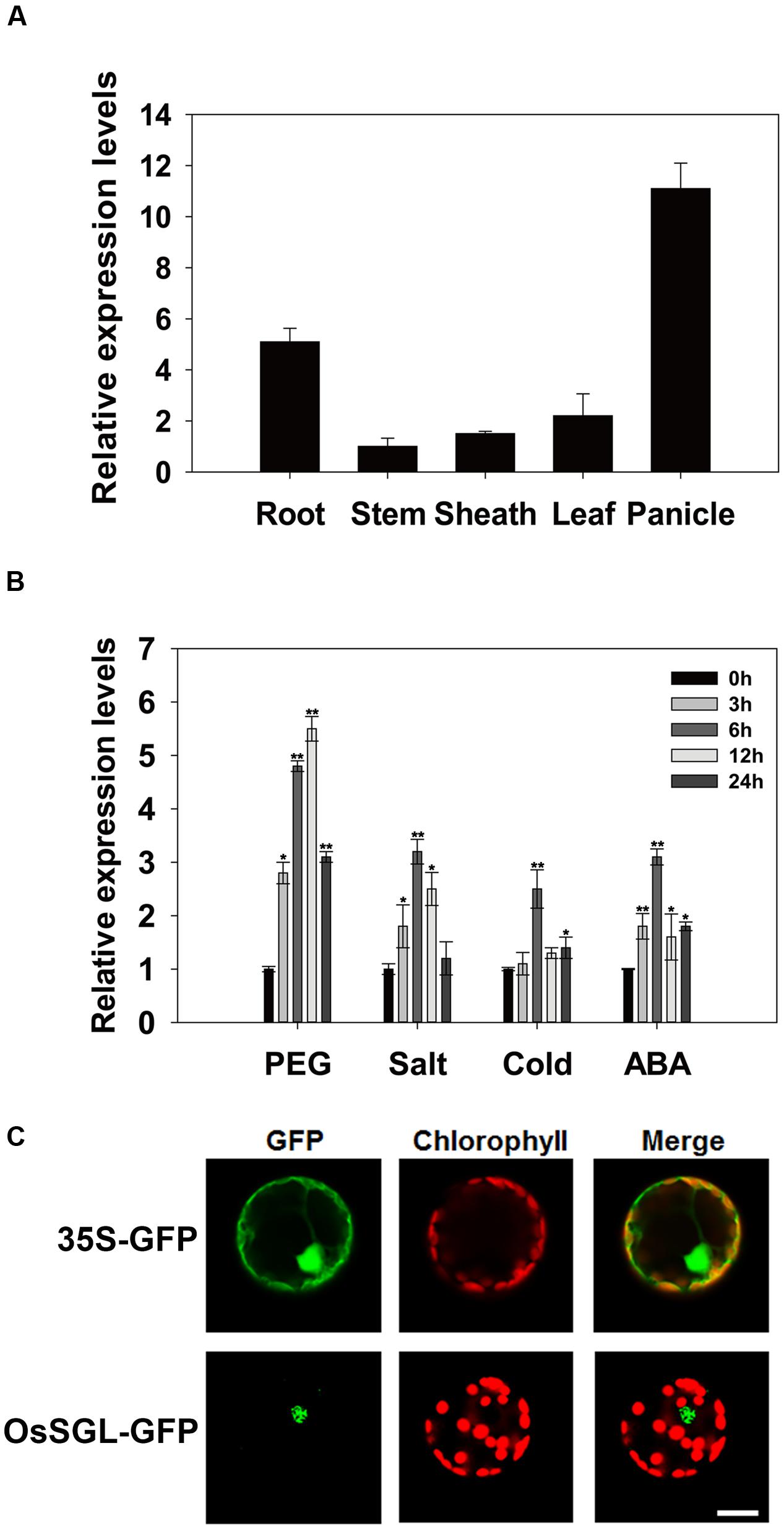
FIGURE 2. Expression pattern and subcellular localization of OsSGL. (A) Expression profile of OsSGL in organs at different developmental stages. (B) Relative expression of OsSGL under drought, salt, cold, or ABA stress. Materials were harvested at indicated time intervals for qPCR. Error bars represent SE for 3 independent experiments. (C) Subcellular localization of GFP control (upper panel) and OsSGL-GFP fusion protein (lower panel) in rice protoplast cells. CK, controls; Scale bars = 10 μm. ∗ and ∗∗ indicate significant difference from WT at P < 0.05 and < 0.01, respectively, by Student’s t-tests.
Subcellular localization of OsSGL was determined using a gene construct containing a DNA fragment that encodes OsSGL-GFP fusion protein driven by the CaMV35S promoter. Transient expression of OsSGL was analyzed by confocal laser scanning microscopy. As depicted in Figure 2C, cells expressing the control GFP gene showed cytoplasmic and nuclear distribution of the GFP signals. In contrast, cells expressing the OsSGL-GFP fusion gene showed GFP signal only in the nucleus, suggesting that OsSGL is a nucleus-localized protein.
Rice and Arabidopsis Plants Over- or Hetero-Expressing OsSGL Have Enhanced Drought or Osmotic Tolerance
To examine the biological function of OsSGL, we generated transgenic rice and Arabidopsis plants with a vector containing its full-length ORF, driven by the CaMV 35S promoter. As we had expected, this gene was variably expressed in the transgenic lines. For further phenotypic analysis, we selected homozygous T3 transgenic Arabidopsis lines B2 and B4 and rice lines L3 and L5, all of which demonstrated higher levels of expression (Supplementary Figures S1 and S2). Strong induction of OsSGL expression by stresses suggested that the gene might be involved in stress tolerance. No obvious difference was observed in shoots between transgenic rice lines and WT under normal condition, while transgenic rice lines exhibited longer shoots than the WT under osmotic condition at the post-germination stage. In contrast, roots from transgenic lines were longer than the WT 9311 roots under both normal and osmotic conditions (Figures 3A–C). To assess drought tolerance at the seedling stage, we withheld irrigation for a short period before returning to a normal watering regimen. By Day 5 of this simulated-drought treatment, all WT plants were severely affected whereas the transgenic lines showed less leaf rolling and wilting. Following the recovery period, the transgenic plants also grew more vigorously (Figure 3D). After recovery, only 37.9% of the WT plants still contained green tissues while an average of 78.7% of the transgenic rice had survived the drought stress (Figure 3E).
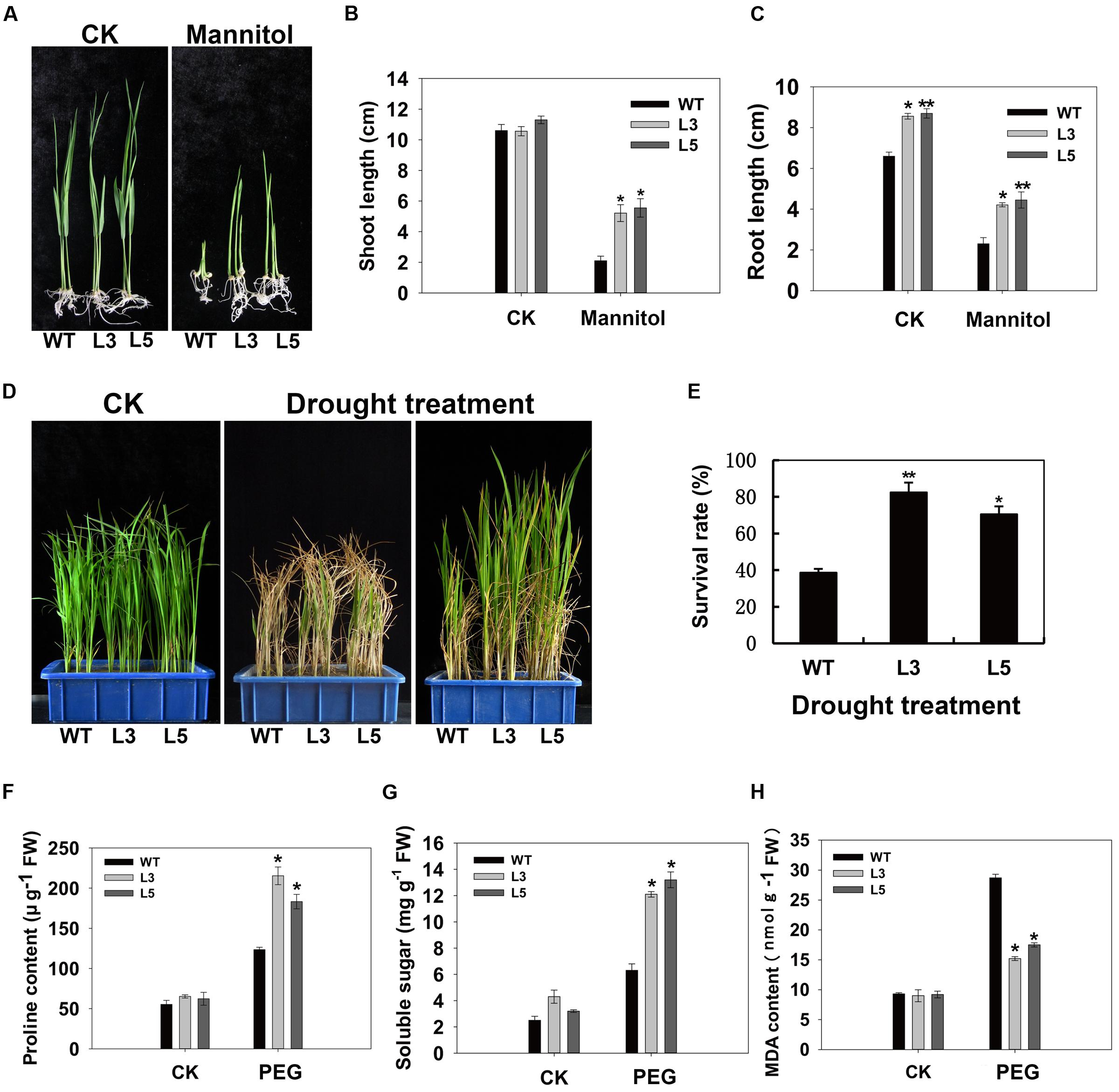
FIGURE 3. Osmotic and drought tolerances in OsSGL transgenic plants. (A) Phenotypic comparison of WT and transgenic plants under normal and osmotic stress conditions at post-germination stage. Shoot (B) and root (C) lengths were measured after 11 days of mannitol treatment. (D) OsSGL improves drought tolerance in transgenic rice. Water was withheld from four-leaf-stage seedlings (Left panel) and irrigation resumed 5 d later. After 3 weeks of recovery (right panel), survival rates were determined. Middle panel represents 1 week of recovery after re-watering. (E) Survival rates of wild type (WT) and OsSGL transgenic rice plants after drought treatment. (F) Comparison of the contents of proline in WT and OsSGL transgenic plants. (G) Measurement of soluble sugars in WT and OsSGL transgenic plants. (H) Comparison of MDA content in WT and OsSGL transgenic plants. For the measurement of proline, soluble sugars and MDA contents, 4-week-old WT and transgenic plants were treated with 20% PEG for 2 d before measuring. Data are means ± SE (n = 3). ∗ and ∗∗ indicate significant difference from WT at P < 0.05 and < 0.01, respectively, by Student’s t-tests. CK, controls; WT, 9311; L3 and L5, transgenic lines L3 and L5, respectively.
The possible physiological mechanism responsible for improved drought tolerance was investigated by noting changes in stress-relevant parameters, i.e., proline, soluble sugar, and MDA contents. Under normal growing conditions, proline and MDA contents did not differ significantly between WT and OsSGL transgenic rice plants, but the level of soluble sugars was slightly higher in the latter. Upon exposure to osmotic stress, proline and soluble sugar contents were significantly elevated in all genotypes, but those increases were much more evident in the transgenic lines than in the WT (Figures 3F,G). Moreover, the MDA contents detected in transgenic plants were significantly lower than that in the WT (Figure 3H).
Consistent with the results obtained with the transgenic rice, similar osmotic-tolerant phenotypes were observed in transgenic Arabidopsis plants hetero-expressing OsSGL. Under normal growing conditions, cotyledon opening and the rate of greening did not differ significantly among genotypes (data not shown). However, under osmotic conditions, the transgenic plants had higher survival rates and produced longer roots (Figures 4A–D). These results demonstrated that OsSGL plays a positive role in osmotic tolerance.
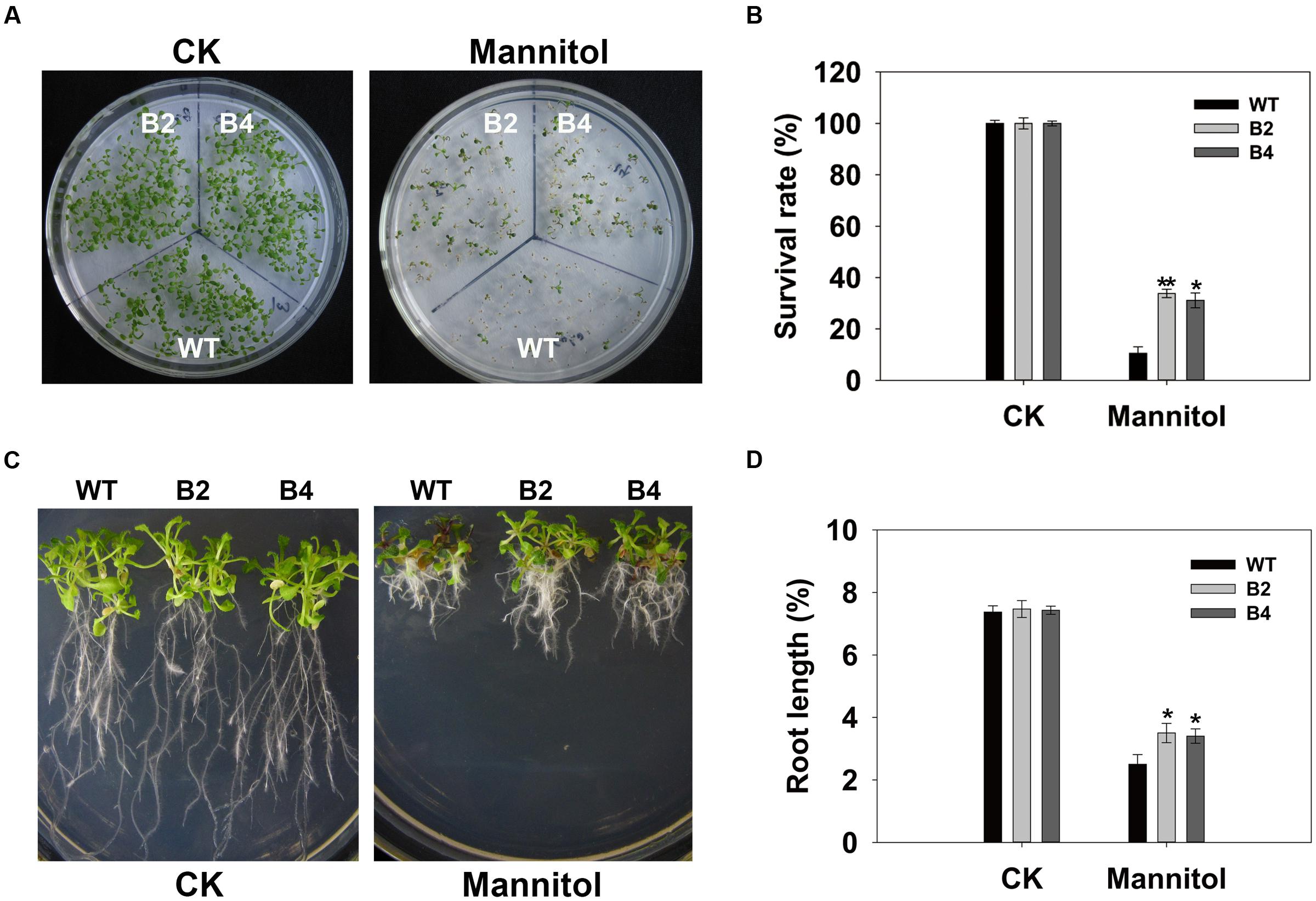
FIGURE 4. Overexpression of OsSGL enhances osmotic tolerance in transgenic Arabidopsis. (A) Performance of WT and transgenic lines under normal and osmotic stress conditions, respectively. (B) Survival rates after osmotic stress. Vernalized seeds of WT and OsSGL transgenic plants were sown in triplicate on ½12 MS medium or ½12 MS supplemented with 300 mM mannitol, then monitored for 9 days. Seedlings with green cotyledons were considered survivors. (C) After seeds were germinated on ½12 MS medium for 3 days, seedlings were transferred to medium containing 300 mM mannitol and grown vertically for another 12 days. (D) Root lengths of WT and transgenic plants after imposition of osmotic stress. Error bars represent SD for 3 independent experiments. ∗ and ∗∗ indicate significant difference from WT at P < 0.05 and <0.01, respectively, by Student’s t-test. CK, controls; WT, wild-type Arabidopsis; B2 and B4, transgenic Lines 2 and 4, respectively.
Up-Regulated Expression of Stress-Responsive Genes in OsSGL-Overexpressing Plants
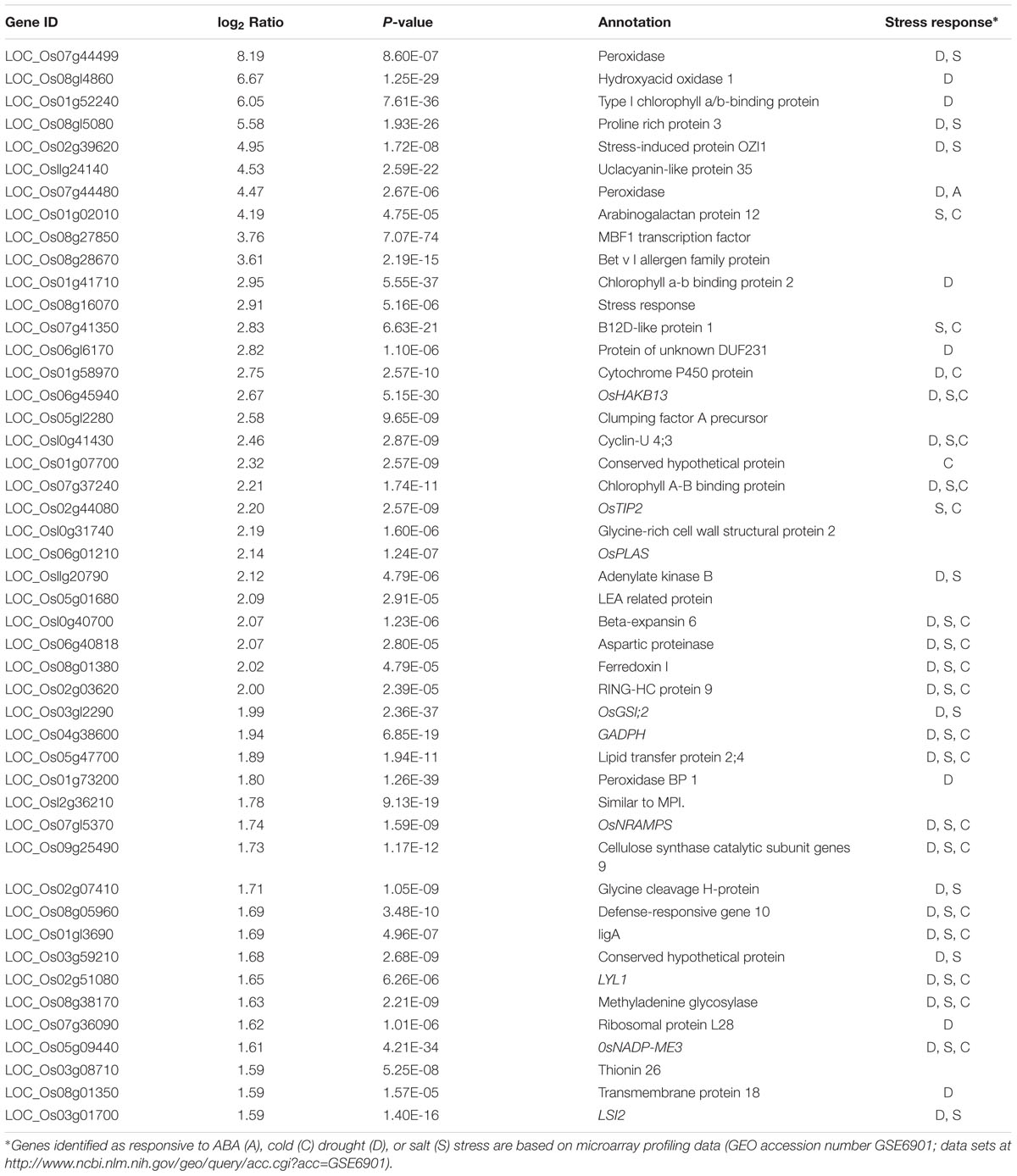
To gain further insights into the mechanism of the improved drought tolerance in OsSGL-transgenic plants, RNA-seq analysis was performed with RNA samples extracted from 15-day-old seedlings after osmotic stress. Among the 47 genes that were up-regulated in transgenic plants, 33 were drought-inducible, as had been revealed by previous microarray experiments (GEO accession number GSE6901). In all, 41 target genes showed changes in transcript levels of more than three-fold in OsSGL-transgenic plants when compared with the WT (P-value < 0.05). These genes encode both functional and regulatory proteins, including stress response proteins, antioxidants, protein kinases, transcription factors, and enzymes involved in metabolism (Table 1). Some reactive oxygen species (ROS)-responsive genes also showed higher expression in the transgenic line, including hydroxyacid oxidase (LOC_Os08g14860), peroxidase (LOC_Os07g44499, LOC_Os07g44480), peroxidase binding protein (LOC_Os01g73200), proline-rich protein (LOC_Os08g15080), and LEA-related protein (LOC_Os05g01680), suggesting that the improved drought tolerance in transgenic plants partially resulted from ROS pathways modulation. We then selected six stress-responsive genes and compared their expression between transgenic and WT plants grown under osmotic stress conditions. qPCR analysis showed an overall pattern of expression similar to that revealed by RNA-seq analysis, confirming the reliability of RNA-seq analysis (Supplementary Figure S3). These data indicated that overexpression of OsSGL in rice affects the expression of a set of stress-responsive genes, thereby conferring greater drought tolerance in transgenic plants.
OsSGL Alters Root Architecture and Transgenic Rice Plants Produce Deeper Root Systems
Overexpression of OsSGL in rice yielded altered phenotypes in root architecture. At the seedling stage, the roots of transgenic plants were approximately 50% longer than those of the WT. Root fresh weights were also significantly higher for those transgenic plants (Figures 5A,B, and Supplementary Table S2). To test whether the root enhancement was maintained during subsequent plant development stages, transgenic and WT plants were further cultivated in the same bucket. Figure 5C shows that transgenic plants still possessed enlarged root systems. The dry biomasses of the root systems of the two tested transgenic lines were 10.3 and 13.8% higher than that of WT, respectively (Figure 5D). In accordance with seedling and vegetative stages, transgenic plants also had larger root systems at the reproductive stage, displaying the increased root dry weight (Figures 5E,F).
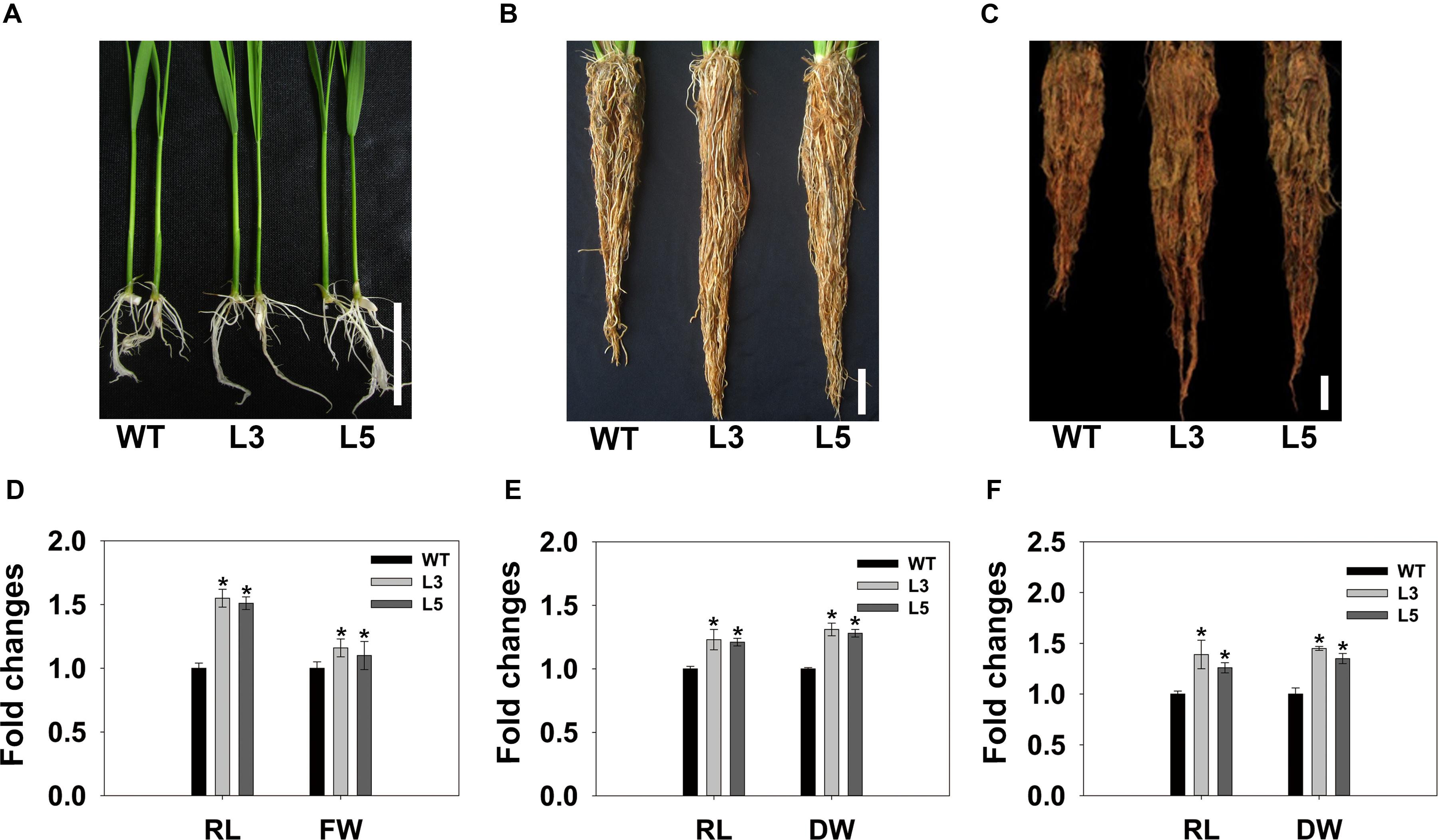
FIGURE 5. Comparisons of root growth between WT and transgenic rice plants at seedling, vegetative, and reproductive stages. (A) Seeds were germinated on ½12 MS medium and plants were grown for 7 days. (B,C) Transgenic and WT plants were grouped in individual buckets and grown for 6 and 12 weeks, respectively. (D–F) Lengths, fresh weights, or dry weights of transgenic plant roots were normalized to WT plants at seedling, vegetative, and reproductive stage, respectively. Scale bars = 5 cm. Data are means ± SD of 30 roots (10 per growth stage). ∗ and ∗∗ indicate significant difference from WT at P < 0.05 and <0.01, respectively, by Student’s t-tests. RL, root length; FW, fresh weight; DW, dry weight. WT, 9311; L3 and L5, transgenic lines L3 and L5, respectively.
OsSGL Functions in Regulating Expression of Genes Responsible for Root Development and Hormone Responses
The regulatory pathways for root initiation and growth involve CRL1, CRL4/GNOM1, DR01, CKX4, NAC5, NAC9, and WOX11 (Liu et al., 2005; Zhao et al., 2009; Redillas et al., 2012; Jeong et al., 2013; Uga et al., 2013; Gao et al., 2014). To determine whether OsSGL regulates these genes, we performed qPCR analysis with root samples from transgenic plants and found that CRL1, CRL4, DR01, CKX4, and NAC9 were highly expressed. This implied that OsSGL directly or indirectly influences the expression of those genes. In contrast, transcripts of NAC5 and WOX11 were not clearly affected in the transgenic lines (Figure 6A).
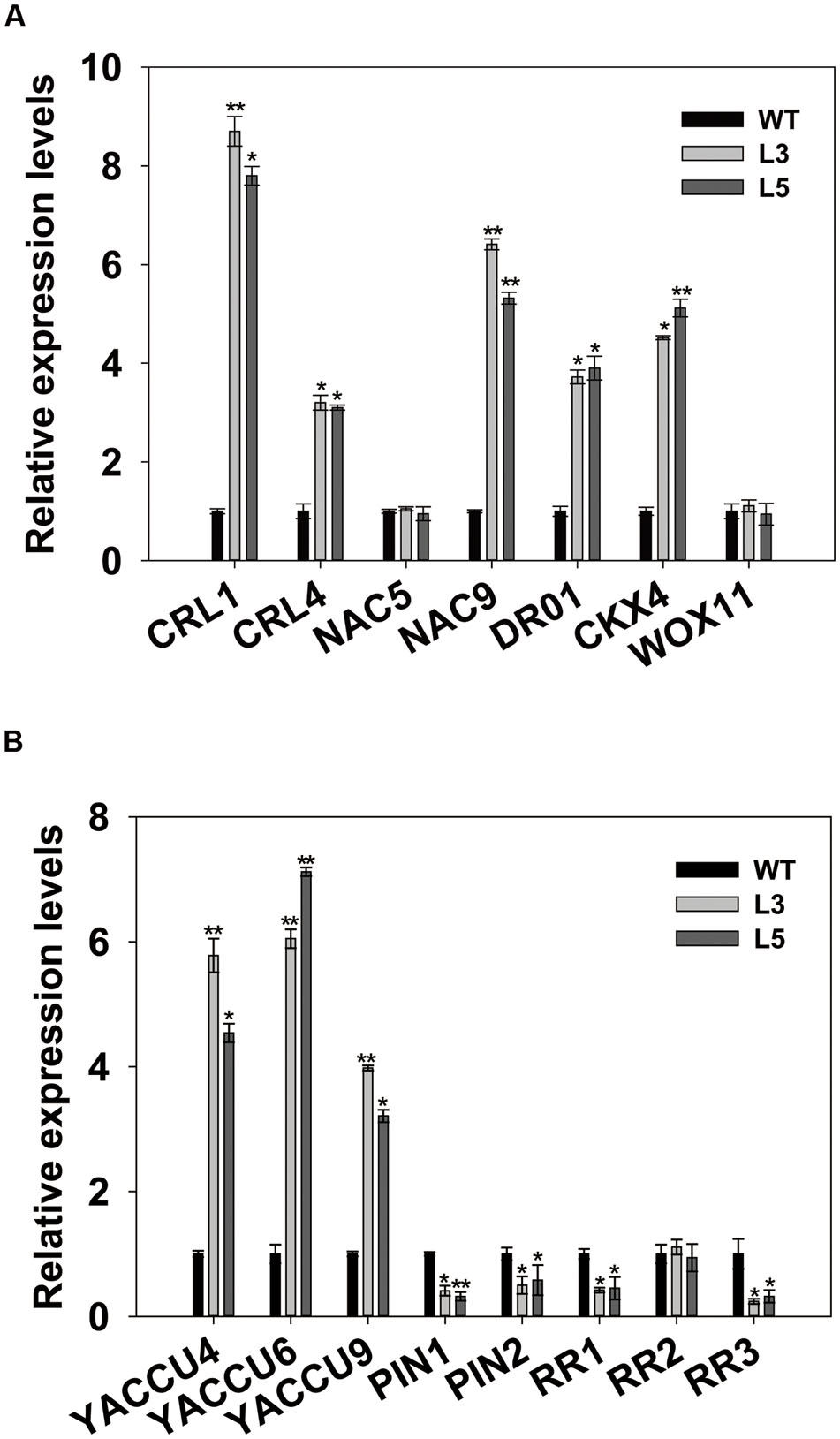
FIGURE 6. Expression analyses. (A) Relative expression levels of root growth-related genes in wild-type (WT) and OsSGL-transgenic plants. (B) Relative expression levels of auxin- and cytokinin-related genes in WT and transgenic plants. ∗ and ∗∗ indicate significant difference from WT at P < 0.05 and < 0.01, respectively, by Student’s t-tests.
To determine whether OsSGL is also involved in auxin and cytokinin signaling, we analyzed the expression of two auxin transport genes (PIN1 and PIN2), three auxin biosynthesis genes (YUCCA4, YUCCA6, YUCCA9), and three cytokinin-responsive Type-A RR genes (RR1-RR3) in roots from 14-day-old seedlings of WT transgenic rice. While transcript levels of YUCCA4, YUCCA6, and YUCCA9 were elevated in the OsSGL transgenic plants, expression of PIN1, PIN2, RR1, and RR3 was repressed, and that of RR2 was not changed (Figure 6B). These results suggested that OsSGL regulates genes for root development by modulating the signaling pathways of cytokinin and auxin.
Discussion
Drought stress, or water deficit, is a primary inhibitor of crop productivity and distribution (Lobell et al., 2014). Global climate change will likely make this situation more serious in the near future (Ahuja et al., 2010). Thus, it is extremely urgent that researchers dissect the functions and mechanisms underlying the activities of genes associated with drought tolerance if we are to develop new drought-tolerant crop varieties through molecular breeding. In this study, the rice gene OsSGL was analyzed and evaluated in transgenic rice and Arabidopsis. Its expression was induced significantly by osmotic stress, salt, cold, or ABA treatment. This indicated that the gene can respond to multiple environmental cues. To investigate the expression pattern of OsSGL orthologs in Arabidopsis, we performed a meta-analysis of publicly available microarray data in the Genevestigator database2 (Hruz et al., 2008). One DUF1645 member, AT1G23710, is induced by salt, cold, drought, ABA, and oxidative stresses (Supplementary Figure S4), which is consistent with the expression pattern we detected for OsSGL. Therefore, AT1G23710 and OsSGL may have similar functions in stress responses. The former has not been functionally characterized, and only a few studies have shown that AT1G23710 expression is altered during cold acclimation, pollen germination, and pollen tube growth (Fowler and Thomashow, 2002; Wang et al., 2008). In the further study, it is fascinating to investigate the possibility of AT1G23710 involved in the drought tolerance.
Our findings demonstrated that both over- and hetero-expression of OsSGL improves tolerance to drought in transgenic plants. Therefore, we can reasonably conclude that this enhanced tolerance might be a result of several beneficial changes at the morphological, physiological, and molecular levels. First, the OsSGL transgenic plants produce a more extensive root system that may contribute to greater stress tolerance at the morphological level. The size and architecture of a root system determine plant capacity to access water and nutrients, all factors that can limit growth and yield in many agricultural ecosystems. Therefore, optimization of root system architecture is positively correlated with drought tolerance (Price et al., 1997). Many transcription factors can help improve stress tolerance by altering this architecture (Jeong et al., 2010, 2013; Redillas et al., 2012). We found here that overexpression of OsSGL in rice plants led to larger root systems, as evidenced by longer roots and increased biomass. Similar results have been described for transgenic rice plants that over-express OsEXPA8 (Ma et al., 2013). It is reasonable to assume that longer roots and increased root biomasses may facilitate the absorption of water in deep soil layers, behavior that is considered a drought avoidance strategy (Fukai and Cooper, 1995; Yue et al., 2006). However, in contrast to rice, roots of OsSGL transgenic Arabidopsis plants were comparable with that of WT under normal conditions (Figure 4A). This was probably due to the different genetic backgrounds of monocot and dicot plants.
Phytohormones such as auxin and cytokinin act as necessary regulators of root architecture. Auxin signaling is required for the initiation of rice crown roots, while cytokinin inhibits root growth (Inukai et al., 2005). Auxin–cytokinin crosstalk signaling plays key roles in root development and can coordinately regulate a series of genes (Dello Ioio et al., 2008; Müller and Sheen, 2008). For example, OsCKX4, a putative cytokinin oxidase/dehydrogenase gene, mediates crown root development in rice by integrating the interaction between cytokinin and auxin (Gao et al., 2014). Utilizing ren1-D mutants of rice, molecular and genetic analyses have revealed that the mutant phenotype is caused by activation of OsCKX4. Thus, increased expression of CRL1 and CRL4 may be responsible for our observed enhanced-root formation phenotype (Figure 6A). Both auxin transport genes, OsPIN1 and OsPIN2, were down-regulated in OsSGL transgenic plants, consistent with the results obtained in ren1-D mutants (Figure 6B). These results were further verified by the fact that auxin biosynthesis genes YUCCA4, YUCCA6, YUCCA7, and YUCCA9 are repressed in the roots of ren1-D seedlings when compared with the WT. Overall, these findings suggest that both auxin distribution and auxin biosynthesis are regulated by cytokinin, which is consistent with conclusions from previous studies (Pernisová et al., 2009; Jones et al., 2010). Another auxin- and cytokinin-responsive gene, WOX11, is expressed primarily in regions of cell division in both root and shoot meristems (Zhao et al., 2009). In WOX11-overexpression lines, OsRR2, a Type-A cytokinin-responsive gene, is repressed. Expression of both auxin- and cytokinin-responsive genes is affected by the mutation or overexpression of WOX11. Those data suggest that WOX11 may integrate both auxin and cytokinin signaling to stimulate cell division during the process of crown root development. Our data also showed that overexpression of OsSGL either activates or represses some auxin- and cytokinin-responsive genes, which suggests that this gene influences auxin-cytokinin signaling to regulate root growth and development.
A second beneficial change due to OsSGL overexpression is the higher accumulation of osmoprotectants and lower MDA contents, which may contribute to the improved stress tolerance in our transgenic plants. Osmotic stress can be a common consequence of drought, high salt, or cold stresses (Apse and Blumwald, 2002; Yamaguchi-Shinozaki and Shinozaki, 2006). In response to osmotic conditions, plants accumulate more osmoprotectants, e.g., proline and soluble sugars, to maintain turgor pressure and to protect enzymes and macro molecules of cells against the damaging effects of ROS (Farooq et al., 2009; Kiani et al., 2007). Proline is an important compatible solute that accumulates in plants exposed to dehydration (Perez-Perez et al., 2007). High levels of free proline allow plants to maintain low water potentials and derive water from the environment. Our OsSGL transgenic plants also accumulated more proline and soluble sugars under stress conditions, and they also had less MDA than the WT under osmotic stress. As a product of ROS-stimulated lipid peroxidation, the measuring of MDA contents can be used to evaluate the extent of ROS-mediated injuries in plants (Moore and Roberts, 1998). The higher accumulation of proline in OsSGL transgenic plants might contribute somewhat to their lower MDA levels measured under stress conditions. Therefore, the significant increases in amounts of osmoprotectants in our plants may represent one of the main mechanisms underlying improved drought tolerance.
Thirdly, RNA-seq analysis showed a correlation between gene expression profiles and the stress tolerance phenotype observed with our transgenic rice plants. We found it is interesting that some peroxidases were highly expressed in those plants. Peroxidases are involved in many processes, including defense responses to biotic and abiotic stresses. The exposure of plants to unfavorable environmental conditions increases ROS production. Because peroxidases oxidize various substrates that utilize H2O2 or organic hydroperoxides, they can help in ROS-scavenging. In fact, several plant peroxidases have important roles in the ROS-detoxification process (Miller, 2002; Apel and Hirt, 2004). Two novel pepper peroxidase genes, CaPO2 and CaPOD, are involved in abiotic stress tolerance and pathogen resistance (Choi and Hwang, 2012; Wang et al., 2013). OsAPX2, a cytosolic ascorbate peroxidase in rice, is critical for growth and development because it protects seedlings from abiotic stresses through ROS-scavenging (Zhang et al., 2013). In addition to these peroxidases, we also found that some stress-responsive genes were up-regulated in our transgenic plants. Therefore, we might speculate that elevated expression of these anti-oxidative and stress-responsive genes is essential to the development of stress tolerance in OsSGL transgenic plants.
In summary, we have demonstrated that over- and hetero-expression of OsSGL in rice and Arabidopsis plants increases their level of drought tolerance. Some related traits were improved in those transgenic plants, including formation of a more extensive root system, altered transcript levels of stress-responsive genes, and higher accumulations of osmolytes. Additionally, a newly study from our group revealed that OsSGL also controlled the grain length in rice (Wang et al., 2016). These findings provide a good foundation for using OsSGL as a target gene for future crop improvement strategies.
Author Contributions
GX and XX designed the experiments; YC and MW performed the experiments with assistance from XY, GZ, ML, and LH; GX, HZ, and FL analyzed and discussed the results; YC and GX wrote the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grants No. 31401314, 31301253, 31201012, 31501393) and the China Postdoctoral Science Foundation (Grant No. 2015M580634).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.02001/full#supplementary-material
Footnotes
References
Ahuja, I., de Vos, R. C. H., Bones, A. M., and Hall, R. D. (2010). Plant molecular stress responses face climate change. Trends Plant Sci. 15, 664–674. doi: 10.1016/j.tplants.2010.08.002
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Apse, M. P., and Blumwald, E. (2002). Engineering salt tolerance in plants. Curr. Opin. Biotechnol. 13, 146–150. doi: 10.1016/S0958-1669(02)00298-7
Asch, F., Dingkuhn, M., Sow, A., and Audebert, A. (2005). Drought-induced changes in rooting patterns and assimilate partitioning between root and shoot in upland rice. Field Crops Res. 93, 223–236. doi: 10.1016/j.fcr.2004.10.002
Bateman, A., Coggill, P., and Finn, R. D. (2010). DUFs: families in search of function. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1148–1152. doi: 10.1107/S1744309110001685
Bischoff, V., Nita, S., Neumetzler, L., Schindelasch, D., Urbain, A., Eshed, R., et al. (2010). TRICHOME BIREFRINGENCE and its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Physiol. 153, 590–602. doi: 10.1104/pp.110.153320
Choi, H. W., and Hwang, B. K. (2012). The pepper extracellular peroxidase CaPO2 is required for salt, drought and oxidative stress tolerance as well as resistance to fungal pathogens. Planta 235, 1369–1382. doi: 10.1007/s00425-011-1580-z
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dello Ioio, R., Linhares, F. S., and Sabatini, S. (2008). Emerging role of cytokinin as a regulator of cellular differentiation. Curr. Opin. Plant Biol. 11, 23–27. doi: 10.1016/j.pbi.2007.10.006
Farooq, M., Wahid, A., Ito, O., Lee, D. J., and Siddique, K. H. M. (2009). Advances in drought resistance of rice. Crit. Rev. Plant Sci. 28, 199–217. doi: 10.1080/07352680902952173
Fowler, S., and Thomashow, M. F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. doi: 10.1105/tpc.003483
Fukai, S., and Cooper, M. (1995). Development of drought-resistant cultivars using physiomorphological traits in rice. Field Crops Res. 40, 67–86. doi: 10.1016/0378-4290(94)00096-U
Gao, S., Fang, J., Xu, F., Wang, W., Sun, X., Chu, J., et al. (2014). CYTOKININ OXIDASE/DEHYDROGENASE4 integrates cytokinin and auxin signaling to control rice crown root formation. Plant Physiol. 165, 1035–1046. doi: 10.1104/pp.114.238584
Golldack, D., Lüking, I., and Yang, O. (2011). Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 30, 1383–1391. doi: 10.1007/s00299-011-1068-0
Henry, A., Gowda, V. R. P., Torres, R. O., McNally, K. L., and Serraj, R. (2011). Variation in root system architecture and drought response in rice (Oryza sativa): phenotyping of the OryzaSNP panel in rainfed lowland fields. Field Crops Res. 120, 205–214. doi: 10.1016/j.fcr.2010.10.003
Ho, M. D., Rosas, J. C., Brown, K. M., and Lynch, J. P. (2005). Root architectural tradeoffs for water and phosphorus acquisition. Funct. Plant Biol. 32, 737–748. doi: 10.1071/FP05043
Hruz, T., Laule, O., Szabo, G., Wessendorp, F., Bleuler, S., Oertle, L., et al. (2008). Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics 2008:420747. doi: 10.1155/2008/420747
Hund, A., Ruta, N., and Liedgens, M. (2009). Rooting depth and water use efficiency of tropical maize inbred lines, differing in drought tolerance. Plant Soil 318, 311–325. doi: 10.1007/s11104-008-9843-6
Inukai, Y., Sakamoto, T., Ueguchi-Tanaka, M., Shibata, Y., Gomi, K., Umemura, I., et al. (2005). Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17, 1387–1396. doi: 10.1105/tpc.105.030981
Jeong, J., Kim, Y., Redillas, M., Jang, G., Jung, H., Bang, S., et al. (2013). OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 11, 101–114. doi: 10.1111/pbi.12011
Jeong, J. S., Kim, Y. S., Baek, K. H., Jung, H., Ha, S. H., Do Choi, Y., et al. (2010). Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153, 185–197. doi: 10.1104/pp.110.154773
Jones, B., Gunnerås, S. A., Petersson, S. V., Tarkowski, P., Graham, N., May, S., et al. (2010). Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell 22, 2956–2969. doi: 10.1105/tpc.110.074856
Kiani, S. P., Talia, P., Maury, P., Grieu, P., Heinz, R., Perrault, A., et al. (2007). Genetic analysis of plant water status and osmotic adjustment in recombinant inbred lines of sunflower under two water treatments. Plant Sci. 172, 773–787. doi: 10.1016/j.plantsci.2006.12.007
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. doi: 10.1186/gb-2013-14-4-r36
Kim, M., Sato, S., Sasaki, K., Saburi, W., Matsui, H., and Imai, R. (2013). COLD SHOCK DOMAIN PROTEIN 3 is involved in salt and drought stress tolerance in Arabidopsis. FEBS Open Biol. 3, 438–442. doi: 10.1016/j.fob.2013.10.003
Kim, S. J., Ryu, M. Y., and Kim, W. T. (2012). Suppression of Arabidopsis RINGDUF1117 E3 ubiquitin ligases, AtRDUF1 and AtRDUF2, reduces tolerance to ABA-mediated drought stress. Biochem. Biophys. Res. Commun. 420, 141–147. doi: 10.1016/j.bbrc.2012.02.131
Kramer, G. F., Norman, H. A., Krizek, D. T., and Mirecki, R. M. (1991). Influence of UV-B radiation on polyamines, lipid peroxidation and membrane lipids in cucumber. Phytochemistry 30, 2101–2108. doi: 10.1016/0031-9422(91)83595-C
Li, C., Chang, P., Ghebremariam, K. M., Qin, L., and Liang, Y. (2014). Overexpression of tomato SpMPK3 gene in Arabidopsis enhances the osmotic tolerance. Biochem. Biophys. Res. Commun. 443, 357–362. doi: 10.1016/j.bbrc.2013.11.061
Li, X., Sun, L., Tan, L., Liu, F., Zhu, Z., Fu, Y., et al. (2012). TH1, a DUF640 domain-like gene controls lemma and palea development in rice. Plant. Mol. Biol. 78, 351–359. doi: 10.1007/s11103-011-9868-8
Liu, H., Wang, S., Yu, X., Yu, J., He, X., Zhang, S., et al. (2005). ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 43, 47–56. doi: 10.1111/j.1365-313X.2005.02434.x
Lobell, D. B., Roberts, M. J., Schlenker, W., Braun, N., Little, B. B., Rejesus, R. M., et al. (2014). Greater sensitivity to drought accompanies maize yield increase in the U.S. Midwest. Science 344, 516–519. doi: 10.1126/science.1251423
Lopes, M. S., and Reynolds, M. P. (2010). Partitioning of assimilates to deeper roots is associated with cooler canopies and increased yield under drought in wheat. Funct. Plant Biol. 37, 147–156. doi: 10.1071/FP09121
Lu, Y., Li, Y., Zhang, J., Xiao, Y., Yue, Y., Duan, L., et al. (2013). Overexpression of Arabidopsis molybdenum cofactor sulfurase gene confers drought tolerance in maize (Zea mays L.). PLoS ONE 8:e52126. doi: 10.1371/journal.pone.0052126
Lynch, J. P. (2013). Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 112, 347–357. doi: 10.1093/aob/mcs293
Ma, N., Wang, Y., Qiu, S., Kang, Z., Che, S., Wang, G., et al. (2013). Overexpression of OsEXPA8, a root-specific gene, improves rice growth and root system architecture by facilitating cell extension. PLoS ONE 8:e75997. doi: 10.1371/journal.pone.0075997
Masclaux-Daubresse, C., Daniel-Vedele, F., Dechorgnat, J., Chardon, F., Gaufichon, L., and Suzuki, A. (2010). Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. 105, 1141–1157. doi: 10.1093/aob/mcq028
Miller, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. doi: 10.1016/S1360-1385(02)02312-9
Moore, K., and Roberts, L. J. (1998). Measurement of lipid peroxidation. Free Radic. Res. 28, 659–671. doi: 10.3109/10715769809065821
Morris, D. L. (1948). Quantitative determination of carbohydrates with Dreywood’s anthrone reagent. Science 107, 254–255. doi: 10.1126/science.107.2775.254
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Müller, B., and Sheen, J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453, 1094–1097. doi: 10.1038/nature06943
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nelson, D. E., Repetti, P. P., Adams, T. R., Creelman, R. A., Wu, J., Warner, D. C., et al. (2007). Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yield on water-limited acres. Proc. Natl. Acad. Sci. U.S.A. 104, 16450–16455. doi: 10.1073/pnas.0707193104
Perez-Perez, J. G., Syvertsen, J. P., Botfa, P., and Garcia-Sanchez, F. (2007). Leaf water relations and net gas exchange responses of salinized Carrizo citrange seedlings during drought stress and recovery. Ann. Bot. 100, 335–345. doi: 10.1093/aob/mcm113
Pernisová, M., Klíma, P., Horák, J., Válková, M., Malbeck, J., Souçek, P., et al. (2009). Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc. Natl. Acad. Sci. U.S.A. 106, 3609–3614. doi: 10.1073/pnas.0811539106
Price, A. H., Tomos, A. D., and Virk, D. S. (1997). Genetic dissection of root growth in rice (Oryza sativa L). I: a hydroponic screen. Theor. Appl. Genet. 95, 132–142. doi: 10.1007/s001220050541
Punta, M., Coggill, P. C., Eberhardt, R. Y., Mistry, J., Tate, J., Boursnell, C., et al. (2012). The Pfam protein families database. Nucleic Acids Res. 40, D290–D301. doi: 10.1002/047001153X.g306303
Redillas, M., Jeong, J., Kim, Y., Jung, H., Bang, S., Choi, Y., et al. (2012). The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 10, 792–805. doi: 10.1111/j.1467-7652.2012.00697.x
Schenk, H. J., and Jackson, R. B. (2005). Mapping the global distribution of deep roots in relation to climate and soil characteristics. Geoderma 126, 129–140. doi: 10.1016/j.geoderma.2004.11.018
Schultz, J., Milpetz, F., Bork, P., and Ponting, C. P. (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864. doi: 10.1073/pnas.95.11.5857
Serraj, R., Kumar, A., McNally, K. L., Slamet-Loedin, I., Bruskiewich, R., Mauleon, R., et al. (2009). Improvement of drought resistance in rice. Adv. Agron. 103, 41–99. doi: 10.1016/S0065-2113(09)03002-8
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. doi: 10.1093/jxb/erl164
Troll, W., and Lindsley, J. (1955). A photometric method for the determination of proline. J. Biol. Chem. 215, 655–660.
Turan, S., Cornish, K., and Kumar, S. (2012). Salinity tolerance in plants: breeding and genetic engineering. Aust. J. Crop Sci. 6, 1337–1348.
Uga, Y., Sugimoto, K., Ogawa, S., Rane, J., Ishitani, M., Hara, N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102. doi: 10.1038/ng.2725
Wang, J. E., Liu, K. K., Li, D. W., Zhang, Y. L., Zhao, Q., He, Y. M., et al. (2013). A novel peroxidase CanPOD gene of pepper is involved in defense responses to Phytophtora capsici Infection as well as abiotic stress tolerance. Int. J. Mol. Sci. 14, 3158–3177. doi: 10.3390/ijms14023158
Wang, M., Lu, X., Xu, G., Yin, X., Cui, Y., Huang, L., et al. (2016). OsSGL, a novel pleiotropic stress-related gene enhances grain length and yield in rice. Sci. Rep. 6:38157. doi: 10.1038/srep38157
Wang, Y. L., Zhang, W. Z., Song, L. F., Zou, J. J., Su, Z., and Wu, W. H. (2008). Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 148, 1201–1211. doi: 10.1104/pp.108.126375
Xiao, Y., Li, L. Y., Xu, M. L., and Xia, X. J. (2008). Establishment of agrobacterium-mediated transformation system of indica rice 93-11. J. Nat. Sci. Hunan Norm. Univ. 31, 77–82.
Xin, Z., Mandaokar, A., Chen, J., Last, R. L., and Browse, J. (2007). Arabidopsis ESK1 encodes a novel regulator of freezing tolerance. Plant J. 49, 786–799. doi: 10.1111/j.1365-313X.2006.02994.x
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yoshimura, K., Masuda, A., Kuwano, M., Yokota, A., and Akashi, K. (2008). Programmed proteome response for drought avoidance/tolerance in the root of a C3 xerophyte (wild watermelon) under water deficits. Plant Cell Physiol. 49, 226–241. doi: 10.1093/pcp/pcm180
Yue, B., Xue, W., Xiong, L., Yu, X., Luo, L., Cui, K., et al. (2006). Genetic basis of drought resistance at reproductive stage in rice: separation of drought tolerance from drought avoidance. Genetics 172, 1213–1228. doi: 10.1534/genetics.105.045062
Zhang, Q. F. (2007). Strategies for developing Green Super Rice. Proc. Natl. Acad. Sci. U.S.A. 104, 16402–16409. doi: 10.1073/pnas.0708013104
Zhang, Y., Su, J. B., Duan, S., Ao, Y., Dai, J. R., Liu, J., et al. (2011). A highly and efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related process. Plant Methods 7:30. doi: 10.1186/1746-4811-7-30
Zhang, Z., Zhang, Q., Wu, J., Zheng, X., Zheng, S., Sun, X., et al. (2013). Gene knockout study reveals that cytosolic ascorbate peroxidase 2 (OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS ONE 8:e57472. doi: 10.1371/journal.pone.0057472
Keywords: OsSGL, drought tolerance, rice, DUF1645 domain, root, overexpression
Citation: Cui Y, Wang M, Zhou H, Li M, Huang L, Yin X, Zhao G, Lin F, Xia X and Xu G (2016) OsSGL, a Novel DUF1645 Domain-Containing Protein, Confers Enhanced Drought Tolerance in Transgenic Rice and Arabidopsis. Front. Plant Sci. 7:2001. doi: 10.3389/fpls.2016.02001
Received: 20 July 2016; Accepted: 16 December 2016;
Published: 27 December 2016.
Edited by:
Olivier Lamotte, CNRS, UMR Agroécologie, FranceReviewed by:
Wayne Snedden, Queen’s University, CanadaDavid John Burritt, University of Otago, New Zealand
Copyright © 2016 Cui, Wang, Zhou, Li, Huang, Yin, Zhao, Lin, Xia and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoyun Xu, gyxu@isa.ac.cn Xinjie Xia, jxxia@isa.ac.cn
†These authors have contributed equally to this work.
 Yanchun Cui
Yanchun Cui Manling Wang
Manling Wang Huina Zhou2
Huina Zhou2 Mingjuan Li
Mingjuan Li Guoyun Xu
Guoyun Xu