- The Sainsbury Laboratory, University of Cambridge, Cambridge, UK
Epigenetic regulation can be altered by environmental cues including abiotic and biotic stresses. In most cases, environmentally-induced epigenetic changes are transient, but in some cases they are maintained for extensive periods of time and may even be transmitted to the next generation. However, the underlying mechanisms of transgenerational transmission of environmentally-induced epigenetic states remain largely unknown. Such traits can be adaptive, but also can have negative consequences if the parentally inherited epigenetic memory interferes with canonical environmental responses of the progeny. This review highlights recent insights into the mechanisms preventing transgenerational transmission of environmentally-induced epigenetic states in plants, which resemble those of germline reprogramming in mammals.
Introduction
Epigenetic marks such as DNA methylation, histone modifications, or histone variants influence the chromatin structure and transcriptional states. These epigenetic marks can be stably maintained, but also can be dynamically altered during development or in response to environmental stimuli.
In most cases, environmentally-induced epigenetic changes are transient. However, in some cases they are stably maintained trough mitotic cell divisions and therefore can be regarded as a type of long-term cellular memory. The best understood example of such epigenetic memory in plants is that of vernalization, which involves the epigenetic silencing of FLOWERING LOCUS C (FLC) gene by prolonged cold (Kim and Sung, 2012; Song et al., 2012). During embryogenesis, the FLC epigenetic state is reset thus allowing the next generation to respond to vernalization signals (further described below).
Epigenetic changes can be meiotically inherited (i.e., transgenerationally transmitted). The inheritance of epigenetic changes is especially well documented in plants where DNA methylation is stably maintained mitotically and meiotically (Law and Jacobsen, 2010). In Arabidopsis, DNA METHYLTRANSFERASE1 (MET1) and chromatin remodeling factor DECREASE IN DNA METHYLATION 1 (DDM1) are essential for global maintenance of DNA methylation as shown by the whole genome hypomethylation occurring in met1 and ddm1 mutants (Finnegan and Dennis, 1993; Vongs et al., 1993; Jeddeloh et al., 1998; Saze et al., 2003). Many of these hypomethylated loci are stably inherited for many generations even after reintroduction of a functional MET1 or DDM1 allele (Johannes et al., 2009; Reinders et al., 2009). Furthermore, transgenes, viral infection, or specific plant tissue culture conditions can also alter DNA methylation patterns in some genes, thus inducing so-called “epialleles” (Vaucheret et al., 1998; Baulcombe, 1999; Vaucheret and Fagard, 2001; Krizova et al., 2009; Rhee et al., 2010).
Besides experimentally-induced epialleles, there are many examples of naturally occurring epialleles inducing visible phenotypes such as flower shape/color, sex determination and genetic incompatibility (Cubas et al., 1999; Iida et al., 2004; Martin et al., 2009; Durand et al., 2012). All natural epialleles reported so far involve changes in DNA methylation. The differences in DNA methylation in the natural epialleles are often associated with transposable elements (TEs) or TE-related sequences located near the genes forming epialleles, suggesting that TE-derived cis-regulatory elements contribute to epiallele formation (Weigel and Colot, 2012). TEs are major components of most eukaryotic genomes, and usually silenced with repressive chromatin marks, which are considered a defense mechanism against TE activity since TE transpositions are frequently deleterious to the host. In some cases these epigenetic marks spread to neighboring genes thus altering their expression (Ahmed et al., 2011). Thus, TEs can contribute to epiallele formation.
Interestingly, TEs can be activated transcriptionally and transpositionally by stress in a wild type genetic background. McClintock (1984) suggested that TE activation could be a genomic response to challenge. In support of this view, several reports have described examples of TEs playing roles in gene regulation and genome evolution (Slotkin and Martienssen, 2007; Fedoroff, 2012).
Experimental induction of epialleles and TE mobilization in epigenetic mutants leading to heritable genetic changes has been well documented. However, the occurrence of stable inheritance induced by environmentally induced epigenetic changes has met some controversy (Boyko and Kovalchuk, 2011; Mirouze and Paszkowski, 2011; Paszkowski and Grossniklaus, 2011; Pecinka and Mittelsten Scheid, 2012). The inheritance of environmentally induced-epigenetic changes could be adaptive, but also could be deleterious given that environmental epigenetic memory of parent might impair canonical responses in the progeny.
Recent studies approached the issue from a different perspective, and described the mechanisms preventing transgenerational inheritance of environmentally-induced epigenetic traits.
In this review, I summarize these findings and discuss their implications on the inheritance of environmentally-induced epigenetic changes.
Resetting Vernalized State
Vernalization is the acquisition of ability to flower by exposure of plants with prolonged cold. In Arabidopsis, vernalization involves epigenetic silencing of the floral repressor FLC, which encodes a MADS box transcription factor (Michaels and Amasino, 1999; Sheldon et al., 1999). FLC is expressed throughout the early vegetative development in vernalization-requiring Arabidopsis accessions. In response to prolonged cold, FLC is epigenetically silenced allowing flowering to be promoted according to other environmental cues such as photoperiod (Figure 1A). This silencing of FLC is associated with chromatin modifications including increased levels of H3K27me3 at the FLC locus, which is mediated by polycomb repressive complex 2 (PRC2; Bastow et al., 2004; De Lucia et al., 2008; Figure 1B). After the cold exposure, the silenced epigenetic state of FLC is stably maintained throughout the rest of the life of the plant until the FLC chromatin state is reset during embryogenesis thus reestablishing vernalization requirement to promote flowering in the progeny (Sheldon et al., 2008; Choi et al., 2009; Figure 1A). Whereas the mechanisms leading to FLC silencing in response to vernalization have been extensively studied, the mechanisms responsible for resetting FLC was less understood.
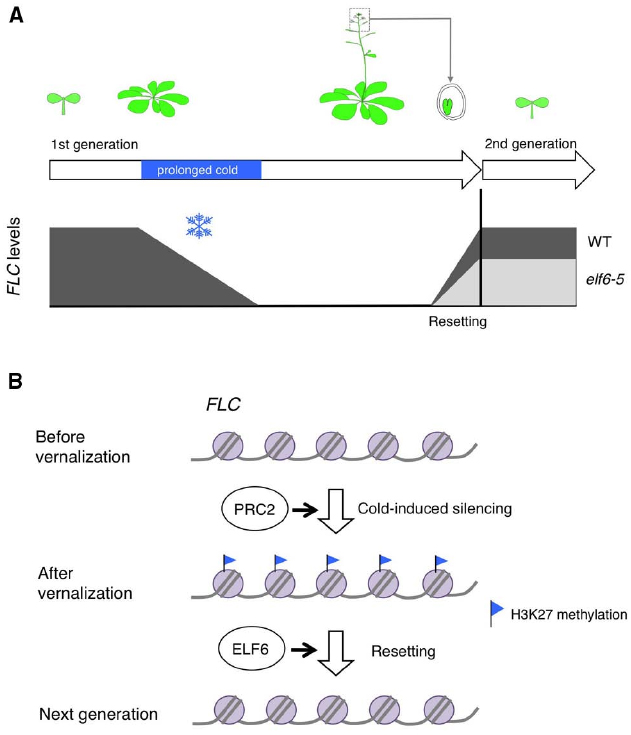
Figure 1. (A) Schematic representation of the experimental data in Crevillén et al. (2014). FLC is epigenetically silenced by prolonged exposure to cold. Thereafter, the silenced epigenetic state of FLC is stably maintained until embryogenesis. In developing embryos the epigenetic state of FLC is reset, thus allowing vernalization responses during the new vegetative phase. In the elf6 hypomorphic-resetting mutant (elf6-5), resetting of FLC is impaired therefore a partially vernalized state is inherited. (B) Simplified model for FLC resetting. This silencing of FLC is associated with H3K27 methylation mediated by PRC2. Removal of H3K27 methylation by ELF6 is required for restoring the pre-vernalization chromatin state of FLC.
Recently, Crevillén et al. (2014) screened for mutants in which resetting of FLC is impaired so that the vernalized states is inherited in the next generation. This led to the discovery of the histone demethylase EARLY FLOWERING 6 (ELF6) as a component required for FLC resetting (Crevillén et al., 2014). In the elf6 hypomorphic mutants, the progeny from vernalized plants flowered earlier and had reduced FLC expression compared to that of the progeny from non-vernalized plants, indicating that vernalized states were transmitted to the elf6 mutant progeny (Figure 1A).
ELF6 is a jumonji-C-domain-containing protein, and has H3K27me3 demethylase activity. In the elf6 hypomorphic resetting mutant, an alanine was substituted with a valine in conserved residues of the jumonji C domain, which leads to a reduction in demethylase activity. ChIP analysis showed the H3K27me3 levels were higher in the progeny of vernalized plants than the progeny of non-vernalized plants in the mutants, suggesting that removal of H3K27me3 by ELF6 is required for restoring the pre-vernalization chromatin state associated with FLC (Figure 1B). Intriguingly, different ELF6 alleles are associated with distinct phenotypic responses. Loss-of-function elf6 alleles are early flowering irrespective of the occurrence of vernalization due to the increased expression of the flowering regulator gene FT (Noh et al., 2004). Thus, it was suggested that ELF6 has a broader function, and that the particular hypomorphic mutation reveals a specific aspect of ELF6’s activity to restore the pre-vernalization chromatin state of FLC during embryogenesis.
In mammals, reprogramming of epigenetic marks, including H3K27me3, occurs in germ cells and early embryo (Cantone and Fisher, 2013). The reprogramming mediated by ELF6 would be a relevant mechanism conserved in evolution. In Arabidopsis genome, H3K27me3 is found in 15% of all genes (Zhang et al., 2007). It would be interesting to address whether other genes are reprogrammed by ELF6 or related proteins.
Resetting Chromatin Changes Induced by Heat Stress
In 2010, three independent research groups reported the influence of environmental stresses on epigenetically silenced loci in Arabidopsis (Lang-Mladek et al., 2010; Pecinka et al., 2010; Tittel-Elmer et al., 2010). They exposed plants to various stress conditions such as temperature shift, drought, elevated salinity, or UV radiation, and examined activities of transcriptionally silenced reporter genes. It was found that heat stress (37 or 42°C) or UV-B radiation releases silencing and activates reporter genes. The release of transcriptional silencing induced by stress occurs at various endogenous loci (Tittel-Elmer et al., 2010). However, this activation is transient since the loci are re-silenced within a few days after stress (Lang-Mladek et al., 2010; Pecinka et al., 2010; Tittel-Elmer et al., 2010). The rapid re-silencing appears to involve nucleosome loading since it is delayed in mutants with impaired chromatin assembly (Pecinka et al., 2010). These results suggest that chromatin non-permissive to transcription displays plasticity in response to stress, but also that there is a robust buffering system that resets chromatin changes to the initial ground state. Interestingly, stress-induced transcriptional activation occurs in differentiated tissues but not in meristematic tissues, suggesting the existence of a mechanism protecting germline cells from epigenetic damage (Pecinka et al., 2010).
Interestingly, the LTR-type retroelement ONSEN was found to behave rather exceptionally in response to heat stress. Unlike other heterochromatic loci destabilized by heat, the transcriptional activation persists for longer periods of time (Pecinka et al., 2010; Tittel-Elmer et al., 2010). The heat stress-induced transcriptional activation is enhanced in siRNA defective mutants, however, eventually ONSEN transcripts gradually decay as the plant pursues its growth, and no transpositions can be detected in vegetative tissues. Surprisingly, high frequency of transposition is observed in the progeny of siRNA defective mutants subjected to heat stress, suggesting that the siRNA pathway prevents transgenerational transposition of ONSEN (Ito et al., 2011).
Recently, Iwasaki and Paszkowski (2014) identified factors preventing transgenerational transmission of stress-induced chromatin changes by forward genetic screen in Arabidopsis. A silenced luciferase (LUC) reporter gene, whose transcription is transiently activated by heat stress, was used to isolate mutants that retain high or prolonged LUC activity after heat stress. This led to the identification of the epigenetic regulators DDM1 and MORPHEUS’ MOLECULE1 (MOM1) as components of a mechanism resetting stress-induced chromatin changes. In the ddm1 mutant, the heat stress-induced LUC activation is stronger and persists longer than WT, but the activated state is not transmitted to the progeny. In the mom1 mutant, stress-induced activation and subsequent extinction is similar to that of WT. However, and remarkably, in ddm1 mom1 double mutants, the activation persists in the next generation (Figure 2A). Genome-wide transcriptional profiles revealed that stress-induced transcriptional alterations at various heterochromatic loci were transmitted to next generation in ddm1 mom1 double mutants. These results indicate that DDM1 and MOM1 redundantly reset chromatin states destabilized by heat stress in order to prevent transgenerational propagation of transcriptional stress memory (Figure 2B).
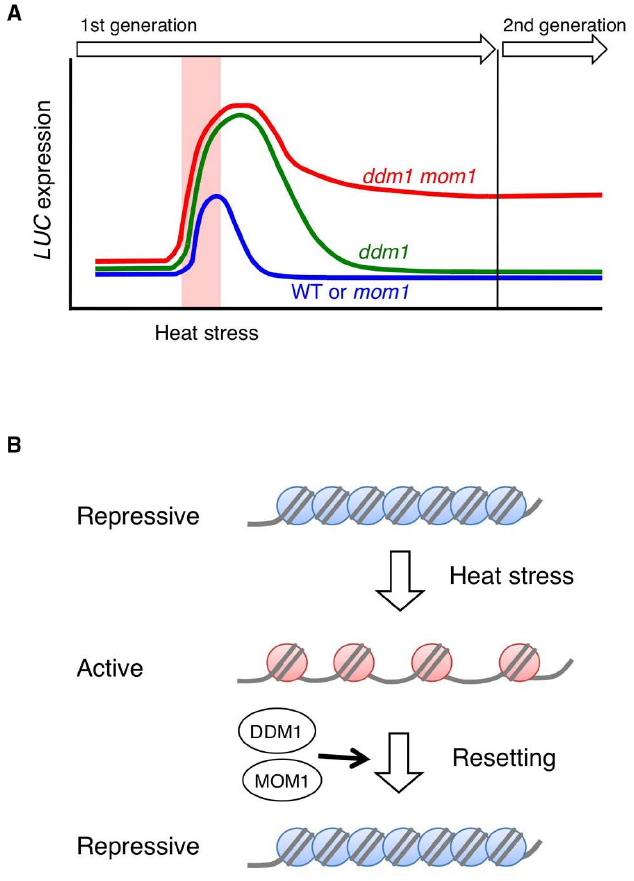
Figure 2. (A) Schematic representation of the experimental data in Iwasaki and Paszkowski (2014). A silenced LUC transgene is transiently transcriptionally activated in response to a heat stress. In ddm1 mutants, the heat stress-induced LUC activation is stronger and persists longer relative to WT plants. However, the activated state is not transmitted to the progeny. In contrast, in ddm1 mom1 double mutants, the activation persists in the next generation. (B) Schematic illustration heterochromatin states upon heat stress. Heterochromatic loci are transcriptionally activated by heat stress, however, they are rapidly resilenced after stress. DDM1 and MOM1 redundantly reset chromatin states destabilized by heat stress thus preventing transgenerational transmission of transcriptional stress memory.
Both DDM1 and MOM1 are required to maintain transcriptional gene silencing (TGS) since mutations in either genes cause release of silencing of heterochromatic loci (Jeddeloh et al., 1998; Amedeo et al., 2000; Steimer et al., 2000). Although DDM1 and MOM1 share a number of common target loci for silencing, the regulation mechanism seems to be different for each gene.
DDM1, which is conserved between plants and animals, is a chromatin remodeling factor of the SWI2/SNF2 family (Jeddeloh et al., 1999; Bourc’his and Bestor, 2002; Tao et al., 2011). ddm1 mutants show progressive global loss of DNA methylation during inbreeding (Kakutani et al., 1996; Jeddeloh et al., 1998). It has been suggested that DDM1 facilitates access of DNA methyltransferases to histone H1-containing heterochromatin (Zemach et al., 2013).
MOM1 is a plant-specific protein with limited homology to the SWI2/SNF2 family whose function remains poorly understood. Mutations in MOM1 cause release of TGS without major changes in DNA methylation levels, suggesting that MOM1 exerts its silencing function through pathways that are either independent or downstream of those of DNA methylation (Amedeo et al., 2000; Vaillant et al., 2006). Structural and genetic studies indicate that a conserved domain of MOM1 forms a homodimer, which may provide a binding platform for additional silencing factors (Yokthongwattana et al., 2010; Nishimura et al., 2012).
The transient release of transcriptional silencing induced by heat stress is not associated with significant changes in DNA methylation or histone modifications (Pecinka et al., 2010; Tittel-Elmer et al., 2010). Likewise, high levels of DNA methylation were maintained on the promoter of the LUC reporter gene in the progeny of heat stressed ddm1 mom1 mutants despite the occurrence of high LUC expression (Iwasaki and Paszkowski, 2014). This strongly suggests that epigenetic marks other than DNA methylation are transmitted to the next generation in the ddm1 mom1 mutants. The nature of these epigenetic marks necessary for the persistence of stress induced activation of heterochromatic transcription remains to be elucidated.
In summary, this study revealed a previously unidentified function of DDM1 and MOM1 to reset stress-induced chromatin changes. Future studies should address whether similar mechanisms occur in other species given that DDM1 is conserved in yeast and animals.
Concluding Remarks
The recent progress in our understanding of the mechanisms preventing transgenerational transmission of environmentally-induced epigenetic states opens new avenues for the study of epigenetic inheritance while raising new questions such as that of redundancy of the system. It was reported that DDM1 and MOM1 act redundantly to reset chromatin destabilized by heat stress. Furthermore, although about 3,000 loci on the Arabidopsis genome are activated by heat stress (Tittel-Elmer et al., 2010), only about one-tenth remain active in the progeny of heat stressed ddm1 mom1 mutants, suggesting that other factors act in parallel in the same silencing pathway (Iwasaki and Paszkowski, 2014). Similarly, in the elf6 hypomorphic mutants, the vernalized state of FLC is partially restored. FLC expression in the progeny of vernalized elf6 hypomorphic mutants is lower than in the non-vernalized plants, but still higher than in fully vernalized plants (Crevillén et al., 2014). Thus these observations suggest that other factors act redundantly to reset FLC.
These redundancies, essential to confer robustness to the system, would be crucial to ensure erasure of parental memory in order to permit progeny to respond appropriately to current environmental conditions. They could also account for the difficulty in documenting the occurrence of transgenerational transmission of environmentally induced epigenetic traits.
It remains possible that certain environmentally induced epigenetic changes could be inherited and become adaptive as in the case of some TEs which contributed to genome evolution. Further investigations would clarify the issue.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I thank Luis Lopez-Molina for critical reading. This work was supported by the Gatsby Charitable Foundation and the European Research Council.
References
Ahmed, I., Sarazin, A., Bowler, C., Colot, V., and Quesneville, H. (2011). Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis. Nucleic Acids Res. 39, 6919–6931. doi: 10.1093/nar/gkr324
Amedeo, P., Habu, Y., Afsar, K., Mittelsten Scheid, O., and Paszkowski, J. (2000). Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature 405, 203–206. doi: 10.1038/35012108
Bastow, R., Mylne, J. S., Lister, C., Lippman, Z., Martienssen, R. A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167. doi: 10.1038/nature02269
Baulcombe, D. C. (1999). Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol. 2, 109–113. doi: 10.1016/S1369-5266(99)80022-3
Bourc’his, D., and Bestor, T. H. (2002). Helicase homologues maintain cytosine methylation in plants and mammals. Bioessays 24, 297–299. doi: 10.1002/bies.10078
Boyko, A., and Kovalchuk, I. (2011). Genome instability and epigenetic modification-heritable responses to environmental stress? Curr. Opin. Plant Biol. 14, 260–266. doi: 10.1016/j.pbi.2011.03.003
Cantone, I., and Fisher, A. G. (2013). Epigenetic programming and reprogramming during development. Nat. Struct. Mol. Biol. 20, 282–289. doi: 10.1038/nsmb.2489
Choi, J., Hyun, Y., Kang, M. J., In Yun, H., Yun, J. Y., Lister, C., et al. (2009). Resetting and regulation of Flowering Locus C expression during Arabidopsis reproductive development. Plant J. 57, 918–931. doi: 10.1111/j.1365-313X.2008.03776.x
Crevillén, P., Yang, H., Cui, X., Greeff, C., Trick, M., Qiu, Q., et al. (2014). Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515, 587–590. doi: 10.1038/nature13722
Cubas, P., Vincent, C., and Coen, E. (1999). An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401, 157–161. doi: 10.1038/43657
De Lucia, F., Crevillén, P., Jones, A. M., Greb, T., and Dean, C. (2008). A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. U.S.A. 105, 16831–16836. doi: 10.1073/pnas.0808687105
Durand, S., Bouche, N., Perez Strand, E., Loudet, O., and Camilleri, C. (2012). Rapid establishment of genetic incompatibility through natural epigenetic variation. Curr. Biol. 22, 326–331. doi: 10.1016/j.cub.2011.12.054
Fedoroff, N. V. (2012). Presidential address. Transposable elements, epigenetics, and genome evolution. Science 338, 758–767. doi: 10.1126/science.338.6108.758
Finnegan, E. J., and Dennis, E. S. (1993). Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 21, 2383–2388. doi: 10.1093/nar/21.10.2383
Iida, S., Morita, Y., Choi, J. D., Park, K. I., and Hoshino, A. (2004). Genetics and epigenetics in flower pigmentation associated with transposable elements in morning glories. Adv. Biophys. 38, 141–159. doi: 10.1016/S0065-227X(04)80136-9
Ito, H., Gaubert, H., Bucher, E., Mirouze, M., Vaillant, I., and Paszkowski, J. (2011). An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–119. doi: 10.1038/nature09861
Iwasaki, M., and Paszkowski, J. (2014). Identification of genes preventing transgenerational transmission of stress-induced epigenetic states. Proc. Natl. Acad. Sci. U.S.A. 111, 8547–8552. doi: 10.1073/pnas.1402275111
Jeddeloh, J. A., Bender, J., and Richards, E. J. (1998). The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev. 12, 1714–1725. doi: 10.1101/gad.12.11.1714
Jeddeloh, J. A., Stokes, T. L., and Richards, E. J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22, 94–97. doi: 10.1038/8803
Johannes, F., Porcher, E., Teixeira, F. K., Saliba-Colombani, V., Simon, M., Agier, N., et al. (2009). Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 5:e1000530. doi: 10.1371/journal.pgen.1000530
Kakutani, T., Jeddeloh, J. A., Flowers, S. K., Munakata, K., and Richards, E. J. (1996). Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. U.S.A. 93, 12406–12411. doi: 10.1073/pnas.93.22.12406
Kim, D. H., and Sung, S. (2012). Environmentally coordinated epigenetic silencing of FLC by protein and long noncoding RNA components. Curr. Opin. Plant Biol. 15, 51–56. doi: 10.1016/j.pbi.2011.10.004
Krizova, K., Fojtova, M., Depicker, A., and Kovarik, A. (2009). Cell culture-induced gradual and frequent epigenetic reprogramming of invertedly repeated tobacco transgene epialleles. Plant Physiol. 149, 1493–1504. doi: 10.1104/pp.108.133165
Lang-Mladek, C., Popova, O., Kiok, K., Berlinger, M., Rakic, B., Aufsatz, W., et al. (2010). Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in Arabidopsis. Mol. Plant 3, 594–602. doi: 10.1093/mp/ssq014
Law, J. A., and Jacobsen, S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. doi: 10.1038/nrg2719
Martin, A., Troadec, C., Boualem, A., Rajab, M., Fernandez, R., Morin, H., et al. (2009). A transposon-induced epigenetic change leads to sex determination in melon. Nature 461, 1135–1138. doi: 10.1038/nature08498
McClintock, B. (1984). The significance of responses of the genome to challenge. Science 226, 792–801. doi: 10.1126/science.15739260
Michaels, S. D., and Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. doi: 10.1105/tpc.11.5.949
Mirouze, M., and Paszkowski, J. (2011). Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 14, 267–274. doi: 10.1016/j.pbi.2011.03.004
Nishimura, T., Molinard, G., Petty, T. J., Broger, L., Gabus, C., Halazonetis, T. D., et al. (2012). Structural basis of transcriptional gene silencing mediated by Arabidopsis MOM1. PLoS Genet. 8:e1002484. doi: 10.1371/journal.pgen.1002484
Noh, B., Lee, S. H., Kim, H. J., Yi, G., Shin, E. A., Lee, M., et al. (2004). Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16, 2601–2613. doi: 10.1105/tpc.104.025353
Paszkowski, J., and Grossniklaus, U. (2011). Selected aspects of transgenerational epigenetic inheritance and resetting in plants. Curr. Opin. Plant Biol. 14, 195–203. doi: 10.1016/j.pbi.2011.01.002
Pecinka, A., Dinh, H. Q., Baubec, T., Rosa, M., Lettner, N., and Mittelsten Scheid, O. (2010). Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22, 3118–3129. doi: 10.1105/tpc.110.078493
Pecinka, A., and Mittelsten Scheid, O. (2012). Stress-induced chromatin changes: a critical view on their heritability. Plant Cell Physiol. 53, 801–808. doi: 10.1093/pcp/pcs044
Reinders, J., Wulff, B. B., Mirouze, M., Mari-Ordonez, A., Dapp, M., Rozhon, W., et al. (2009). Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 23, 939–950. doi: 10.1101/gad.524609
Rhee, Y., Sekhon, R. S., Chopra, S., and Kaeppler, S. (2010). Tissue culture-induced novel epialleles of a Myb transcription factor encoded by pericarp color1 in maize. Genetics 186, 843–855. doi: 10.1534/genetics.110.117929
Saze, H., Mittelsten Scheid, O., and Paszkowski, J. (2003). Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 34, 65–69. doi: 10.1038/ng1138
Sheldon, C. C., Burn, J. E., Perez, P. P., Metzger, J., Edwards, J. A., Peacock, W. J., et al. (1999). The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458. doi: 10.1105/tpc.11.3.445
Sheldon, C. C., Hills, M. J., Lister, C., Dean, C., Dennis, E. S., and Peacock, W. J. (2008). Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc. Natl. Acad. Sci. U.S.A. 105, 2214–2219. doi: 10.1073/pnas.0711453105
Slotkin, R. K., and Martienssen, R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285. doi: 10.1038/nrg2072
Song, J., Angel, A., Howard, M., and Dean, C. (2012). Vernalization—a cold-induced epigenetic switch. J. Cell Sci. 125, 3723–3731. doi: 10.1242/jcs.084764
Steimer, A., Amedeo, P., Afsar, K., Fransz, P., Mittelsten Scheid, O., and Paszkowski, J. (2000). Endogenous targets of transcriptional gene silencing in Arabidopsis. Plant Cell 12, 1165–1178. doi: 10.1105/tpc.12.7.1165
Tao, Y., Xi, S., Shan, J., Maunakea, A., Che, A., Briones, V., et al. (2011). Lsh, chromatin remodeling family member, modulates genome-wide cytosine methylation patterns at nonrepeat sequences. Proc. Natl. Acad. Sci. U.S.A. 108, 5626–5631. doi: 10.1073/pnas.1017000108
Tittel-Elmer, M., Bucher, E., Broger, L., Mathieu, O., Paszkowski, J., and Vaillant, I. (2010). Stress-induced activation of heterochromatic transcription. PLoS Genet. 6:e1001175. doi: 10.1371/journal.pgen.1001175
Vaillant, I., Schubert, I., Tourmente, S., and Mathieu, O. (2006). MOM1 mediates DNA-methylation-independent silencing of repetitive sequences in Arabidopsis. EMBO Rep. 7, 1273–1278. doi: 10.1038/sj.embor.7400791
Vaucheret, H., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J. B., et al. (1998). Transgene-induced gene silencing in plants. Plant J. 16, 651–659. doi: 10.1046/j.1365-313x.1998.00337.x
Vaucheret, H., and Fagard, M. (2001). Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet. 17, 29–35. doi: 10.1016/S0168-9525(00)02166-1
Vongs, A., Kakutani, T., Martienssen, R. A., and Richards, E. J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260, 1926–1928. doi: 10.1126/science.8316832
Weigel, D., and Colot, V. (2012). Epialleles in plant evolution. Genome Biol. 13, 249. doi: 10.1186/gb-2012-13-10-249
Yokthongwattana, C., Bucher, E., Caikovski, M., Vaillant, I., Nicolet, J., Mittelsten Scheid, O., et al. (2010). MOM1 and Pol-IV/V interactions regulate the intensity and specificity of transcriptional gene silencing. EMBO J. 29, 340–351. doi: 10.1038/emboj.2009.328
Zemach, A., Kim, M. Y., Hsieh, P. H., Coleman-Derr, D., Eshed-Williams, L., Thao, K., et al. (2013). The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153, 193–205. doi: 10.1016/j.cell.2013.02.033
Keywords: transgenerational epigenetic memory, chromatin regulation, vernalization, FLC, ELF6, DDM1, MOM1
Citation: Iwasaki M (2015) Chromatin resetting mechanisms preventing transgenerational inheritance of epigenetic states. Front. Plant Sci. 6:380. doi: 10.3389/fpls.2015.00380
Received: 31 March 2015; Accepted: 12 May 2015;
Published: 27 May 2015.
Edited by:
Mahmoud W. Yaish, Sultan Qaboos University, OmanReviewed by:
Zuhua He, Chinese Academy of Sciences, ChinaDavid Secco, University of Western Australia, Australia
Copyright © 2015 Iwasaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mayumi Iwasaki, The Sainsbury Laboratory, University of Cambridge, Bateman Street, Cambridge CB2 1LR, UK, mayumi.iwasaki@unige.ch
†Present address: Mayumi Iwasaki, Department of Botany and Plant Biology, University of Geneva, Sciences III, 30 Quai Ernest-Ansermet, CH-1211 Geneva 4, Switzerland
 Mayumi Iwasaki
Mayumi Iwasaki