- 1 MSU-DOE Plant Research Laboratory, Michigan State University, East Lansing, MI, USA
- 2 Plant Biology Department, Michigan State University, East Lansing, MI, USA
Peroxisomes are small subcellular organelles mediating a multitude of processes in plants. Proteomics studies over the last several years have yielded much needed information on the composition of plant peroxisomes. In this review, the status of peroxisome proteomics studies in Arabidopsis and other plant species and the cumulative advances made through these studies are summarized. A reference Arabidopsis peroxisome proteome is generated, and some unique aspects of Arabidopsis peroxisomes that were uncovered through proteomics studies and hint at unanticipated peroxisomal functions are also highlighted. Knowledge gained from Arabidopsis was utilized to compile a tentative list of peroxisome proteins for the model monocot plant, rice. Differences in the peroxisomal proteome between these two model plants were drawn, and novel facets in rice were expounded upon. Finally, we discuss about the current limitations of experimental proteomics in decoding the complete and dynamic makeup of peroxisomes, and complementary and integrated approaches that would be beneficial to defining the peroxisomal metabolic and regulatory roadmaps. The synteny of genomes in the grass family makes rice an ideal model to study peroxisomes in cereal crops, in which these organelles have received much less attention, with the ultimate goal to improve crop yield.
Introduction
Eukaryotic cells compartmentalize specific biochemical reactions in membrane-bound subcellular organelles. Peroxisomes are small and dynamic single membrane-delimited organelles found in nearly all eukaryotic cells and perform a wide array of functions, which differ in different organisms and even vary depending on the tissue type and prevailing environmental conditions. Although peroxisomes in different organisms exhibit significant functional heterogeneities, two peroxisomal functions, i.e., β-oxidation of fatty acids and hydrogen peroxide (H2O2) catabolism, are universal. Strong defects in peroxisome biogenesis or core peroxisome metabolic functions lead to fatal disorders in humans and embryonic lethality in plants (Schrader and Fahimi, 2008; Kaur et al., 2009).
In the absence of a genome, the entire peroxisome protein complement is comprised of proteins that are nuclear encoded and translated on cytosolic ribosomes prior to import into the organelle. Further, peroxisomes are distinguished from other organelles by their ability to import fully folded proteins into the organelle matrix. The presence of conserved peroxisome targeting signals (PTSs) in the peroxisome matrix proteins facilitates their recognition by cytosolic receptors. These PTSs comprise of two types: the C-terminal tripeptide PTS1 (SKL and derivatives thereof), and the N-terminal nonapeptide PTS2 (RLX5HL and derivatives) that is cleaved post-import in mammals and plants. Once bound by the cytosolic receptors, the PTS-containing proteins are transported to the peroxisome membrane and delivered into the matrix, aided by several peroxisome membrane-associated proteins that form the docking complex and the importomer (Rucktaschel et al., 2011).
Peroxisomes serve as essential “nodes” in a number of metabolic networks within the cell through physical and metabolic links with other cellular compartments such as mitochondria, chloroplasts, and oil bodies. Besides their roles in β-oxidation of fatty acids and degradation of H2O2, plant peroxisomes also mediate pathways such as photorespiration, jasmonic acid biosynthesis, indole 3-butyric acid (IBA) metabolism, glyoxylate cycle, purine degradation, and further contribute toward pathogen defense and essential developmental processes such as embryogenesis and photomorphogenesis (Hayashi and Nishimura, 2003; Baker et al., 2006; Reumann and Weber, 2006; Kaur et al., 2009; Palma et al., 2009). The major protein constituents of plant peroxisomes had been well characterized, yet the complete makeup of these organelles was far from being decoded. Understanding the metabolic and regulatory networks in these vital organelles in model plant systems, and furthermore, crop species, will be highly beneficial to modern agriculture.
Peroxisome Proteomics Studies in Plants
Innovations in protein identification techniques coupled with high sensitivity instrumentation facilities have fueled the increased use of mass spectrometry-based proteomics to map subcellular (organelle) proteomes (Yates et al., 2005; Au et al., 2007; Yan et al., 2009; Wiederhold et al., 2010). Likewise, peroxisome proteomics have also been undertaken by various groups in diverse organisms encompassing yeasts, mammals, plants, and trypanosomes (Colasante et al., 2006; Saleem et al., 2006). Given its completely sequenced and annotated genome and a wide suite of readily available molecular genetic resources, Arabidopsis was naturally the top choice for this approach to decipher the plant peroxisome proteome. However, early peroxisome proteome analysis in Arabidopsis was hindered by the lack of good peroxisome purification protocols. A combination of factors, such as the fragility of peroxisomes, elevated secondary metabolite levels in Arabidopsis, adherence of peroxisomes to mitochondria and chloroplasts, and the low peroxisome number in leaf mesophyll cells, made high purity isolation of Arabidopsis peroxisomes a challenging task. Thus, initial proteomic studies of Arabidopsis peroxisomes from greening and etiolated cotyledons only identified a small number of known and putative novel peroxisomal proteins (Fukao et al., 2002, 2003). The development of an isolation protocol for Arabidopsis leaf peroxisomes, which uses a two successive density gradient centrifugation method, resulted in the successful identification of 36 known peroxisome proteins and dozens of candidate novel proteins, some of which were later confirmed to be peroxisome localized (Reumann et al., 2007). A parallel large scale experiment, which used density centrifugation followed by free-flow electrophoresis, purified peroxisomes from Arabidopsis suspension cultured cells and discovered over 20 possible novel peroxisomal proteins by mass spectrometry (Eubel et al., 2008). Apart from Arabidopsis, soybean (Glycine max), and spinach (Spinacia oleracea) peroxisomes were also subjected to proteome analysis. About 30 peroxisomal proteins were identified from purified peroxisomes from etiolated cotyledons of soybean, among them is an adenine nucleotide transporter (Arai et al., 2008a,b). A few new peroxisomal proteins, including two enzymes in phylloquinone (vitamin K1) biosynthesis, were discovered by spinach leaf peroxisome proteomics (Babujee et al., 2010).
The NSF-funded Arabidopsis peroxisome 2010 project was initiated in late 2006 and is near its completion. The major goal for this project was to discover novel peroxisomal components and reveal new peroxisomal functions in Arabidopsis. Using one-dimensional gel electrophoresis (1-DE) followed by liquid chromatography and tandem mass spectrometry (LC-MS/MS), in-depth proteome analysis of three Arabidopsis peroxisomal subtypes, i.e., those from green leaves, etiolated germinating seedlings, and senescent leaves, respectively, was performed. Using fluorescence microscopy, the subcellular localization of over 100 putative novel peroxisomal proteins identified from proteomics and in silico PTS searches of the Arabidopsis genome was tested, and peroxisomal targeting for about 50 of them was confirmed (Reumann et al., 2009; Quan et al., unpublished). From leaves of 4-week-old plants, 85 known peroxisomal proteins were detected, and another 14 novel proteins were assigned to peroxisomes after subcellular targeting validations (Reumann et al., 2009; Quan et al., 2010). From peroxisomes of etiolated seedlings, another 15 novel peroxisomal proteins were discovered by proteomics combined with subcellular targeting assays (Quan et al., unpublished).
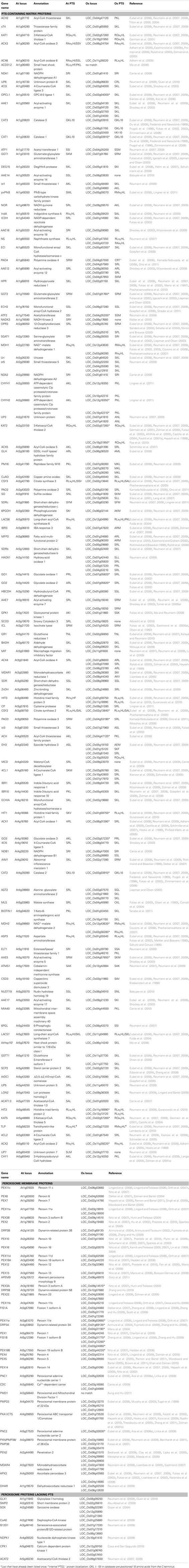
The recent proteomic studies followed by subcellular targeting verifications significantly expanded the list of bona-fide plant peroxisome proteins. Using data from published studies of plant peroxisomes, including the proteome analyses mentioned above, we have compiled a reference proteome for Arabidopsis peroxisomes (Table 1). For proteins identified by mass spectrometry-based experiments, we only included those that carry obvious PTS, unless they were later confirmed to be peroxisomal by a second approach, e.g., fluorescent protein subcellular targeting assay or genetic analysis. This list of Arabidopsis peroxisomal proteins currently stands at 163, which can be divided into the following categories: 117 PTS-containing matrix proteins, 38 membrane proteins, and eight proteins lacking recognizable PTS information. The 98 PTS1-containing proteins carry 23 diverse PTS1s, and the 19 PTS2-containing proteins harbor seven different PTS2s. Six proteins in the PTS2-containing protein category also bear C-terminal PTS1 or PTS1-like sequences.
Some Novel Aspects of Plant Peroxisomes Revealed by Experimental Proteomics
Results from mass spectrometry-based proteomics studies in plants suggested novel metabolic and regulatory functions of peroxisomes in processes such as auxiliary β-oxidation, detoxification, nucleic acid metabolism, protein degradation, plant defense, and other metabolic processes (Kaur et al., 2009; Reumann, 2011). Several examples that represent plant-specific features of peroxisomes are described here. More examples can be found in a recent review (Reumann, 2011).
Histidine Triad Family
Histidine Triad (HIT) proteins belong to an evolutionarily conserved superfamily of nucleotide binding proteins whose defining feature is the H-φ-H-φ-H-φ−φ motif, where φ represents a hydrophobic amino acid. HIT proteins act as hydrolases or transferases on a multitude of nucleotide conjugate substrates (Brenner, 2002). The consequences of loss of HIT activity have various effects, from tumor formation in mammals, high temperature sensitive growth on galactose in yeast, to a reduction of bacterial growth in the presence of D-alanine (Bieganowski et al., 2002; Bardaweel et al., 2011; Martin et al., 2011), yet the exact roles for HIT proteins in these processes remain unclear. Animal HIT proteins have been reported to be cytosolic, nuclear, or mitochondrial (Huber and Weiske, 2008), and none was shown to be associated with peroxisomes. In contrast, three out of the five Arabidopsis HIT proteins (HIT1, 2, and 3) were detected in proteomics experiments (Reumann et al., 2007, 2009; Eubel et al., 2008) and later confirmed to be localized in peroxisomes (Reumann et al., 2009). HIT2 carries a characteristic PTS2, and the PTS1-like sequence on HIT1 (SKV>) and PTS2-like sequence on HIT3 (RVx5HF) were later confirmed to be functional PTSs (Reumann et al., 2009; Quan et al., 2010). The occurrence of these proteins in peroxisomes appears to be a plant-specific phenomenon, as the Arabidopsis HIT PTSs are also conserved in their homologs in other plant species such as rice (Table 1). A recent in vitro study showed that most of the Arabidopsis HIT proteins can function as sulfohydrolases by catalyzing the conversion of adenosine 5′-phosphosulfate (APS) to AMP and sulfate (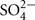 ; Guranowski et al., 2010). This study also showed that, in the presence of orthophosphate (Pi), HIT1/Hint4 exhibited APS phosphorylase activity as well, resulting in the formation of ADP. Moreover, this activity was determined to be pH dependent, with HIT1 exclusively (and more efficiently) catalyzing this reaction at acidic pHs. Considering their enzymatic activity, it is possible that the three peroxisomal HIT proteins are involved in recycling the pool of adenosine nucleotides in the peroxisome. Based on the pH specific activity of HIT1, we speculate that this protein may serve to buffer peroxisomal pH. Phenotypic and functional characterization of these HIT proteins will be needed to address their role in the peroxisome and in plant physiology.
; Guranowski et al., 2010). This study also showed that, in the presence of orthophosphate (Pi), HIT1/Hint4 exhibited APS phosphorylase activity as well, resulting in the formation of ADP. Moreover, this activity was determined to be pH dependent, with HIT1 exclusively (and more efficiently) catalyzing this reaction at acidic pHs. Considering their enzymatic activity, it is possible that the three peroxisomal HIT proteins are involved in recycling the pool of adenosine nucleotides in the peroxisome. Based on the pH specific activity of HIT1, we speculate that this protein may serve to buffer peroxisomal pH. Phenotypic and functional characterization of these HIT proteins will be needed to address their role in the peroxisome and in plant physiology.
Peroxisomal NADPH Production
Many peroxisomal enzymes consume NADPH during reductive reactions. For instance, several enzymes involved in JA biosynthesis (OPR3), auxiliary β-oxidation (ECRs, SDRa), and detoxification (MDAR, GR, NQR), require NADPH for their activity, underscoring the critical need for this cofactor in the peroxisome (Nyathi and Baker, 2006; Kaur et al., 2009). Several NADPH dehydrogenases are also found in the peroxisome (Carrie et al., 2008). To maintain optimal activities of these enzymes, the diminishing NADPH pool needs to be continuously replenished. In addition to providing reducing equivalents, NAPDH is also essential in oxidative damage response and peroxisome protein import (Juhnke et al., 1996; Pool et al., 1998; Pollak et al., 2007).
In plants, the plastid localized oxidative pentose phosphate pathway (OPPP) is a primary source of NADPH. OPPP consists of a three-enzyme cascade comprising of glucose-6-phosphate dehydrogenase (G6PD), 6-phosphogluconolactonase (6PGL), and 6-phosphogluconate dehydrogenase (6PGDH). These enzymes act sequentially to convert glucose-6-phosphate to ribose-5-phosphate with the concomitant production of NADPH (Kruger and von Schaewen, 2003). Besides the OPPP, NADPH is also generated by NADP-dependent isocitrate dehydrogenase (ICDH), malic enzyme (ME), aldehyde dehydrogenase (ALDH), and NAD kinase (NADK; Pollak et al., 2007). Some earlier studies indicated that the NADPH generating enzymes in OPPP and ICDH are compartmentalized in plant peroxisomes (Donaldson, 1982; Corpas et al., 1998; del Rio et al., 2002; Mateos et al., 2003). In yeasts, the presence of peroxisomal ICDH is also necessary for β-oxidation of unsaturated fatty acids and NADPH is critical to dissipate H2O2 (Henke et al., 1998; van Roermund et al., 1998; Minard and McAlister-Henn, 1999). Arabidopsis peroxisome proteomics studies detected ICDH and 6PGDH and subcellular targeting studies validated their peroxisomal localization (Fukao et al., 2002, 2003; Reumann et al., 2007, 2009; Eubel et al., 2008). Although not identified in proteomics experiments, 6PGL was shown to be a dual localized protein carrying an N-terminal transit peptide directing it to chloroplasts and a C-terminal PTS1 targeting it to peroxisomes (Reumann et al., 2007). Interestingly, although the chloroplast targeting of 6PGL is indispensable for viability, plants were unaffected in the absence of a peroxisome localized isoform (Xiong et al., 2009), strengthening the notion that plant peroxisomes have alternative sources for NADPH generation. The only enzyme missing from the set of peroxisomal OPPP was recently found to be the plastidic G6PD1, which was selectively recruited to the peroxisomes under conditions that promote transient oxidation events. This study further demonstrated the cysteine dependent heterodimer formation between the plastid targeted G6PD1 isoform and a catalytically inactive isoform G6PD4, and the import of this heterodimer into peroxisomes (Meyer et al., 2011). As such, peroxisomes seem to have a complete set of OPPP enzymes.
De novo biosynthesis of NADPH also occurs in peroxisomes by the preferential phosphorylation of NADH by NADK3 (Turner et al., 2005a,b). Though not found in proteomics studies, NADK3 has been localized to peroxisomes via a novel PTS1, SRY> (Waller et al., 2010). Lastly, ALDHs are classified as NADP-dependent enzymes that detoxify aldehyde substrates (Kirch et al., 2004). Mammals possess multiple isoforms of ALDHs, which were found to be induced by oxidative stress; one particular isoform was linked to the prevention of oxidative damage (Pappa et al., 2003; Vasiliou and Nebert, 2005). Peroxisome proteomic analysis in Arabidopsis has repeatedly identified an ALDH, BADH, which has a putative role in polyamine degradation (Reumann et al., 2007, 2009; Eubel et al., 2008). Whether this protein contributes to the maintenance of peroxisomal redox homeostasis/NADP turnover needs to be ascertained. Thus, plant peroxisomes seem to have several routes to maintain a supply of NADPH within the organelle, possibly as a countermeasure for the dangers posed by oxidative stress emanating from unchecked/continuous H2O2 generated within the peroxisome.
Metabolite Transporters
Proteomic analysis of the soybean peroxisomal membrane proteins led to the identification of a soybean peroxisomal adenine nucleotide transporter and later on two homologous proteins (PNC1, PNC2) from Arabidopsis (Arai et al., 2008b). An independent study of peroxisome membrane proteins isolated from Arabidopsis suspension cultured cells also identified PNC2 as a novel constituent of the peroxisome membrane (Eubel et al., 2008). A bioinformatics based approach followed by cell biological and biochemical validations found PNC1 and PNC2 as adenine nucleotide transporters in Arabidopsis as well (Linka et al., 2008). Consistent with the ATP transporter activity of the PNCs, their RNAi lines were sucrose dependent for seedling establishment and impaired in post-germinative lipid mobilization. Functional analysis of the PNCs highlights the critical need for ATP within peroxisomes and reinforces the notion that these proteins are the sole purveyors of ATP transport into peroxisomes (Arai et al., 2008b; Linka et al., 2008).
PMP38/PXN, like the PNCs, is also a member of the mitochondrial carrier family. It was initially identified as an integral peroxisome membrane protein in pumpkin cotyledons and considered to be a potential ATP/ADP transporter (Fukao et al., 2001). The presence of PMP38 in Arabidopsis peroxisomes was subsequently detected in isolated peroxisomes from proteomics works using suspension cultured cells and adult leaves (Eubel et al., 2008; Reumann et al., 2009). However, this protein failed to complement a yeast adenine nucleotide mutant (Linka et al., 2008) but instead was found to serve as an NAD+ carrier involved in peroxisomal β-oxidation (Bernhardt et al., 2011).
Peroxisome Proteins Lacking PTS
Dehydroascorbate reductase (DHAR) is part of the peroxisomal ascorbate-glutathione (PAG) cycle, which encompasses the enzymes MDAR1, MDAR4, APX3, and glutathione reductase (GR) and plays a key role in antioxidant metabolism (Kaur et al., 2009). DHAR was found to be peroxisomal in pea and tomato (Jimenez et al., 1997; Mittova et al., 2003), but was missing from the Arabidopsis PAG pathway until its proteomic identification and subcellular localization validation (Reumann et al., 2009). Glyoxylase I (GLXI) is another protein without obvious PTS identified from the leaf peroxisome proteome (Reumann et al., 2009). GLX system disposes of toxic byproducts such as methylglyoxal in two consecutive steps catalyzed by GLXI and GLXII (Mannervik, 2008; Yadav et al., 2008; Inoue et al., 2011). The verification of GLXI in the peroxisome expanded the suite of detoxification related proteins found in the peroxisome and indicated that half of the GLX pathway is compartmentalized in peroxisomes in Arabidopsis (Quan et al., 2010).
The plant-specific, senescence-associated B12D gene encodes for a small protein lacking any recognizable PTS. It was only detected in the proteome of leaf peroxisomes, and as a C-terminal YFP fusion was found to target to peroxisomes (Reumann et al., 2009). B12D genes in monocots such as barley and wheat are expressed during seed development but cease to be transcribed at seed maturity (Aalen et al., 1994, 2001; McIntosh et al., 2007). The germinating seed is purported to induce the expression of these genes via a putative gibberellic acid (GA) responsive promoter element. Consistent with this notion, the expression of this gene was found to be induced by GA but suppressed by abscisic acid (ABA; Steinum et al., 1998). Although no function has been attributed to B12D, the gene expression pattern suggests its role in seed germination/dormancy.
Dephospho-CoA kinase (CoAE) was identified in leaf peroxisome proteomics and localized to the periphery of the peroxisome membrane as a YFP fusion (Reumann et al., 2009). Coenzyme A (CoA) and derivatives thereof are the major currency behind many cellular metabolic pathways. CoA biosynthesis is accomplished in five successive enzymatic steps, the last of which is carried out by CoAE (Leonardi et al., 2005). Though the in planta effects of some of the enzymes catalyzing the preceding steps in CoA biosynthesis have been studied, the physiological role of CoAE in plants has not been analyzed (Rubio et al., 2006, 2008; Tilton et al., 2006). Given the number of β-oxidation reactions that require CoA, it will be necessary to determine the effect of CoAE on peroxisome metabolism.
Nucleoside diphosphate kinase type 1 (NDPK1) was another unexpected peroxisomal protein to be found through proteomics, and was seen to localize to peroxisomes as well as the cytosol and nucleus (Reumann et al., 2009). This multi-localization is perhaps not so surprising in light of the even more promiscuous localization reported for the mammalian NDPKs (Bosnar et al., 2009). NDPKs catalyze the interconversion of nucleoside diphosphates by transferring the phosphate group from a nucleoside triphosphates (NTP) to any other nucleoside diphosphates (NDP) except for ADP (Yegutkin, 2008). In view of this, NDPK1 might serve to regulate the concentration of different nucleotide phosphates within the peroxisome.
Peroxisome Proteome in Rice
The Need to Study Peroxisomes in the Monocot Crop Plant, Rice
The pivotal roles of peroxisomes in plant development and stress responses make it highly necessary to study these organelles in crop plants, with the goal to improve the quality and yield of crop species. Rice (Oryza sativa) is one the three major staple food crops in the world and a model system for basic research in monocot plants. Traditionally, plant peroxisome studies have mainly focused on dicot species such as cucumber, pumpkin, watermelon, and pea (Beevers, 1979), and lately, Arabidopsis (Kaur et al., 2009); however, very little research has been carried out with these organelles in monocots, which differ significantly from dicots in architecture and physiology. Having a completely sequenced and well annotated genome, well developed transformation methods, and rich genetic and mutant resources, rice is deemed to be the logical choice for a model system to study peroxisome functions in monocot plants.
Despite the fact that rice has a larger genome than Arabidopsis, the advantages of studying rice orthologs of Arabidopsis genes have been exemplified by a number of cases, in which mutant phenotypes were revealed in rice but not in Arabidopsis mutant due to functional redundancy among gene family members in the latter. The best example is the identification of the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1) from rice, in which the gid1 mutant shows a strong GA insensitive dwarf phenotype (Ueguchi-Tanaka et al., 2005). In contrast, three GID1 homologs exist in Arabidopsis and as a result, phenotypes could only be shown in higher order mutants whereas single mutant is indistinguishable from the wild-type plants (Griffiths et al., 2006). T-DNA insertion mutants for many of the new peroxisome genes we recently identified have no apparent phenotypes (Cassin and Hu, unpublished). Studying mutants of their orthologs in rice may be an easier way to decipher the functions of some of these proteins.
Studying peroxisomes in rice may also have applications in engineering the more efficient C4 photosynthetic pathway into C3 crops for yield increase. Rice is a C3 plant growing in warm environment, which favors photorespiration and thus reduces photosynthesis. However, in rice cells chloroplasts and stromules occupy 95% of the cell periphery, whereas peroxisomes and mitochondria, players in photorespiration, are present in the interior of the cell and lined up along the chloroplast walls (Sage and Sage, 2009). This interesting anatomy of rice mesophyll cells, which is atypical for a C3 plant, was suggested to be significant in scavenging photorespiratory CO2 to enhance the carboxylation capability of Rubisco for CO2 refixation. In C3 plants, peroxisomes were found to be responsible for the vast majority of the H2O2 produced during photorespiration (Foyer and Noctor, 2003). It would be interesting to investigate whether there are any changes to the activity of rice peroxisomal photorespiratory enzymes in association with these organelle rearrangements.
In silico Analysis of the Rice Peroxisome Proteome
As a first step toward exploring the peroxisome proteome in cereal crop species, we performed an in silico analysis of the rice peroxisome proteome by searching the rice genome for proteins with sequence similarities with the Arabidopsis peroxisomal proteins (Table 1). Remarkably, with the exception of two proteins, every Arabidopsis peroxisome protein seems to have at least one homolog in rice. One exception is ACD31.2, a small heat shock protein. It is interesting that this protein as well as its PTS2 peptide are well conserved in most plant species (Ma et al., 2006) but entirely missing from rice. Another protein without an apparent homolog in rice is peroxisome and mitochondrial division factor 1 (PMD1), a plant-specific dual localized membrane protein involved in the proliferation of peroxisomes and mitochondria in Arabidopsis (Aung and Hu, 2011). Altogether, the putative rice peroxisome proteome consists of 133 matrix proteins, 47 membrane proteins, and 14 proteins with no apparent PTSs. Among the candidate peroxisome matrix proteins in rice, most proteins contain PTS1 or PTS2-like sequences just like their Arabidopsis counterparts, suggesting that these proteins are highly likely to be peroxisomal. Proteins which appear to have lost PTS1 signal include snowy cotyledon3 (SCO3), the dual localized malonyl-CoA decarboxylase (MCD; Carrie et al., 2008), a protein of unknown function (UP7), NADK3 (Waller et al., 2010), AAE14 (Babujee et al., 2010), and macrophage migration inhibitory factor, MIF (Li et al., 2009). Despite having a C-terminal PTS1, Arabidopsis SCO3 is actually localized to the periphery of peroxisomes and influences plastid development possibly through interaction with the cytoskeleton (Albrecht et al., 2010). Hence, the loss of the matrix targeting signal PTS1 in its rice homolog is not that surprising. For proteins whose homologs in Arabidopsis contain dual PTSs, CSY2 and LACS7 homologs retain the same PTSs, LACS6 gains a PTS1 (PKL>), ACX3 and ACX6 both lose their PTS1s, while ACD31.2 does not have an apparent rice homolog.
Rice appears to employ a greater diversity of PTSs, with (potentially) 37 assorted PTS1s and 5 PTS2s. Recognition of PTS1 in plants seems fairly plastic and tolerant of non-canonical substitutions (Lingner et al., 2011). These new PTS1s in rice still need to be validated in planta as genuine PTSs. An outright observation is the frequent substitution to Methionine (M) in both PTSs, i.e., at position 3 in the PTS1 in lieu of L or position 2 in PTS2 in lieu of L/I. Examples include ATF, OPR3, AAE7, AAE12, SO, 6PGDH, MFP2, and IBR3 for PTS1 and MDH and TLP for PTS2. Another interesting observation is the change of PTSs in the orthologs. For example, for the two proteins predicted to work sequentially in the pseudouridine catabolism pathway, i.e., Indigoidine synthase A (IndA) and PfkB-type carbohydrate kinase family protein (pxPfkB; Eubel et al., 2008; Reumann, 2011), IndA harbors a PTS2 (RIX5HL), and pxPfkb has a PTS1 (SML>) in Arabidopsis, whereas in rice IndA contains a PTS1 (SAL>) and pxPfkb has a PTS2 (RMX5HL). In a similar case, the Arabidopsis HIT2 and HIT3 proteins both have PTS2 (RLX5HL and RVX5HF respectively) and HIT1 has a PTS1 (SKV>), whereas in rice all three putative peroxisomal HIT proteins have PTS2s (RLX5HL). A third example is the acquisition of a minor PTS2 (RQX4HL) in one of the putative homologs of epoxide hydrolase (EH) in rice.
Expansion and Contraction of Peroxisomal Protein Families in Rice
Rice is considered to be an ancient polyploid, as evidenced by remnants of duplicated blocks in its genome (Paterson et al., 2004). Its genome is predicted to have 1.5 times as many protein coding genes as those in Arabidopsis (Sasaki et al., 2008). Thus, many Arabidopsis peroxisome proteins or protein families have expanded in number in the rice genome, augmenting the number of rice peroxisome proteins.
Among the rice peroxisome proteins, PEX11a, MDAR4, polyamine oxidase (PAO), a protein of unknown function (UP3), AAE1, AAE7, and PM16 seem to have undergone tandem duplications. Duplication of genes is a recurrent evolutionary strategy that drives genetic diversity, and divergent expression of duplicated genes has shaped functional evolution of proteins and is crucial for their retention in the genome (Gu, 2003; Pal et al., 2006; Innan and Kondrashov, 2010). Consistent with this, the duplicated genes in each pair of the tandem duplicates of PAO, UP3, AAE7, and MDAR4 show highly dissimilar expressions (http://evolver.psc.riken.jp/seiken/OS/index.html).
The peroxisomal ATP-binding cassette (ABC) transporter PXA1/CTS/PED3 appears to have two homologs in rice. PXA1/CTS/PED3 has been attributed with a role in transporting β-oxidation substrates into the peroxisome, and mutant analysis in Arabidopsis uncovered a plethora of plant phenotypes associated with its malfunction (Zolman et al., 2001b; Hayashi et al., 2002; Theodoulou et al., 2006; Footitt et al., 2007b). In yeasts and mammals, the PXA1 function is executed by two proteins (each being a half transporter), which heterodimerize to form a functional complex (Hettema and Tabak, 2000; Wanders et al., 2007). However, the rice PXA1 homologs, like Arabidopsis PXA1, encode for full transporters and presumably have full activity. It will be worthwhile to explore as to whether the two rice homologs have different substrate specificities and whether this feature is unique to monocots.
Genes encoding several enzymes associated with the major peroxisomal functions, such as β-oxidation and related functions and detoxification, also increased in numbers in rice. Examples include fatty acid multifunctional protein (MFP), acyl-CoA oxidase (ACX), small thioesterase (sT), esterase/lipase/thioesterase family 1 (ELT1), OPR3 in JA biosynthesis, and napthoate synthase (NS), which has a predicted role in benzoate or phylloquinone metabolism. It will be interesting to investigate whether the acquisition of these additional copies of genes resulted in diversification of the enzymatic activities. β-oxidation activities are the primary source of H2O2 generation in peroxisomes. MDAR and APX are two major enzymes in the glutathione–ascorbate cycle, which serves to eliminate toxic H2O2. In line with the expanded core of β-oxidation enzymes, which are capable of generating H2O2, the antioxidative enzyme complement, including MDAR1, APX3, and GSTT, has also undergone concomitant expansion. EHs are involved in removal of toxic metabolites and enzymatic byproducts, thus fulfilling an important role in peroxisomal detoxification. In rice, there seems to have four peroxisome EHs, in contrast to the presence of a single EH in the Arabidopsis peroxisome. A study in Nicotiana suggests that peroxisomal EH may play a role in basal resistance during fungal infection (Wijekoon et al., 2011). It will be worthwhile to analyze if they are involved in pathogen response in rice as well.
Biotin is a vitamin and an important cofactor of enzymes in both decarboxylation and carboxylation reactions; their biosynthetic enzymes have been shown to be mostly mitochondrial (Smith et al., 2007; Asensi-Fabado and Munne-Bosch, 2010). Biotin F (7-keto-8-aminopelargonic acid synthase), an enzyme implicated in the first step of biotin biosynthesis, has been a recent and surprising addition to peroxisome localized proteins in Arabidopsis (Tanabe et al., 2011). Biotin F has two rice homologs, both of which contain the canonical PTS1, SKL. Production of vitamins is an economically important agricultural trait that is often exploited to enhance nutritional value of food crops (Potrykus, 2001; Beyer et al., 2002; Datta et al., 2003). Investigations into the impact of these proteins on the biotin content of the crops would define new functions of peroxisomes and be vital in understanding the contributions of peroxisomes in this process.
Betaine aldehyde dehydrogenases (BADH) metabolize 4-aminobutyraldehyde/Δ1-pyrroline and probably function in polyamine catabolism in peroxisomes. Rice has two copies of this gene, and the genetic basis of fragrance in rice was linked to the BADH2 locus. Interestingly, the non-functional BADH2 allele causes fragrance production, because the accumulation of the substrate, 4-aminobutyraldehyde/Δ1-pyrroline, in this allele enhances the synthesis of 2-acetyl-1-pyrroline, a major volatile responsible for aroma in rice (Bradbury et al., 2008; Chen et al., 2008). BADH1, on the other hand, was reported to oxidize acetaldehyde and might be important to relieve oxidative stress related to the submergence and re-aeration of rice plants (Mitsuya et al., 2009).
Two peroxisome membrane proteins, PEX22 and FIS1, which serve as scaffolds to recruit downstream proteins (PEX4 and DRP3, respectively) in peroxisome biogenesis (Zolman et al., 2005; Scott et al., 2006; Lingard et al., 2008; Zhang and Hu, 2008, 2009), both seem to have an additional homolog in rice. However, subcellular targeting analysis needs to be done to verify this observation, as targeting signals for peroxisome membrane proteins are hard to define.
Conversely, some Arabidopsis multigene family proteins only have a single equivalent gene in rice. PEX19 and PEX3 are involved in peroxisome membrane protein import and both have two isoforms in Arabidopsis (Kaur et al., 2009); yet only one copy each is found in rice. The two peroxisomal adenosine nucleotide transporters PNC1 and PNC2 were suggested to have arisen from genomic chromosomal rearrangements (Palmieri et al., 2011), therefore the occurrence of a single-copy PNC in rice is not unreasonable. Lastly, citrate synthase (CSY), glutamate:glyoxylate aminotransferase (GGT), 3-ketoacyl-CoA thiolase (KAT), and acetyltransferase (ATF) have multiple isoforms in Arabidopsis but seem to be encoded by a single gene in rice. Mutants for these single-copy genes may be promising candidates to bypass gene redundancy problems encountered in Arabidopsis to unveil the function of their protein products in plants.
Future Perspectives
Proteomics provide a wealth of information regarding organelle protein constituents, yet there are limitations to this approach. Many membrane proteins along with low-abundance proteins and proteins peripherally associated with peroxisomes tend to escape detection. In fact, none of the plant mass spectrometry-based proteome studies were successful in identifying most known peroxisomal membrane proteins, including those involved in various aspects of peroxisome biogenesis. However, many of their counterparts were successfully discovered in previous peroxisomal proteomics experiments in yeasts and mammals (Schafer et al., 2001; Kikuchi et al., 2004; Wiese et al., 2007). As such, there is still a lot of space for technology improvement to maximize the coverage of plant peroxisomal proteins, especially those associated with the membrane. In addition, peroxisomes and other organelles are not static entities within the cell. Some proteins may be accumulated or redistributed in the peroxisome in response to varied stimuli in a tissue-, environment-, or development-specific manner, and as a result, underrepresented in the analyzed proteome. Therefore, sampling of the peroxisome proteome at various developmental stages and under different environmental conditions may uncover proteins with provisional presence in these organelles.
Since experimental proteomics has its limitations in detecting low-abundance and transient peroxisomal proteins, this approach needs to be complemented by in silico protein prediction studies in order to completely decode the peroxisome proteome. Bioinformatics approaches have been very powerful in predicting peroxisomal proteins based on the presence of PTSs on them, allowing researchers to verify the predictions by subcellular localization studies (Kamada et al., 2003; Reumann et al., 2004; Lingner et al., 2011; Reumann, 2011). Furthermore, to assign functions to each newly identified peroxisomal protein, reverse genetics analysis and biochemical characterizations need to be conducted. In the Arabidopsis peroxisome 2010 project, we have analyzed more than 90 sequence-indexed T-DNA insertion mutants of over 50 novel peroxisomal genes through a series of physiological, biochemical, and cell biological assays to assess the roles of the corresponding proteins in peroxisomes. This systematic approach revealed the involvement of more peroxisomal proteins in embryogenesis, peroxisome protein import, and defense response (Cassin and Hu, unpublished). Other physiological assays coupled with metabolic profiling will need to be employed to elucidate the novel roles of plant peroxisomes, as our current tool box for analyzing peroxisome-related functions is only limited to the well known peroxisomal functions such as β-oxidation and photorespiration.
Knowledge gained through organelle proteomics can help us build increasingly complex models regarding the functions and regulation of these compartments, and map metabolic fluxes in relation to other organelles. Cross comparison of global environmental stress proteomics data using known peroxisome proteins identified proteins with changed abundance under salt (6PGL, NDPK1), cadmium (MDAR1, ATMS1, CAT3), and cold (CAT3) stresses (Taylor et al., 2009). Post-translational modification (PTM) of plant peroxisomal proteins is a field that has been unexplored, except for the phosphorylation of PMP38/PXN reported by Eubel et al. (2008). The list of Arabidopsis peroxisomal proteins can be used to query preexisting databases that compile data of global PTM events. Information extracted from the databases can then be used to formulate hypothesis, followed by experimental testing.
A systems biology approach, which combines functional genomics, proteomics, and computational tools, may help to establish a global network of peroxisome function in plants. Studies in yeasts at the systems level defined the network dynamics that control the response of yeast cells to fatty acids at multiple levels, including signaling, transcription, chromatin dynamics, and peroxisome biogenesis (Saleem et al., 2010a,b). Three transcription factors in yeast have been demonstrated to be directly responsible for transcriptional regulation of peroxisome biogenesis genes as well as metabolic enzymes therein during response to oleic acids (Gurvitz and Rottensteiner, 2006). Likewise, the mammalian nuclear receptor, PPARα, controls the activation of peroxisomal genes in response to metabolic stimuli (Desvergne and Wahli, 1999). Plant peroxisome proteins far outnumber those found in yeasts or mammals, i.e., over 160 in Arabidopsis and (putatively) more than 190 in rice, vs. 61 in Saccharomyces cerevisiae and 85 in humans (Schrader and Fahimi, 2008). Yet how transcriptional regulation of peroxisomal genes is accomplished in plants is largely unknown. Direct binding of two transcriptional factors to promoters of peroxisome genes in plants has been reported. The first is the bZIP transcription factor, HY5 homolog (HYH), which binds to the promoter of the peroxisome proliferator gene PEX11b and controls its light specific activation in a phytochrome A-dependent manner, resulting in light-induced peroxisome proliferation (Desai and Hu, 2008). In addition, using chromatin immunoprecipitation-on-chip analysis, PEX11b and a glyoxalase I homolog (GLX1) were found to be direct targets of the bHLH transcription factor POPEYE (PYE) under iron deplete conditions (Long et al., 2010). We can now use the inventory of Arabidopsis peroxisome proteins in combination with available global expression datasets to build transcriptional regulatory networks based on co-expression analysis. Mining such data should also enable us to connect common expression patterns to possible regulatory factors. This knowledge would be instrumental to broadening our understanding of what factors govern peroxisome protein composition in plants and how they relate to global environmental or developmental changes.
Given the agronomical importance of plant peroxisomes, extending the large scale proteome study of these organelles into crop plants will be highly beneficial to improving the quality, yield, and stress response of crop species. In addition to the in silico proteome analysis of rice peroxisomes performed in this study, experimental proteomics should be employed to understand the dynamic rice peroxisomal proteome in different tissues and developmental stages, and under various environmental cues. Comparison of the peroxisome proteomes in rice and Arabidopsis will shed light onto the evolution of peroxisomal functions in diverse plant lineages. Grass (Poaceae) genomes display extensive synteny (Devos and Gale, 1997), thus information gained from rice could be applied to other cereal crops such as maize and wheat, which are also prominent food crops worldwide.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to those colleagues whose works are not covered in this review. Work in the Hu lab was supported by grants from the National Science Foundation (MCB 0618335) and the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (DE-FG02-91ER20021) to Jianping Hu.
References
Aalen, R. B., Opsahl-Ferstad, H. G., Linnestad, C., and Olsen, O. A. (1994). Transcripts encoding an oleosin and a dormancy-related protein are present in both the aleurone layer and the embryo of developing barley (Hordeum vulgare L.) seeds. Plant J. 5, 385–396.
Aalen, R. B., Salehian, Z., and Steinum, T. M. (2001). Stability of barley aleurone transcripts: dependence on protein synthesis, influence of the starchy endosperm and destabilization by GA3. Physiol Plant 112, 403–413.
Abu-Abied, M., Avisar, D., Belausov, E., Holdengreber, V., Kam, Z., and Sadot, E. (2009). Identification of an Arabidopsis unknown small membrane protein targeted to mitochondria, chloroplasts, and peroxisomes. Protoplasma 236, 3–12.
Adham, A. R., Zolman, B. K., Millius, A., and Bartel, B. (2005). Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in beta-oxidation. Plant J. 41, 859–874.
Afitlhile, M. M., Fukushige, H., Nishimura, M., and Hildebrand, D. F. (2005). A defect in glyoxysomal fatty acid beta-oxidation reduces jasmonic acid accumulation in Arabidopsis. Plant Physiol. Biochem. 43, 603–609.
Albrecht, V., Simková, K., Carrie, C., Delannoy, E., Giraud, E., Whelan, J., Small, I. D., Apel, K., Badger, M. R., and Pogson, B. J. (2010). The cytoskeleton and the peroxisomal-targeted snowy cotyledon3 protein are required for chloroplast development in Arabidopsis. Plant Cell 22, 3423–3438.
Arai, Y., Hayashi, M., and Nishimura, M. (2008a). Proteomic analysis of highly purified peroxisomes from etiolated soybean cotyledons. Plant Cell Physiol. 49, 526–539.
Arai, Y., Hayashi, M., and Nishimura, M. (2008b). Proteomic identification and characterization of a novel peroxisomal adenine nucleotide transporter supplying ATP for fatty acid beta-oxidation in soybean and Arabidopsis. Plant Cell 20, 3227–3240.
Arent, S., Christensen, C. E., Pye, V. E., Norgaard, A., and Henriksen, A. (2010). The multifunctional protein in peroxisomal beta-oxidation: structure and substrate specificity of the Arabidopsis thaliana protein MFP2. J. Biol. Chem. 285, 24066–24077.
Arimura, S.-I., Aida, G. P., Fujimoto, M., Nakazono, M., and Tsutsumi, N. (2004). Arabidopsis dynamin-like protein 2a (ADL2a), like ADL2b, is involved in plant mitochondrial division. Plant Cell Physiol. 45, 236–242.
Arimura, S.-I., and Tsutsumi, N. (2002). A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc. Natl. Acad. Sci. U.S.A. 99, 5727–5731.
Asensi-Fabado, M. A., and Munne-Bosch, S. (2010). Vitamins in plants: occurrence, biosynthesis and antioxidant function. Trends Plant Sci. 15, 582–592.
Au, C. E., Bell, A. W., Gilchrist, A., Hiding, J., Nilsson, T., and Bergeron, J. J. (2007). Organellar proteomics to create the cell map. Curr. Opin. Cell Biol. 19, 376–385.
Aung, K., and Hu, J. (2011). The Arabidopsis tail-anchored protein peroxisomal and mitochondrial division factor1 is involved in the morphogenesis and proliferation of peroxisomes and mitochondria. Plant Cell doi: 10.1105/tpc.111.090142
Babujee, L., Wurtz, V., Ma, C., Lueder, F., Soni, P., van Dorsselaer, A., and Reumann, S. (2010). The proteome map of spinach leaf peroxisomes indicates partial compartmentalization of phylloquinone (vitamin K1) biosynthesis in plant peroxisomes. J. Exp. Bot. 61, 1441–1453.
Baker, A., Graham, I. A., Holdsworth, M., Smith, S. M., and Theodoulou, F. L. (2006). Chewing the fat: beta-oxidation in signalling and development. Trends Plant Sci. 11, 124–132.
Bardaweel, S., Ghosh, B., Chou, T. F., Sadowsky, M. J., and Wagner, C. R. (2011). E. coli histidine triad nucleotide binding protein 1 (ecHinT) is a catalytic regulator of D-alanine dehydrogenase (DadA) activity in vivo. PLoS ONE 6, e20897. doi: 10.1371/journal.pone.0020897
Bednarek, P. Ç., Pislewska-Bednarek, M., Svatos, A., Schneider, B., Doubsky, J., Mansurova, M., Humphry, M., Consonni, C., Panstruga, R., Sanchez-Vallet, A., Molina, A., and Schulze-Lefert, P. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106.
Bernhardt, K., Wilkinson, S., Weber, A. P., and Linka, N. (2011). A peroxisomal carrier delivers NAD(+) and contributes to optimal fatty acid degradation during storage oil mobilization. Plant J. doi: 10.1111/j.1365-313X.2011.04775.x
Beyer, P., Al-Babili, S., Ye, X., Lucca, P., Schaub, P., Welsch, R., and Potrykus, I. (2002). Golden rice: introducing the beta-carotene biosynthesis pathway into rice endosperm by genetic engineering to defeat vitamin A deficiency. J. Nutr. 132, 506S–510S.
Bieganowski, P., Garrison, P. N., Hodawadekar, S. C., Faye, G., Barnes, L. D., and Brenner, C. (2002). Adenosine monophosphoramidase activity of Hint and Hnt1 supports function of Kin28, Ccl1, and Tfb3. J. Biol. Chem. 277, 10852–10860.
Boisson-Dernier, A., Frietsch, S., Kim, T.-H., Dizon, M. B., and Schroeder, J. I. (2008). The peroxin loss-of-function mutation abstinence by mutual consent disrupts recognition between male and female gametophytes. Curr. Biol. 18, 63–68.
Bosnar, M. H., Bago, R., and Cetkovic, H. (2009). Subcellular localization of Nm23/NDPK A and B isoforms: a reflection of their biological function? Mol. Cell. Biochem. 329, 63–71.
Bradbury, L. M., Gillies, S. A., Brushett, D. J., Waters, D. L., and Henry, R. J. (2008). Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice. Plant Mol. Biol. 68, 439–449.
Brenner, C. (2002). Hint, Fhit, and GalT: function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases. Biochemistry 41, 9003–9014.
Brown, L. A., O’Leary-Steele, C., Brookes, P., Armitage, L., Kepinski, S., Warriner, S. L., and Baker, A. (2011). A small molecule with differential effects on the PTS1 and PTS2 peroxisome matrix import pathways. Plant J. 65, 980–990.
Carrie, C., Giraud, E., Duncan, O., Xu, L., Wang, Y., Huang, S., Clifton, R., Murcha, M., Filipovska, A., Rackham, O., Vrielink, A., and Whelan, J. (2010). Conserved and novel functions for Arabidopsis thaliana MIA40 in assembly of proteins in mitochondria and peroxisomes. J. Biol. Chem. 285, 36138–36148.
Carrie, C., Giraud, E., and Whelan, J. (2009). Protein transport in organelles: dual targeting of proteins to mitochondria and chloroplasts. FEBS J. 276, 1187–1195.
Carrie, C., Murcha, M., Millar, A., Smith, S., and Whelan, J. (2007). Nine 3-ketoacyl-CoA thiolases (KATs) and acetoacetyl-CoA thiolases (ACATs) encoded by five genes in Arabidopsis thaliana are targeted either to peroxisomes or cytosol but not to mitochondria. Plant Mol. Biol. 63, 97–108.
Carrie, C., Murcha, M. W., Kuehn, K., Duncan, O., Barthet, M., Smith, P. M., Eubel, H., Meyer, E., Day, D. A., Millar, A. H., and Whelan, J. (2008). Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett. 582, 3073–3079.
Castillo, M. C., Martinez, C., Buchala, A., Metraux, J.-P., and Leon, J. (2004). Gene-specific involvement of β-oxidation in wound-activated responses in Arabidopsis. Plant Physiol. 135, 85–94.
Charlton, W. L., Johnson, B., Graham, I. A., and Baker, A. (2005). Non-coordinate expression of peroxisome biogenesis, β-oxidation and glyoxylate cycle genes in mature Arabidopsis plants. Plant Cell Rep. 23, 647–653.
Chen, S., Yang, Y., Shi, W., Ji, Q., He, F., Zhang, Z., Cheng, Z., Liu, X., and Xu, M. (2008). Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance. Plant Cell 20, 1850–1861.
Clay, N. K., Adio, A. M., Denoux, C., Jander, G., and Ausubel, F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323, 95–101.
Coca, M., and San Segundo, B. (2010). AtCPK1 calcium-dependent protein kinase mediates pathogen resistance in Arabidopsis. Plant J. 63, 526–540.
Colasante, C., Ellis, M., Ruppert, T., and Voncken, F. (2006). Comparative proteomics of glycosomes from bloodstream form and procyclic culture form Trypanosoma brucei brucei. Proteomics 6, 3275–3293.
Cornah, J. E., Germain, V., Ward, J. L., Beale, M. H., and Smith, S. M. (2004). Lipid utilization, gluconeogenesis, and seedling growth in Arabidopsis mutants lacking the glyoxylate cycle enzyme malate synthase. J. Biol. Chem. 279, 42916–42923.
Corpas, F. J., Barroso, J. B., Sandalio, L. M., Distefano, S., Palma, J. M., Lupianez, J. A., and Del Rio, L. A. (1998). A dehydrogenase-mediated recycling system of NADPH in plant peroxisomes. Biochem. J. 330(Pt 2), 777–784.
Cousins, A. B., Pracharoenwattana, I., Zhou, W., Smith, S. M., and Badger, M. R. (2008). Peroxisomal malate dehydrogenase is not essential for photorespiration in Arabidopsis but its absence causes an increase in the stoichiometry of photorespiratory CO2 release. Plant Physiol. 148, 786–795.
Datta, K., Baisakh, N., Oliva, N., Torrizo, L., Abrigo, E., Tan, J., Rai, M., Rehana, S., Al-Babili, S., Beyer, P., Potrykus, I., and Datta, S. K. (2003). Bioengineered ‘golden’ indica rice cultivars with beta-carotene metabolism in the endosperm with hygromycin and mannose selection systems. Plant Biotechnol. J. 1, 81–90.
del Rio, L. A., Corpas, F. J., Sandalio, L. M., Palma, J. M., Gomez, M., and Barroso, J. B. (2002). Reactive oxygen species, antioxidant systems and nitric oxide in peroxisomes. J. Exp. Bot. 53, 1255–1272.
Delker, C., Zolman, B. K., Miersch, O., and Wasternack, C. (2007). Jasmonate biosynthesis in Arabidopsis thaliana requires peroxisomal [beta]-oxidation enzymes – additional proof by properties of pex6 and aim1. Phytochemistry 68, 1642–1650.
Desai, M., and Hu, J. (2008). Light induces peroxisome proliferation in Arabidopsis seedlings through the photoreceptor phytochrome A, the transcription factor HY5 homolog, and the peroxisomal protein PEROXIN11b. Plant Physiol. 146, 1117–1127.
Desvergne, B., and Wahli, W. (1999). Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev. 20, 649–688.
Devos, K. M., and Gale, M. D. (1997). Comparative genetics in the grasses. Plant Mol. Biol. 35, 3–15.
Dixon, D. P., Hawkins, T., Hussey, P. J., and Edwards, R. (2009). Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J. Exp. Bot. 60, 1207–1218.
Donaldson, R. P. (1982). Nicotinamide cofactors (NAD and NADP) in glyoxysomes, mitochondria, and plastids isolated from castor bean endosperm. Arch. Biochem. Biophys. 215, 274–279.
Du, Y.-Y., Wang, P.-C., Chen, J., and Song, C.-P. (2008). Comprehensive functional analysis of the catalase gene family in Arabidopsis thaliana. J. Integr. Plant Biol. 50, 1318–1326.
Eastmond, P. J. (2007). Monodehyroascorbate reductase4 is required for seed storage oil hydrolysis and postgerminative growth in Arabidopsis. Plant Cell 19, 1376–1387.
Eastmond, P. J., Germain, V., Lange, P. R., Bryce, J. H., Smith, S. M., and Graham, I. A. (2000a). Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc. Natl. Acad. Sci. U.S.A. 97, 5669–5674.
Eastmond, P. J., Hooks, M., and Graham, I. A. (2000b). The Arabidopsis acyl-CoA oxidase gene family. Biochem. Soc. Trans. 28, 755–757.
Eastmond, P. J., and Graham, I. A. (2000). The multifunctional protein AtMFP2 is co-ordinately expressed with other genes of fatty acid beta-oxidation during seed germination in Arabidopsis thaliana (L.) Heynh. Biochem. Soc. Trans. 28, 95–99.
Edqvist, J., Rönnberg, E., Rosenquist, S., Blomqvist, K., Viitanen, L., Salminen, T. A., Nylund, M., Tuuf, J., and Mattjus, P. (2004). Plants express a lipid transfer protein with high similarity to mammalian sterol carrier protein-2. J. Biol. Chem. 279, 53544–53553.
Eilers, T., Schwarz, G., Brinkmann, H., Witt, C., Richter, T., Nieder, J., Koch, B., Hille, R., Hänsch, R., and Mendel, R. R. (2001). Identification and biochemical characterization of Arabidopsis thaliana sulfite oxidase. A new player in plant sulfur metabolism. J. Biol. Chem. 276, 46989–46994.
Eubel, H., Meyer, E. H., Taylor, N. L., Bussell, J. D., O’Toole, N., Heazlewood, J. L., Castleden, I., Small, I. D., Smith, S. M., and Millar, A. H. (2008). Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol. 148, 1809–1829.
Fan, J., Quan, S., Orth, T., Awai, C., Chory, J., and Hu, J. (2005). The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol. 139, 231–239.
Flynn, C. R., Heinze, M., Schumann, U., Gietl, C., and Trelease, R. N. (2005). Compartmentalization of the plant peroxin, AtPex10p, within subdomain(s) of ER. Plant Sci. 168, 635–652.
Footitt, S., Cornah, J. E., Pracharoenwattana, I., Bryce, J. H., and Smith, S. M. (2007a). The Arabidopsis 3-ketoacyl-CoA thiolase-2 (kat2-1) mutant exhibits increased flowering but reduced reproductive success. J. Exp. Bot. 58, 2959–2968.
Footitt, S., Dietrich, D., Fait, A., Fernie, A. R., Holdsworth, M. J., Baker, A., and Theodoulou, F. L. (2007b). The COMATOSE ATP-binding cassette transporter is required for full fertility in Arabidopsis. Plant Physiol. 144, 1467–1480.
Footitt, S., Slocombe, S. P., Larner, V., Kurup, S., Wu, Y., Larson, T., Graham, I., Baker, A., and Holdsworth, M. (2002). Control of germination and lipid mobilization by comatose, the Arabidopsis homologue of human ALDP. EMBO J. 21, 2912–2922.
Foyer, C. H., and Noctor, G. (2003). Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol. Plant 119, 355–364.
Frederick, S. E., and Newcomb, E. H. (1969). Cyochemical localization of catalase in leaf microbodies (peroxisomes). J. Cell Biol. 43, 343–353.
Froman, B. E., Edwards, P. C., Bursch, A. G., and Dehesh, K. (2000). ACX3, a novel medium-chain acyl-coenzyme A oxidase from Arabidopsis. Plant Physiol. 123, 733–742.
Frugoli, J. A., Zhong, H. H., Nuccio, M. L., McCourt, P., McPeek, M. A., Thomas, T. L., and McClung, C. R. (1996). Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 112, 327–336.
Fujimoto, M., Arimura, S., Mano, S., Kondo, M., Saito, C., Ueda, T., Nakazono, M., Nakano, A., Nishimura, M., and Tsutsumi, N. (2009). Arabidopsis dynamin-related proteins DRP3A and DRP3B are functionally redundant in mitochondrial fission, but have distinct roles in peroxisomal fission. Plant J. 58, 388–400.
Fukao, Y., Hayashi, M., Hara-Nishimura, I., and Nishimura, M. (2003). Novel glyoxysomal protein kinase, GPK1, identified by proteomic analysis of glyoxysomes in etiolated cotyledons of Arabidopsis thaliana. Plant Cell Physiol. 44, 1002–1012.
Fukao, Y., Hayashi, M., and Nishimura, M. (2002). Proteomic analysis of leaf peroxisomal proteins in greening cotyledons of Arabidopsis thaliana. Plant Cell Physiol. 43, 689–696.
Fukao, Y., Hayashi, Y., Mano, S., Hayashi, M., and Nishimura, M. (2001). Developmental analysis of a putative ATP/ADP carrier protein localized on glyoxysomal membranes during the peroxisome transition in pumpkin cotyledons. Plant Cell Physiol. 42, 835–841.
Fulda, M., Schnurr, J., Abbadi, A., Heinz, E., and Browse, J. (2004). Peroxisomal Acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana. Plant Cell 16, 394–405.
Germain, V., Rylott, E. L., Larson, T. R., Sherson, S. M., Bechtold, N., Carde, J. P., Bryce, J. H., Graham, I. A., and Smith, S. M. (2001). Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid β-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. Plant J. 28, 1–12.
Goepfert, S., Hiltunen, J. K., and Poirier, Y. (2006). Identification and functional characterization of a monofunctional peroxisomal enoyl-CoA hydratase 2 that participates in the degradation of even cis-unsaturated fatty acids in Arabidopsis thaliana. J. Biol. Chem. 281, 35894–35903.
Goepfert, S., Vidoudez, C., Rezzonico, E., Hiltunen, J. K., and Poirier, Y. (2005). Molecular identification and characterization of the Arabidopsis {delta}3,5,{delta}2,4-Dienoyl-coenzyme a isomerase, a peroxisomal enzyme participating in the {beta}-oxidation cycle of unsaturated fatty acids. Plant Physiol. 138, 1947–1956.
Goepfert, S., Vidoudez, C., Tellgren-Roth, C., Delessert, S., Hiltunen, J. K., and Poirier, Y. (2008). Peroxisomal delta(3), delta(2)-enoyl CoA isomerases and evolution of cytosolic paralogues in embryophytes. Plant J. 56, 728–742.
Goto, S., Mano, S., Nakamori, C., and Nishimura, M. (2011). Arabidopsis aberrant peroxisome morphology9 is a peroxin that recruits the PEX1-PEX6 complex to peroxisomes. Plant Cell 23, 1573–1587.
Goyer, A., Johnson, T. L., Olsen, L. J., Collakova, E., Shachar-Hill, Y., Rhodes, D., and Hanson, A. D. (2004). Characterization and metabolic function of a peroxisomal sarcosine and pipecolate oxidase from Arabidopsis. J. Biol. Chem. 279, 16947–16953.
Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z. L., Powers, S. J., Gong, F., Phillips, A. L., Hedden, P., Sun, T. P., and Thomas, S. G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18, 3399–3414.
Gu, X. (2003). Evolution of duplicate genes versus genetic robustness against null mutations. Trends Genet. 19, 354–356.
Guranowski, A., Wojdyla, A. M., Zimny, J., Wypijewska, A., Kowalska, J., Jemielity, J., Davis, R. E., and Bieganowski, P. (2010). Dual activity of certain HIT-proteins: A. thaliana Hint4 and C. elegans DcpS act on adenosine 5′-phosphosulfate as hydrolases (forming AMP) and as phosphorylases (forming ADP). FEBS Lett. 584, 93–98.
Gurvitz, A., and Rottensteiner, H. (2006). The biochemistry of oleate induction: transcriptional upregulation and peroxisome proliferation. Biochim. Biophys. Acta 1763, 1392–1402.
Hadden, D. A., Phillipson, B. A., Johnston, K. A., Brown, L. A., Manfield, I. W., El-Shami, M., Sparkes, I. A., and Baker, A. (2006). Arabidopsis PEX19 is a dimeric protein that binds the peroxin PEX10. Mol. Membr. Biol. 23, 325–336.
Hayashi, H., De Bellis, L., Ciurli, A., Kondo, M., Hayashi, M., and Nishimura, M. (1999). A novel acyl-CoA oxidase that can oxidize short-chain acyl-CoA in plant peroxisomes. J. Biol. Chem. 274, 12715–12721.
Hayashi, M., and Nishimura, M. (2003). Entering a new era of research on plant peroxisomes. Curr. Opin. Plant Biol. 6, 577–582.
Hayashi, M., Nito, K., Takei-Hoshi, R., Yagi, M., Kondo, M., Suenaga, A., Yamaya, T., and Nishimura, M. (2002). Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid {beta}-oxidation. Plant Cell Physiol. 43, 1–11.
Hayashi, M., Nito, K., Toriyama-Kato, K., Kondo, M., Yamaya, T., and Nishimura, M. (2000). AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J. 19, 5701–5710.
Hayashi, M., Toriyama, K., Kondo, M., and Nishimura, M. (1998). 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid ss-oxidation. Plant Cell 10, 183–196.
Hayashi, M., Yagi, M., Nito, K., Kamada, T., and Nishimura, M. (2005). Differential contribution of two peroxisomal protein receptors to the maintenance of peroxisomal functions in Arabidopsis. J. Biol. Chem. 280, 14829–14835.
Helm, M., Lück, C., Prestele, J., Hierl, G., Huesgen, P. F., Fröhlich, T., Arnold, G. J., Adamska, I., Görg, A., Lottspeich, F., and Gietl, C. (2007). Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc. Natl. Acad. Sci. U.S.A. 104, 11501–11506.
Henke, B., Girzalsky, W., Berteaux-Lecellier, V., and Erdmann, R. (1998). IDP3 encodes a peroxisomal NADP-dependent isocitrate dehydrogenase required for the beta-oxidation of unsaturated fatty acids. J. Biol. Chem. 273, 3702–3711.
Hettema, E. H., and Tabak, H. F. (2000). Transport of fatty acids and metabolites across the peroxisomal membrane. Biochim. Biophys. Acta 1486, 18–27.
Hooks, M. A., Kellas, F., and Graham, I. A. (1999). Long-chain acyl-CoA oxidases of Arabidopsis. Plant J. 20, 1–13.
Hooks, M. A., Turner, J. E., Murphy, E. C., Johnston, K. A., Burr, S., and Jaroslawski, S. (2007). The Arabidopsis ALDP protein homologue COMATOSE is instrumental in peroxisomal acetate metabolism. Biochem. J. 406, 399–406.
Hu, J., Aguirre, M., Peto, C., Alonso, J., Ecker, J., and Chory, J. (2002). A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297, 405–409.
Hunt, J. E., and Trelease, R. N. (2004). Sorting pathway and molecular targeting signals for the Arabidopsis peroxin 3. Biochem. Biophys. Res. Commun. 314, 586–596.
Igarashi, D., Miwa, T., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K., and Ohsumi, C. (2003). Identification of photorespiratory glutamate:glyoxylate aminotransferase (GGAT) gene in Arabidopsis. Plant J. 33, 975–987.
Innan, H., and Kondrashov, F. (2010). The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. 11, 97–108.
Inoue, Y., Maeta, K., and Nomura, W. (2011). Glyoxalase system in yeasts: structure, function, and physiology. Semin. Cell Dev. Biol. 22, 278–284.
Jimenez, A., Hernandez, J. A., Del Rio, L. A., and Sevilla, F. (1997). Evidence for the presence of the ascorbate-glutathione cycle in mitochondria and peroxisomes of pea leaves. Plant Physiol. 114, 275–284.
Juhnke, H., Krems, B., Kotter, P., and Entian, K. D. (1996). Mutants that show increased sensitivity to hydrogen peroxide reveal an important role for the pentose phosphate pathway in protection of yeast against oxidative stress. Mol. Gen. Genet. 252, 456–464.
Kamada, T., Nito, K., Hayashi, H., Mano, S., Hayashi, M., and Nishimura, M. (2003). Functional differentiation of peroxisomes revealed by expression profiles of peroxisomal genes in Arabidopsis thaliana. Plant Cell Physiol. 44, 1275–1289.
Kamada-Nobusada, T., Hayashi, M., Fukazawa, M., Sakakibara, H., and Nishimura, M. (2008). A putative peroxisomal polyamine oxidase, AtPAO4, is involved in polyamine catabolism in Arabidopsis thaliana. Plant Cell Physiol. 49, 1272–1282.
Karnik, S. K., and Trelease, R. N. (2005). Arabidopsis peroxin 16 coexists at steady state in peroxisomes and endoplasmic reticulum. Plant Physiol. 138, 1967–1981.
Karnik, S. K., and Trelease, R. N. (2007). Arabidopsis peroxin 16 trafficks through the ER and an intermediate compartment to pre-existing peroxisomes via overlapping molecular targeting signals. J. Exp. Bot. 58, 1677–1693.
Kataya, A., and Reumann, S. (2010). Arabidopsis glutathione reductase 1 is dually targeted to peroxisomes and the cytosol. Plant Signal. Behav. 5, 171–175.
Kaur, N., Reumann, S., and Hu, J. (2009). “Peroxisome biogenesis and function,” in The Arabidopsis Book, (Rockville, MD: American Society of Plant Biologists). doi: 10.1199/tab.0123
Khan, B. R., and Zolman, B. K. (2010). pex5 Mutants that differentially disrupt PTS1 and PTS2 peroxisomal matrix protein import in Arabidopsis. Plant Physiol. 154, 1602–1615.
Kienow, L., Schneider, K., Bartsch, M., Stuible, H. P., Weng, H., Miersch, O., Wasternack, C., and Kombrink, E. (2008). Jasmonates meet fatty acids: functional analysis of a new acyl-coenzyme A synthetase family from Arabidopsis thaliana. J. Exp. Bot. 59, 403–419.
Kikuchi, M., Hatano, N., Yokota, S., Shimozawa, N., Imanaka, T., and Taniguchi, H. (2004). Proteomic analysis of rat liver peroxisome: presence of peroxisome-specific isozyme of Lon protease. J. Biol. Chem. 279, 421–428.
Kirch, H. H., Bartels, D., Wei, Y., Schnable, P. S., and Wood, A. J. (2004). The ALDH gene superfamily of Arabidopsis. Trends Plant Sci. 9, 371–377.
Kliebenstein, D. J., Monde, R.-A., and Last, R. L. (1998). Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 118, 637–650.
Koo, A. J. K., Chung, H. S., Kobayashi, Y., and Howe, G. A. (2006). Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J. Biol. Chem. 281, 33511–33520.
Kruger, N. J., and von Schaewen, A. (2003). The oxidative pentose phosphate pathway: structure and organisation. Curr. Opin. Plant Biol. 6, 236–246.
Kunz, H.-H., Scharnewski, M., Feussner, K., Feussner, I., Flügge, U. I., Fulda, M., and Gierth, M. (2009). The ABC transporter PXA1 and peroxisomal {beta}-oxidation are vital for metabolism in mature leaves of Arabidopsis during extended darkness. Plant Cell 21, 2733–2749.
Lamberto, I., Percudani, R., Gatti, R., Folli, C., and Petrucco, S. (2010). Conserved alternative splicing of Arabidopsis transthyretin-like determines protein localization and S-allantoin synthesis in peroxisomes. Plant Cell 22, 1564–1574.
Lang, C., Popko, J., Wirtz, M., Hell, R., Herschbach, C., Kreuzwieser, J., Rennenberg, H., Mendel, R. R., and Hänsch, R. (2007). Sulphite oxidase as key enzyme for protecting plants against sulphur dioxide. Plant Cell Environ. 30, 447–455.
Lange, P. R., Eastmond, P. J., Madagan, K., and Graham, I. A. (2004). An Arabidopsis mutant disrupted in valine catabolism is also compromised in peroxisomal fatty acid β-oxidation. FEBS Lett. 571, 147–153.
Leonardi, R., Zhang, Y. M., Rock, C. O., and Jackowski, S. (2005). Coenzyme A: back in action. Prog. Lipid Res. 44, 125–153.
Li, J. F., Park, E., von Arnim, A. G., and Nebenfuhr, A. (2009). The FAST technique: a simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 5, 6.
Liepman, A. H., and Olsen, L. J. (2003). Alanine aminotransferase homologs catalyze the glutamate:glyoxylate aminotransferase reaction in peroxisomes of Arabidopsis. Plant Physiol. 131, 215–227.
Lin, Y., Cluette-Brown, J. E., and Goodman, H. M. (2004). The peroxisome deficient Arabidopsis mutant sse1 exhibits impaired fatty acid synthesis. Plant Physiol. 135, 814–827.
Lin, Y., Sun, L., Nguyen, L. V., Rachubinski, R. A., and Goodman, H. M. (1999). The Pex16p homolog SSE1 and storage organelle formation in Arabidopsis seeds. Science 284, 328–330.
Lingard, M. J., and Bartel, B. (2009). Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiol. 151, 1354–1365.
Lingard, M. J., Gidda, S. K., Bingham, S., Rothstein, S. J., Mullen, R. T., and Trelease, R. N. (2008). Arabidopsis peroxin11c-e, fission1B, and dynamin-related protein3A cooperate in cell cycle-associated replication of peroxisomes. Plant Cell 20, 1567–1585.
Lingard, M. J., Monroe-Augustus, M., and Bartel, B. (2009). Peroxisome-associated matrix protein degradation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 4561–4566.
Lingard, M. J., and Trelease, R. N. (2006). Five Arabidopsis peroxin 11 homologs individually promote peroxisome elongation, duplication or aggregation. J. Cell. Sci. 119, 1961–1972.
Lingner, T., Kataya, A. R., Antonicelli, G. E., Benichou, A., Nilssen, K., Chen, X. Y., Siemsen, T., Morgenstern, B., Meinicke, P., and Reumann, S. (2011). Identification of novel plant peroxisomal targeting signals by a combination of machine learning methods and in vivo subcellular targeting analyses. Plant Cell 23, 1556–1572.
Linka, N., Theodoulou, F. L., Haslam, R. P., Linka, M., Napier, J. A., Neuhaus, H. E., and Weber, A. P. (2008). Peroxisomal ATP import is essential for seedling development in Arabidopsis thaliana. Plant Cell 20, 3241–3257.
Lipka, V., Dittgen, J., Bednarek, P., Bhat, R., Wiermer, M., Stein, M., Landtag, J., Brandt, W., Rosahl, S., Scheel, D., Llorente, F., Molina, A., Parker, J., Somerville, S., and Schulze-Lefert, P. (2005). Pre- and post-invasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310, 1180–1183.
Lisenbee, C. S., Heinze, M., and Trelease, R. N. (2003). Peroxisomal ascorbate peroxidase resides within a subdomain of rough endoplasmic reticulum in wild-type Arabidopsis Cells. Plant Physiol. 132, 870–882.
Lisenbee, C. S., Lingard, M. J., and Trelease, R. N. (2005). Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase. Plant J. 43, 900–914.
Long, T. A., Tsukagoshi, H., Busch, W., Lahner, B., Salt, D. E., and Benfey, P. N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22, 2219–2236.
Ma, C., Haslbeck, M., Babujee, L., Jahn, O., and Reumann, S. (2006). Identification and characterization of a stress-inducible and a constitutive small heat-shock protein targeted to the matrix of plant peroxisomes. Plant Physiol. 141, 47–60.
Ma, C., and Reumann, S. (2008). Improved prediction of peroxisomal PTS1 proteins from genome sequences based on experimental subcellular targeting analyses as exemplified for protein kinases from Arabidopsis. J. Exp. Bot. 59, 3767–3779.
Maeda, K., Houjyou, Y., Komatsu, T., Hori, H., Kodaira, T., and Ishikawa, A. (2009). AGB1 and PMR5 contribute to PEN2-mediated preinvasion resistance to Magnaporthe oryzae in Arabidopsis thaliana. Mol. Plant Microbe Interact. 22, 1331–1340.
Mannervik, B. (2008). Molecular enzymology of the glyoxalase system. Drug Metabol. Drug Interact. 23, 13–27.
Mano, S., Hayashi, M., Kondo, M., and Nishimura, M. (1997). Hydroxypyruvate reductase with a carboxy-terminal targeting signal to microbodies is expressed in Arabidopsis. Plant Cell Physiol. 38, 449–455.
Mano, S., Nakamori, C., Kondo, M., Hayashi, M., and Nishimura, M. (2004). An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J. 38, 487–498.
Mano, S., Nakamori, C., Nito, K., Kondo, M., and Nishimura, M. (2006). The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. Plant J. 47, 604–618.
Martin, J., St-Pierre, M. V., and Dufour, J. F. (2011). Hit proteins, mitochondria and cancer. Biochim. Biophys. Acta 1807, 626–632.
Mateos, R. M., Leon, A. M., Sandalio, L. M., Gomez, M., del Rio, L. A., and Palma, J. M. (2003). Peroxisomes from pepper fruits (Capsicum annuum L.): purification, characterisation and antioxidant activity. J. Plant Physiol. 160, 1507–1516.
McIntosh, S., Watson, L., Bundock, P., Crawford, A., White, J., Cordeiro, G., Barbary, D., Rooke, L., and Henry, R. (2007). SAGE of the developing wheat caryopsis. Plant Biotechnol. J. 5, 69–83.
Mettler, I. J., and Beevers, H. (1980). Oxidation of NADH in glyoxysomes by a malate-aspartate shuttle. Plant Physiol. 66, 555–560.
Meyer, T., Holscher, C., Schwoppe, C., and von Schaewen, A. (2011). Alternative targeting of Arabidopsis plastidic glucose-6-phosphate dehydrogenase G6PD1 involves cysteine-dependent interaction with G6PD4 in the cytosol. Plant J. 66, 745–758.
Minard, K. I., and McAlister-Henn, L. (1999). Dependence of peroxisomal beta-oxidation on cytosolic sources of NADPH. J. Biol. Chem. 274, 3402–3406.
Mitsuya, S., Yokota, Y., Fujiwara, T., Mori, N., and Takabe, T. (2009). OsBADH1 is possibly involved in acetaldehyde oxidation in rice plant peroxisomes. FEBS Lett. 583, 3625–3629.
Mittova, V., Tal, M., Volokita, M., and Guy, M. (2003). Up-regulation of the leaf mitochondrial and peroxisomal antioxidative systems in response to salt-induced oxidative stress in the wild salt-tolerant tomato species Lycopersicon pennellii. Plant Cell Environ. 26, 845–856.
Moschou, P. N., Sanmartin, M., Andriopoulou, A. H., Rojo, E., Sanchez-Serrano, J. J., and Roubelakis-Angelakis, K. A. (2008). Bridging the gap between plant and mammalian polyamine catabolism: a novel peroxisomal polyamine oxidase responsible for a full back-conversion pathway in Arabidopsis. Plant Physiol. 147, 1845–1857.
Murphy, M. A., Phillipson, B. A., Baker, A., and Mullen, R. T. (2003). Characterization of the targeting signal of the Arabidopsis 22-kD integral peroxisomal membrane protein. Plant Physiol. 133, 813–828.
Narendra, S., Venkataramani, S., Shen, G., Wang, J., Pasapula, V., Lin, Y., Kornyeyev, D., Holaday, A. S., and Zhang, H. (2006). The Arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for Arabidopsis growth and development. J. Exp. Bot. 57, 3033–3042.
Nito, K., Hayashi, M., and Nishimura, M. (2002). Direct interaction and determination of binding domains among peroxisomal import factors in Arabidopsis thaliana. Plant Cell Physiol. 43, 355–366.
Nito, K., Kamigaki, A., Kondo, M., Hayashi, M., and Nishimura, M. (2007). Functional classification of Arabidopsis peroxisome biogenesis factors proposed from analyses of knockdown mutants. Plant Cell Physiol. 48, 763–774.
Nyathi, Y., and Baker, A. (2006). Plant peroxisomes as a source of signalling molecules. Biochim. Biophys. Acta 1763, 1478–1495.
Nyathi, Y., De Marcos Lousa, C., van Roermund, C. W., Wanders, R. J., Johnson, B., Baldwin, S. A., Theodoulou, F. L., and Baker, A. (2010). The Arabidopsis peroxisomal ABC transporter, comatose, complements the Saccharomyces cerevisiae pxa1 pxa2Delta mutant for metabolism of long-chain fatty acids and exhibits fatty acyl-CoA-stimulated ATPase activity. J. Biol. Chem. 285, 29892–29902.
Olsen, L. J., Ettinger, W. F., Damsz, B., Matsudaira, K., Webb, M. A., and Harada, J. J. (1993). Targeting of glyoxysomal proteins to peroxisomes in leaves and roots of a higher plant. Plant Cell 5, 941–952.
Ono, Y., Kim, D. W., Watanabe, K., Sasaki, A., Niitsu, M., Berberich, T., Kusano, T., and Takahashi, Y. (2011). Constitutively and highly expressed Oryza sativa polyamine oxidases localize in peroxisomes and catalyze polyamine back conversion. Amino Acids. doi: 10.1007/s00726-011-1002-3
Orth, T., Reumann, S., Zhang, X., Fan, J., Wenzel, D., Quan, S., and Hu, J. (2007). The PEROXIN11 protein family controls peroxisome proliferation in Arabidopsis. Plant Cell 19, 333–350.
Pal, C., Papp, B., and Lercher, M. J. (2006). An integrated view of protein evolution. Nat. Rev. 7, 337–348.
Palma, J. M., Corpas, F. J., and del Rio, L. A. (2009). Proteome of plant peroxisomes: new perspectives on the role of these organelles in cell biology. Proteomics 9, 2301–2312.
Palmieri, F., Pierri, C. L., De Grassi, A., Nunes-Nesi, A., and Fernie, A. R. (2011). Evolution, structure and function of mitochondrial carriers: a review with new insights. Plant J. 66, 161–181.
Pappa, A., Chen, C., Koutalos, Y., Townsend, A. J., and Vasiliou, V. (2003). Aldh3a1 protects human corneal epithelial cells from ultraviolet- and 4-hydroxy-2-nonenal-induced oxidative damage. Free Radic. Biol. Med. 34, 1178–1189.
Paterson, A. H., Bowers, J. E., and Chapman, B. A. (2004). Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. U.S.A. 101, 9903–9908.
Pinfield-Wells, H., Rylott, E. L., Gilday, A. D., Graham, S., Job, K., Larson, T. R., and Graham, I. A. (2005). Sucrose rescues seedling establishment but not germination of Arabidopsis mutants disrupted in peroxisomal fatty acid catabolism. Plant J. 43, 861–872.
Pollak, N., Dolle, C., and Ziegler, M. (2007). The power to reduce: pyridine nucleotides – small molecules with a multitude of functions. Biochem. J. 402, 205–218.
Pool, M. R., Lopez-Huertas, E., Horng, J. T., and Baker, A. (1998). NADPH is a specific inhibitor of protein import into glyoxysomes. Plant J. 15, 1–14.
Pracharoenwattana, I., Cornah, J. E., and Smith, S. M. (2005). Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell 17, 2037–2048.
Pracharoenwattana, I., Cornah, J. E., and Smith, S. M. (2007). Arabidopsis peroxisomal malate dehydrogenase functions in β-oxidation but not in the glyoxylate cycle. Plant J. 50, 381–390.
Pracharoenwattana, I., Zhou, W., and Smith, S. (2010). Fatty acid beta-oxidation in germinating Arabidopsis seeds is supported by peroxisomal hydroxypyruvate reductase when malate dehydrogenase is absent. Plant Mol. Biol. 72, 101–109.
Prestele, J., Hierl, G., Scherling, C., Hetkamp, S., Schwechheimer, C., Isono, E., Weckwerth, W., Wanner, G., and Gietl, C. (2010). Different functions of the C3HC4 zinc ring finger peroxins PEX10, PEX2, and PEX12 in peroxisome formation and matrix protein import. Proc. Natl. Acad. Sci. U.S.A. 107, 14915–14920.
Pye, V. E., Christensen, C. E., Dyer, J. H., Arent, S., and Henriksen, A. (2010). Peroxisomal plant 3-ketoacyl-CoA thiolase structure and activity are regulated by a sensitive redox switch. J. Biol. Chem. 285, 24078–24088.
Quan, S., Switzenberg, R., Reumann, S., and Hu, J. (2010). In vivo subcellular targeting analysis validates a novel peroxisome targeting signal type 2 and the peroxisomal localization of two proteins with putative functions in defense in Arabidopsis. Plant Signal. Behav. 5, 151–153.
Ramon, N. M., and Bartel, B. (2011). Interdependence of the peroxisome-targeting receptors in Arabidopsis thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into peroxisomes. Mol. Biol. Cell 21, 1263–1271.
Reumann, S. (2011). Toward a definition of the complete proteome of plant peroxisomes: where experimental proteomics must be complemented by bioinformatics. Proteomics 11, 1764–1779.
Reumann, S., Babujee, L., Ma, C., Wienkoop, S., Siemsen, T., Antonicelli, G. E., Rasche, N., Lüder, F., Weckwerth, W., and Jahn, O. (2007). Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19, 3170–3193.
Reumann, S., Ma, C., Lemke, S., and Babujee, L. (2004). AraPerox. A database of putative Arabidopsis proteins from plant peroxisomes. Plant Physiol. 136, 2587–2608.
Reumann, S., Quan, S., Aung, K., Yang, P., Manandhar-Shrestha, K., Holbrook, D., Linka, N., Switzenberg, R., Wilkerson, C. G., Weber, A. P., Olsen, L. J., and Hu, J. (2009). In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 150, 125–143.
Reumann, S., and Weber, A. P. (2006). Plant peroxisomes respire in the light: some gaps of the photorespiratory C2 cycle have become filled – others remain. Biochim. Biophys. Acta 1763, 1496–1510.
Richmond, T. A., and Bleecker, A. B. (1999). A defect in {beta}-oxidation causes abnormal inflorescence development in Arabidopsis. Plant Cell 11, 1911–1924.
Rubio, S., Larson, T. R., Gonzalez-Guzman, M., Alejandro, S., Graham, I. A., Serrano, R., and Rodriguez, P. L. (2006). An Arabidopsis mutant impaired in coenzyme A biosynthesis is sugar dependent for seedling establishment. Plant Physiol. 140, 830–843.
Rubio, S., Whitehead, L., Larson, T. R., Graham, I. A., and Rodriguez, P. L. (2008). The coenzyme a biosynthetic enzyme phosphopantetheine adenylyltransferase plays a crucial role in plant growth, salt/osmotic stress resistance, and seed lipid storage. Plant Physiol. 148, 546–556.
Rucktaschel, R., Girzalsky, W., and Erdmann, R. (2011). Protein import machineries of peroxisomes. Biochim. Biophys. Acta 1808, 892–900.
Rylott, E. L., Eastmond, P. J., Gilday, A. D., Slocombe, S. P., Larson, T. R., Baker, A., and Graham, I. A. (2006). The Arabidopsis thaliana multifunctional protein gene (MFP2) of peroxisomal β-oxidation is essential for seedling establishment. Plant J. 45, 930–941.
Rylott, E. L., Rogers, C. A., Gilday, A. D., Edgell, T., Larson, T. R., and Graham, I. A. (2003). Arabidopsis mutants in short- and medium-chain acyl-CoA oxidase activities accumulate acyl-CoAs and reveal that fatty acid {beta}-oxidation is essential for embryo development. J. Biol. Chem. 278, 21370–21377.
Sage, T. L., and Sage, R. F. (2009). The functional anatomy of rice leaves: implications for refixation of photorespiratory CO2 and efforts to engineer C4 photosynthesis into rice. Plant Cell Physiol. 50, 756–772.
Saleem, R. A., Rogers, R. S., Ratushny, A. V., Dilworth, D. J., Shannon, P. T., Shteynberg, D., Wan, Y., Moritz, R. L., Nesvizhskii, A. I., Rachubinski, R. A., and Aitchison, J. D. (2010a). Integrated phosphoproteomics analysis of a signaling network governing nutrient response and peroxisome induction. Mol. Cell Proteomics 9, 2076–2088.
Saleem, R. A., Long-O’Donnell, R., Dilworth, D. J., Armstrong, A. M., Jamakhandi, A. P., Wan, Y., Knijnenburg, T. A., Niemisto, A., Boyle, J., Rachubinski, R. A., Shmulevich, I., and Aitchison, J. D. (2010b). Genome-wide analysis of effectors of peroxisome biogenesis. PLoS ONE 5, e11953. doi: 10.1371/journal.pone.0011953
Saleem, R. A., Smith, J. J., and Aitchison, J. D. (2006). Proteomics of the peroxisome. Biochim. Biophys. Acta 1763, 1541–1551.
Sanders, P. M., Lee, P. Y., Biesgen, C., Boone, J. D., Beals, T. P., Weiler, E. W., and Goldberg, R. B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12, 1041–1062.
Sasaki, T., Wu, J., Mizuno, H., Antonio, B. A., and Matsumoto, T. (2008). “The rice genome sequence as an indispensable tool for crop improvement,” in Rice Biology in the Genomics Era, eds H.-Y. Hirano, A. Hirai, Y. Sano, and T. Sasaki (Heidelberg: Springer-Verlag Berlin), 3–12.
Schafer, H., Nau, K., Sickmann, A., Erdmann, R., and Meyer, H. E. (2001). Identification of peroxisomal membrane proteins of Saccharomyces cerevisiae by mass spectrometry. Electrophoresis 22, 2955–2968.
Schaller, F., Biesgen, C., Müssig, C., Altmann, T., and Weiler, E. W. (2000). 12-Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 210, 979–984.
Schilmiller, A. L., Koo, A. J. K., and Howe, G. A. (2007). Functional diversification of acyl-coenzyme A oxidases in jasmonic acid biosynthesis and action. Plant Physiol. 143, 812–824.
Schneider, K., Kienow, L., Schmelzer, E., Colby, T., Bartsch, M., Miersch, O., Wasternack, C., Kombrink, E., and Stuible, H. P. (2005). A new type of peroxisomal acyl-coenzyme A synthetase from Arabidopsis thaliana has the catalytic capacity to activate biosynthetic precursors of jasmonic acid. J. Biol. Chem. 280, 13962–13972.
Schrader, M., and Fahimi, H. D. (2008). The peroxisome: still a mysterious organelle. Histochem. Cell Biol. 129, 421–440.
Schuhmann, H., Huesgen, P. F., Gietl, C., and Adamska, I. (2008). The DEG15 serine protease cleaves peroxisomal targeting signal 2-containing proteins in Arabidopsis. Plant Physiol. 148, 1847–1856.
Schultz, C. J., and Coruzzi, G. M. (1995). The aspartate aminotransferase gene family of Arabidopsis encodes isoenzymes localized to three distinct subcellular compartments. Plant J. 7, 61–75.
Schumann, U., Prestele, J., O’Geen, H., Brueggeman, R., Wanner, G., and Gietl, C. (2007). Requirement of the C3HC4 zinc RING finger of the Arabidopsis PEX10 for photorespiration and leaf peroxisome contact with chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 104, 1069–1074.
Schumann, U., Wanner, G., Veenhuis, M., Schmid, M., and Gietl, C. (2003). AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 9626–9631.
Scott, I., Tobin, A. K., and Logan, D. C. (2006). BIGYIN, an orthologue of human and yeast FIS1 genes functions in the control of mitochondrial size and number in Arabidopsis thaliana. J. Exp. Bot. 57, 1275–1280.
Shockey, J. M., Fulda, M. S., and Browse, J. (2003). Arabidopsis contains a large superfamily of acyl-activating enzymes. Phylogenetic and biochemical analysis reveals a new class of acyl-coenzyme A synthetases. Plant Physiol. 132, 1065–1076.
Singh, T., Hayashi, M., Mano, S., Arai, Y., Goto, S., and Nishimura, M. (2009). Molecular components required for the targeting of PEX7 to peroxisomes in Arabidopsis thaliana. Plant J. 60, 488–498.
Smith, A. G., Croft, M. T., Moulin, M., and Webb, M. E. (2007). Plants need their vitamins too. Curr. Opin. Plant Biol. 10, 266–275.
Sparkes, I. A., Brandizzi, F., Slocombe, S. P., El-Shami, M., Hawes, C., and Baker, A. (2003). An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol. 133, 1809–1819.
Sparkes, I. A., Hawes, C., and Baker, A. (2005). AtPEX2 and AtPEX10 are targeted to peroxisomes independently of known endoplasmic reticulum trafficking routes. Plant Physiol. 139, 690–700.
Steinum, T. M., Berner, H. S., S tacy, R. A. P., Salehian, Z., and Aalen, R. B. (1998). Differential regulation of the barley (Hordeum vulgare) transcripts B12D and B22E in mature aleuron layers. Physiol. Plant 102, 337–345.
Stintzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. U.S.A. 97, 10625–10630.
Strader, L. C., Wheeler, D. L., Christensen, S. E., Berens, J. C., Cohen, J. D., Rampey, R. A., and Bartel, B. (2011). Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 23, 984–999.
Tanabe, Y., Maruyama, J., Yamaoka, S., Yahagi, D., Matsuo, I., Tsutsumi, N., and Kitamoto, K. (2011). Peroxisomes are involved in biotin biosynthesis in Aspergillus and Arabidopsis. J. Biol. Chem. 286, 30455–30461.
Taylor, N. L., Tan, Y. F., Jacoby, R. P., and Millar, A. H. (2009). Abiotic environmental stress induced changes in the Arabidopsis thaliana chloroplast, mitochondria and peroxisome proteomes. J. Proteomics 72, 367–378.
Theodoulou, F. L., Holdsworth, M., and Baker, A. (2006). Peroxisomal ABC transporters. FEBS Lett. 580, 1139–1155.
Theodoulou, F. L., Job, K., Slocombe, S. P., Footitt, S., Holdsworth, M., Baker, A., Larson, T. R., and Graham, I. A. (2005). Jasmonic acid levels are reduced in comatose ATP-binding cassette transporter mutants. Implications for transport of jasmonate precursors into peroxisomes. Plant Physiol. 137, 835–840.
Tilton, G., Shockey, J., and Browse, J. (2000). Two families of acyl-CoA thioesterases in Arabidopsis. Biochem. Soc. Trans. 28, 946–947.
Tilton, G. B., Shockey, J. M., and Browse, J. (2004). Biochemical and Molecular characterization of ACH2, an acyl-CoA thioesterase from Arabidopsis thaliana. J. Biol. Chem. 279, 7487–7494.
Tilton, G. B., Wedemeyer, W. J., Browse, J., and Ohlrogge, J. (2006). Plant coenzyme A biosynthesis: characterization of two pantothenate kinases from Arabidopsis. Plant Mol. Biol. 61, 629–642.
Tugal, H. B., Pool, M., and Baker, A. (1999). Arabidopsis 22-kilodalton peroxisomal membrane protein. nucleotide sequence analysis and biochemical characterization. Plant Physiol. 120, 309–320.
Turner, J. E., Greville, K., Murphy, E. C., and Hooks, M. A. (2005a). Characterization of Arabidopsis fluoroacetate-resistant mutants reveals the principal mechanism of acetate activation for entry into the glyoxylate cycle. J. Biol. Chem. 280, 2780–2787.
Turner, W. L., Waller, J. C., and Snedden, W. A. (2005b). Identification, molecular cloning and functional characterization of a novel NADH kinase from Arabidopsis thaliana (thale cress). Biochem. J. 385, 217–223.
Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T. Y., Hsing, Y. I., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698.
van Roermund, C. W., Hettema, E. H., Kal, A. J., van den Berg, M., Tabak, H. F., and Wanders, R. J. (1998). Peroxisomal beta-oxidation of polyunsaturated fatty acids in Saccharomyces cerevisiae: isocitrate dehydrogenase provides NADPH for reduction of double bonds at even positions. EMBO J. 17, 677–687.
Vasiliou, V., and Nebert, D. W. (2005). Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum. Genomics 2, 138–143.
Waller, J. C., Dhanoa, P. K., Schumann, U., Mullen, R. T., and Snedden, W. A. (2010). Subcellular and tissue localization of NAD kinases from Arabidopsis: compartmentalization of de novo NADP biosynthesis. Planta 231, 305–317.
Wanders, R. J., Visser, W. F., van Roermund, C. W., Kemp, S., and Waterham, H. R. (2007). The peroxisomal ABC transporter family. Pflugers Arch. 453, 719–734.
Westphal, L., Scheel, D., and Rosahl, S. (2008). The coi1-16 mutant harbors a second site mutation rendering PEN2 nonfunctional. Plant Cell 20, 824–826.
Wiederhold, E., Veenhoff, L. M., Poolman, B., and Slotboom, D. J. (2010). Proteomics of Saccharomyces cerevisiae organelles. Mol. Cell Proteomics 9, 431–445.
Wiese, S., Gronemeyer, T., Ofman, R., Kunze, M., Grou, C. P., Almeida, J. A., Eisenacher, M., Stephan, C., Hayen, H., Schollenberger, L., Korosec, T., Waterham, H. R., Schliebs, W., Erdmann, R., Berger, J., Meyer, H. E., Just, W., Azevedo, J. E., Wanders, R. J., and Warscheid, B. (2007). Proteomics characterization of mouse kidney peroxisomes by tandem mass spectrometry and protein correlation profiling. Mol. Cell Proteomics 6, 2045–2057.
Wijekoon, C. P., Goodwin, P. H., Valliani, M., and Hsiang, T. (2011). The role of a putative peroxisomal-targeted epoxide hydrolase of Nicotiana benthamiana in interactions with Colletotrichum destructivum, C. orbiculare or Pseudomonas syringae pv. tabaci. Plant Sci. 181, 177–187.
Wiszniewski, A. A. G., Zhou, W., Smith, S. M., and Bussell, J. D. (2009). Identification of two Arabidopsis genes encoding a peroxisomal oxidoreductase-like protein and an acyl-CoA synthetase-like protein that are required for responses to pro-auxins. Plant Mol. Biol. 69, 503–515.
Woodward, A. W., and Bartel, B. (2005). The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol. Biol. Cell 16, 573–583.
Xiong, Y., DeFraia, C., Williams, D., Zhang, X., and Mou, Z. (2009). Characterization of Arabidopsis 6-phosphogluconolactonase T-DNA insertion mutants reveals an essential role for the oxidative section of the plastidic pentose phosphate pathway in plant growth and development. Plant Cell Physiol. 50, 1277–1291.
Yadav, S. K., Singla-Pareek, S. L., and Sopory, S. K. (2008). An overview on the role of methylglyoxal and glyoxalases in plants. Drug Metabol. Drug Interact. 23, 51–68.
Yan, W., Aebersold, R., and Raines, E. W. (2009). Evolution of organelle-associated protein profiling. J. Proteomics 72, 4–11.
Yates, J. R. III, Gilchrist, A., Howell, K. E., and Bergeron, J. J. (2005). Proteomics of organelles and large cellular structures. Nat. Rev. Mol. Cell Biol. 6, 702–714.
Yegutkin, G. G. (2008). Nucleotide- and nucleoside-converting ectoenzymes: important modulators of purinergic signalling cascade. Biochim. Biophys. Acta 1783, 673–694.
Zhang, X., De Marcos Lousa, C., Schutte-Lensink, N., Ofman, R., Wanders, R. J., Baldwin, S. A., Baker, A., Kemp, S., and Theodoulou, F. L. (2011). Conservation of targeting but divergence in function and quality control of peroxisomal ABC transporters: an analysis using cross-kingdom expression. Biochem. J. 436, 547–557.
Zhang, X., and Hu, J. (2008). Fission1A and fission1B proteins mediate the fission of peroxisomes and mitochondria in Arabidopsis. Mol Plant 1, 1036–1047.
Zhang, X., and Hu, J. (2010). The Arabidopsis chloroplast division protein dynamin-related protein5B also mediates peroxisome division. Plant Cell 22, 431–442.
Zhang, X. C., and Hu, J. P. (2009). Two small protein families, dynamin-related protein3 and fission1, are required for peroxisome fission in Arabidopsis. Plant J. 57, 146–159.
Zheng, B., Rönnberg, E., Viitanen, L., Salminen, T. A., Lundgren, K., Moritz, T., and Edqvist, J. (2008). Arabidopsis sterol carrier protein-2 is required for normal development of seeds and seedlings. J. Exp. Bot. 59, 3485–3499.
Zimmermann, P., Heinlein, C., Orendi, G., and Zentgraf, U. (2006). Senescence-specific regulation of catalases in Arabidopsis thaliana (L.) Heynh. Plant Cell Environ. 29, 1049–1060.
Zolman, B., Nyberg, M., and Bartel, B. (2007). IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol. Biol. 64, 59–72.
Zolman, B. K., and Bartel, B. (2004). An Arabidopsis indole-3-butyric acid-response mutant defective in peroxin6, an apparent ATPase implicated in peroxisomal function. Proc. Natl. Acad. Sci. U.S.A. 101, 1786–1791.
Zolman, B. K., Martinez, N., Millius, A., Adham, A. R., and Bartel, B. (2008). Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 180, 237–251.
Zolman, B. K., Monroe-Augustus, M., Silva, I. D., and Bartel, B. (2005). Identification and functional characterization of Arabidopsis peroxin4 and the interacting protein peroxin22. Plant Cell 17, 3422–3435.
Zolman, B. K., Monroe-Augustus, M., Thompson, B., Hawes, J. W., Krukenberg, K. A., Matsuda, S. P., and Bartel, B. (2001a). chy1, an Arabidopsis mutant with impaired beta-oxidation, is defective in a peroxisomal beta-hydroxyisobutyryl-CoA hydrolase. J. Biol. Chem. 276, 31037–31046.
Zolman, B. K., Silva, I. D., and Bartel, B. (2001b). The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol. 127, 1266–1278.
Keywords: peroxisome, proteomics, Arabidopsis, rice
Citation: Kaur N and Hu J (2011) Defining the plant peroxisomal proteome: from Arabidopsis to rice. Front. Plant Sci. 2:103. doi: 10.3389/fpls.2011.00103
Received: 14 November 2011; Paper pending published: 30 November 2011;
Accepted: 08 December 2011; Published online: 27 December 2011.
Edited by:
Andreas P. M. Weber, University of Duesseldorf, GermanyReviewed by:
Roland Krause, Max Planck Institute of Molecular Cell Biology and Genetics, GermanyRobert Mullen, University of Guelph, Canada
Copyright: © 2011 Kaur and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Jianping Hu, MSU-DOE Plant Research Laboratory, Michigan State University, East Lansing, MI 48824, USA. e-mail: huji@msu.edu