- 1College of Plant Protection, Hunan Agricultural University, Changsha, China
- 2Hunan Plant Protection Institute, Hunan Academy of Agricultural Science, Changsha, China
- 3Long Ping Branch, Graduate School of Hunan University, Changsha, China
Myzus persicae (Sulzer), commonly known as the green peach aphid, is a notorious pest that causes substantial losses to a range of crops and can transmit several plant viruses, including potato virus Y (PVY). Chemical insecticides provide only partial control of this pest and their use is not environmentally sustainable. In recent years, many genes related to growth, development, and reproduction have been used as targets for pest control. These include Gustavus (Gus), a highly conserved gene that has been reported to play an essential part in the genesis of germline cells and, hence, in fecundity in the model insect Drosophila melanogaster. We hypothesized that the Gustavus (Gus) gene was a potential target that could be used to regulate the M. persicae population. In this study, we report the first investigation of an ortholog of Gus in M. persicae, designated MpGus, and describe its role in the fecundity of this insect. First, we identified the MpGus mRNA sequence in the M. persicae transcriptome database, verified its identity with reverse transcription-polymerase chain reaction (RT-PCR), and then evaluated the transcription levels of MpGus in M. persicae nymphs of different instars and tissues with real-time quantitative PCR (RT-qPCR). To investigate its role in regulating the fecundity of M. persicae, we used RNA interference (RNAi) to silence the expression of MpGus in adult insects; this resulted in a significant reduction in the number of embryos (50.6%, P < 0.01) and newborn nymphs (55.7%, P < 0.01) in the treated aphids compared with controls. Interestingly, MpGus was also significantly downregulated in aphids fed on tobacco plants that had been pre-infected with PVYN, concomitant with a significant reduction (34.1%, P < 0.01) in M. persicae fecundity. Collectively, these data highlight the important role of MpGus in regulating fecundity in M. persicae and indicate that MpGus is a promising RNAi target gene for control of this pest species.
Introduction
Myzus persicae (Sulzer) (Hemiptera: Aphididae) is a pest that can infest crops all over the world (van Emden and Harrington, 2017). This aphid is known to extract nutrients from the phloem and xylem of more than 400 plant species in over 50 families (Smith, 1926; van Emden et al., 1969, van Emden and Harrington, 2017; Weber, 1986; Spiller et al., 1990; Bernays and Chapman, 1994; Gupta, 2001; Chen et al., 2020), including both row crops and ornamentals grown in greenhouses. M. persicae reduces crop yields by causing systemic perturbations in nutrient allocation, leaf discoloration and necrosis, and leaf and/or fruit deformation (Zhan et al., 2020). Furthermore, M. persicae is one of the most effective vectors for plant viruses (Ng and Perry, 2004; Ng and Falk, 2006) and can transmit a large number of economically important viruses in a persistent, semi-persistent, or non-persistent manner (Carmo-Sousa et al., 2014). Under certain conditions, M. persicae can produce wings for migration; they can also be transported over long distances by wind and storms. M. persicae breeds in a highly prolific manner, making these particular aphids very difficult to control. Currently, the main form of control for M. persicae is the application of chemical insecticides. However, the efficacy of insecticides can be limited by over-application and the rapid development of resistance (Bass et al., 2014). Furthermore, long-term use of pesticides can cause environmental damage. Consequently, there is a clear need to develop new control strategies that are more selective and have better environmental profiles.
RNA interference (RNAi), mediated by double-stranded RNA (dsRNA), has been shown to be an effective means of controlling a large number of agricultural pests (Upadhyay et al., 2011; Zhang et al., 2015; Niu et al., 2018; Liu et al., 2019; Christiaens et al., 2020). Recent reports have also identified a range of genes that could be used as RNA targets to control aphids. For example, Hou et al. (2018) showed that downregulation of the guanine nucleotide-binding protein G(q) subunit alpha (Gqα) gene resulted in a reduction in the molting rate and fecundity of the grain aphid (Sitobion avenae F.). Similar results were observed by Ye et al. (2019), who used RNAi to target the chitin synthase gene in pea aphids (Acyrthosiphon pisum). In another study, Tariq et al. (2019) demonstrated that knockdown of the voltage-gated sodium channel gene caused an increase in mortality of M. persicae. In addition, silencing of the gene that encodes cuticular protein has been shown to result in impaired fecundity in M. persicae (Bhatia and Bhattacharya, 2018). A range of methods have been used to deliver dsRNA to pests. For example, Zhao et al. (2018) fed S. avanae with transgenic leaves expressing dsRNA targeting the chitin synthase 1 gene, and observed a significant reduction in the size of the insect population. Yan et al. (2019) reported that spraying dsRNA on Aphis glycines also led to a substantial reduction in the expression levels of target genes. Gui et al. (2020) completely inhibited oviposition in Leptinotarsa decemlineata by injecting dsRNA. Most recently, Lü et al. (2020) fed Henosepilachna vigintioctopunctata with leaf discs that had been immersed in dsHvlwr solution, causing an increase in mortality. Although RNAi is considered to be a revolutionary strategy for pest control, the efficacy of this technique is highly dependent on the identification of target genes that are responsive to RNAi-mediated mRNA degradation and on the efficiency of delivery of dsRNA to the target. Genes involved in the reproductive cycle of pests are likely to represent highly promising targets for RNAi.
Gustavus (Gus) is a protein that was first identified in the fruit fly (Drosophila melanogaster) as a positive regulator of insect spawning (Styhler et al., 2002; Gustafson et al., 2011). The Gus protein possesses both a sp1A/ryanodine receptor (SPRY) domain and a suppressor of cytokine signaling (SOCS) box, so it belongs to the SPRY domain-containing SOCS box (Spsb) family of proteins (Styhler et al., 2002; Gustafson et al., 2011). Previous studies have shown that Gus interacts with VASA, a protein in D. melanogaster that has key roles in the specialized translational activity and localization of the pole plasm, and in the specification of germ cells. Mutations in the Gus gene have been reported to cause female sterility in the fruit fly (Styhler et al., 2002). Collectively, these studies indicate that the Gus protein has critical roles in the reproductive success of a diverse number of arthropods.
The rate of reproduction, also known as fecundity, is a common insect trait that can be frequently modified by the plant viruses they transmit (Ziebell et al., 2011; Mauck et al., 2014; Casteel et al., 2015). For example, previous studies showed that potato virus Y (PVY; genus Potyvirus; family Potyviridae), a virus known to be transmitted by M. persicae in a non-persistent manner, affected the fecundity of M. persicae (Srinivasan and Alvarez, 2007; Valkonen et al., 2007; Cervantes and Alvarez, 2011; Ren et al., 2015; Bak et al., 2017). However, it is not yet known whether the Gus gene is involved in the process of PVY affecting aphid fecundity.
Based on these previous findings, we hypothesized that orthologs of the Gus protein may also serve to regulate the fecundity of M. persicae, and that the Gus gene plays a part in the process of PVY affecting aphid fecundity. To test this hypothesis, we identified MpGus, a Gus ortholog in M. persicae, and then determined the expression profile of MpGus in different instars. Next, we used dsRNA-mediated RNAi to silence the expression of MpGus. Finally, we fed PVY-infected tobacco plants to M. persicae and investigated the changes in MpGus expression and fecundity. Our results showed that MpGus has a crucial role in the fecundity of M. persicae, and that its expression and functionality are regulated by the acquisition of PVY. These findings suggest that MpGus could be used as an RNAi target to control M. persicae.
Materials and Methods
Plant and Insect Care
Tobacco (Nicotiana tabacum L. cv. Samsun NN) plants were kept in aphid-proof screen cages in a greenhouse room set to 25 ± 1°C and 75 ± 5% relative humidity, with a 16 h light/8 h dark photoperiod. In order to systemically infect tobacco plants with PVY, we mechanically inoculated plants at the five-true-leaf stage with leaf sap containing the PVY N strain (PVYN) (Shrestha et al., 2014). Symptoms (veinal necrosis) appeared after 10 days and the infection was further confirmed by reverse transcription-polymerase chain reaction (RT-PCR) assay using primers that were specific to the coat protein (CP) gene (PVY-F/R; Table 1; Liu et al., 2010).
The M. persicae individuals used in this study were descendants of a single adult and were reared for more than 10 generations on healthy tobacco plants before being used in our experiments.
MpGus Cloning and Bioinformatics Analysis
Total RNA was extracted from aphids that contain all nymphal and adult stages using TRIzol reagent (Thermo Fisher Scientific, Shanghai, China) and quantified by a NanoDrop spectrophotometer (Model ND-2000, Thermo Fisher Scientific, Waltham, MA, United States). First-strand cDNA was synthesized from 500 ng of total RNA using TransScript All-in-One First-Strand cDNA Synthesis SuperMix for PCR (TransGene Biotech, Beijing, China), in accordance with the manufacturer’s instructions. The resulting cDNA was then used as templates for PCR reactions to amplify the full-length coding sequence of MpGus using the primers MpGus-F1–3/R1–3 (Table 1); these primers were designed to the sequence information provided by the M. persicae genome assembly (NW_019102926.1). The PCR reaction included 1 μL of KOD-Plus-Neo (Toyobo Life Scientific, Shanghai, China), 5 μL of 10 × PCR buffer for KOD-Plus-Neo, 3 μL of 25 mmol/L MgSO4, 5 μL of 2 mmol/L dNTPs, 1 μL of each primer (10 μmol/L), 1 μL of cDNA template, and 33 μL of ultrapure water, in a total volume of 50 μL. The following cycling conditions were used for PCR: 94°C for 2 min, 35 cycles of 15 s at 98°C, 15 s at 58°C, and 90 s at 68°C. The PCR products were purified with an EasyPure PCR Purification Kit (TransGene Biotech, Beijing, China) and then sequenced.
Sequences were analyzed at the nucleotide and protein level using BLAST1 Multiple sequence alignments were generated using Clustal Omega2. A phylogenetic tree was also constructed using the amino acid sequences of Gus orthologs from 14 different species; for this, we used the neighbor-joining method in the MEGA 7 software (Kumar et al., 2016). The bootstra p-values (as percentages) shown at the branch points were calculated from 1,000 replicates3.
The Expression Profile of MpGus in Different Instars and Tissues
To collect the nymphs of different instars, we infested 10 healthy tobacco plants with 10 apterous M. persicae adults per plant. These adults were subsequently removed after 24 h; 150 newborns were collected as the first instar nymphs. The rest of the nymphs were covered individually using home-made mini cages. These cages were made from 0.2 mL PCR tubes with lids and the bottom halves removed, followed by sealing of the bottoms with 100-mesh gauze; they could be pasted onto the leaves with starch dextrin (Figure 1A). Over the next few days, we monitored the nymphs for molting activity and collected 80 s instar nymphs, 40 third instar nymphs, and 20 fourth instar nymphs. To collect cephalothorax, leg, and abdomen of apterous adult aphids, we dissected 50 aphids under a stereoscopic microscope (LEICA S8AP0). The collected samples were placed individually in 1.5 mL RNase-free centrifuge tubes, rapidly frozen in liquid nitrogen, and stored at −80°C until expression profiling of MpGus.
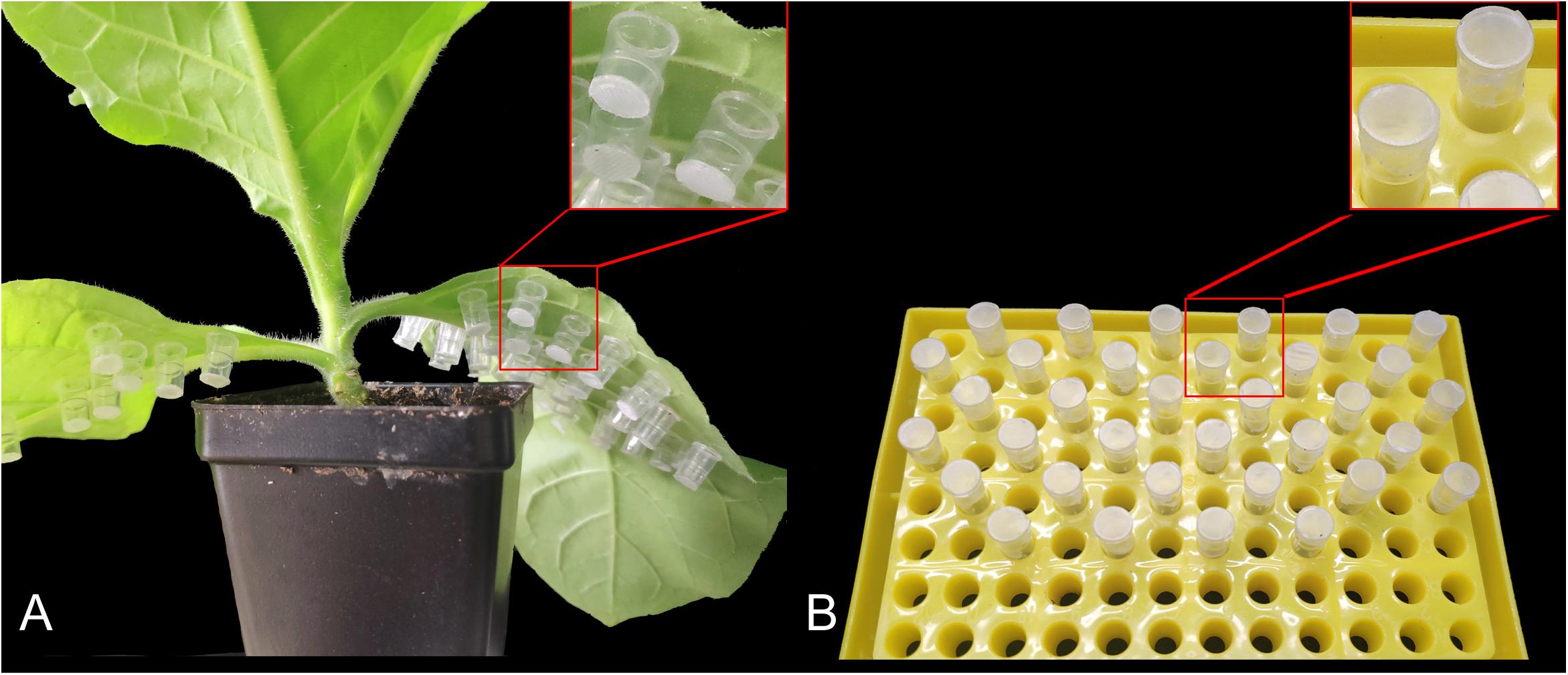
Figure 1. Mini cages used to rear individual Myzus persicae adults. (A) Mounting of the mini cages on tobacco leaves. (B) Mini cages supplied with artificial diets.
The expression levels of MpGus in different instars and tissues were determined by real-time quantitative PCR (RT-qPCR) with specific primers (RT-qPCR-MpGus-F/R; Table 1). The amplification efficiency of these primers was 94.6%. According to Bustin et al. (2009), the relevant information for PCR primer verification is provided in Supplementary Figures S1, S2. RT-qPCR reactions were carried out in a 96-well optical plate in an Analytik Jena AG PCR instrument; the system software was used to normalize and quantify expression data. The target gene was amplified with 2 × TransStar Green qPCR SuperMix UDG (TransGene Biotech, Beijing, China). For each sample, we performed three repetitions of three biologically independent experiments. The relative expression levels of the MpGus gene were then calculated using the 2–△△Ct method (Livak and Schmittgen, 2001). β-actin and GAPDH genes from M. persicae were used as reference genes to normalize gene expression levels (Table 1; Mp-actin-F/R, Mp-GAPDH-F/R).
RNAi Methodology
Primers were designed to amplify a 442 bp PCR fragment from MpGus. Both primers (ds-MpGus-F/R) contained the T7 promoter sequence at their 5’ ends (Table 1). The PCR product was purified using an EasyPure®, PCR Purification Kit (TransGene Biotech, Beijing, China) and used as a template to synthesize dsRNA targeting MpGus (dsMpGus) using a MEGAscript®, RNAi Kit (Thermo Fisher Scientific, Waltham, MA, United States). Once synthesized, dsMpGus was purified and resuspended in nuclease-free water. Next, we verified the molecular weight of dsMpGus by 1.5% agarose gel electrophoresis. Concentrations of purified dsRNAs were determined using a NanoDrop spectrophotometer (Model ND-2000, Thermo Fisher Scientific, Waltham, MA, United States). The purified dsMpGus was stored at −20°C prior to use. We also used specific primers (dsGFP-F/R; Table 1) to synthesize a dsGFP control in the same manner.
For the RNAi assays, we added different concentrations of dsMpGus or dsGFP to an artificial diet, as described previously by Sadeghi et al. (2009). The artificial diet for M. persicae was prepared according to the method described by Dadd and Mittler (1966), with slight modifications. Next, we transferred aphids that had begun to reproduce from healthy plants and fed them on an artificial diet containing 250, 500, or 750 ng/μL (final concentration) of dsRNA. In order to minimize degradation of the dsRNA in the artificial diet or insect bodies (Allen and Walker, 2012; Yu et al., 2013), we replaced the diet every 24 h. To analyze the gene suppression of cognate mRNA, we collected 30 insects as one sample from each treatment group after 24 h; these were then flash-frozen in liquid nitrogen, and stored at −80°C in 1.5 mL centrifuge tubes until extraction of total RNA.
To further clarify the MpGus transcripts levels at different time points under the effective dsMpGus concentration, we treated aphids using the same method as described above with effective concentrations of dsMpGus, collected samples at 6, 12, 24, 48, and 72 h, respectively, and subsequently assayed them individually by RT-qPCR. We also measured the effect of dsRNA on aphid mortality. The experiment included 3 groups: H2O + artificial diet (CK), dsGFP + artificial diet and dsMpGus + artificial diet. Each group contained 75 aphids. The number of dead aphids was recorded every 24 h for 5 consecutive days.
Analysis of the Effects of Tobacco Infected With PVYN on MpGus Gene Expression
To assess whether PVYN could alter the expression levels of MpGus in M. persicae, we compared M. persicae fed on tobacco plants infected with PVYN with those fed on mock-inoculated tobacco plants. Aphids that began to reproduce were taken from healthy plants and transferred to PVYN-infected tobacco. Next, we randomly collected 30 aphids as one sample from the leaves of PVYN-infected/mock-inoculated tobacco plants after 24, 48, and 72 h. The collected aphids were stored at −80°C in 1.5 mL centrifuge tubes and subsequently assayed individually by RT-qPCR.
Fecundity Analysis
Next, we attempted to determine the fecundity of M. persicae exposed to the dsRNA diet. To do this, we collected 150 adult aphids that were just beginning to reproduce and transferred them individually to separate mini cages (one aphid per mini cage). Individual aphids were then fed with dsRNA; a small piece of parafilm, containing 5 μL artificial diet mixed with dsMpGus (final concentration 500 ng/μL) was placed at the top of each mini cage (Figure 1B). Half of these individual aphids (75) were fed with dsMpGus; the other half were fed with dsGFP as a non-specific dsRNA control. Once, the aphids had been fed for 24 h, we began to count newborn nymphs. All counted nymphs were removed to ensure that only new nymphs were counted in subsequent measurements. Counting sessions were carried out once every 24 h for 5 days.
To determine the fecundity of aphids that had been exposed to PVYN, the aphids that had been maintained separately on non-viruliferous tobacco were subsequently transferred to a mini cage mounted on viruliferous tobacco plants; we did this when the aphids first began to reproduce. Each mini cage, containing an individual aphid, was mounted on the lower surface of the third to the sixth blade of the plants. A set of 75 adults were prepared for each experimental group. The newborn nymphs in each mini cage were counted once every 24 h, for 5 days. Once counted, nymphs were removed. Aphids maintained on mock-inoculated tobacco were used as controls.
Quantification of Developing Embryos
Apterous adult aphids treated with dsGFP or dsMpGus for more than 24 h were transferred to clean slides. Their abdomens were then dissected with a needle under a stereomicroscope (LEICA S8AP0). For each female aphid, we only counted developing embryos with obvious red compound. A total of 15 females were inspected per treatment.
Data Analysis
Statistical analyses were performed with the SPSS package (version 22.0; SPSS Inc., Chicago, IL, United States). For normally distributed data (Shapiro–Wilk test: P > 0.05), we used the independent Student’s t-test to compare experimental and control data. Data that did not conform to a normal distribution (Shapiro–Wilk test: P ≤ 0.05), were compared using a non-parametric Mann–Whitney U-test. The expression levels of MpGus in different instars were compared by one-way analysis of variance (ANOVA) and Tukey’s post hoc test. The independent Student’s t-test was used to compare the expression levels of MpGus induced by RNAi and PVYN in different groups. The number of newborn nymphs from different treatment groups was compared using the Mann–Whitney U-test. Means were compared by the least significant difference test at P = 0.05.
Results
Identification of MpGus (the Ortholog of Gus in M. persicae)
Three cDNA sequences that represent three predicted transcript variants of the same M. persicae gene, named MpGus X1, MpGus X2, and MpGus X3, have been annotated as Gus in GenBank (term = + Gustavus + Myzus)4. We thus presumed that these genes were the M. persicae orthologs of Gus and designated these genes as MpGus. To determine whether all three of the splicing variants were expressed in aphid bodies, we designed three pairs of specific primers and used these in RT-PCR to amplify separate fragments. Each primer pair allowed the amplification of a full-length coding sequence. The amplified sequences of MpGus X1, MpGus X2, and MpGus X3 were identical to XM_022314172.1, XM_022314173.1, and XM_022314174.1 in GenBank, respectively. Although there were differences in the N-terminals of the three sequences, the SPRY and SOCS domain were completely identical (Figure 2). In order to study the function of MpGus, dsMpGus was prepared, targeting the conserved regions of the three transcriptional variants, and specific primers RT-qPCR-MpGus-F/R for RT-qPCR detection were also designed according to the conserved regions (Figure 3).
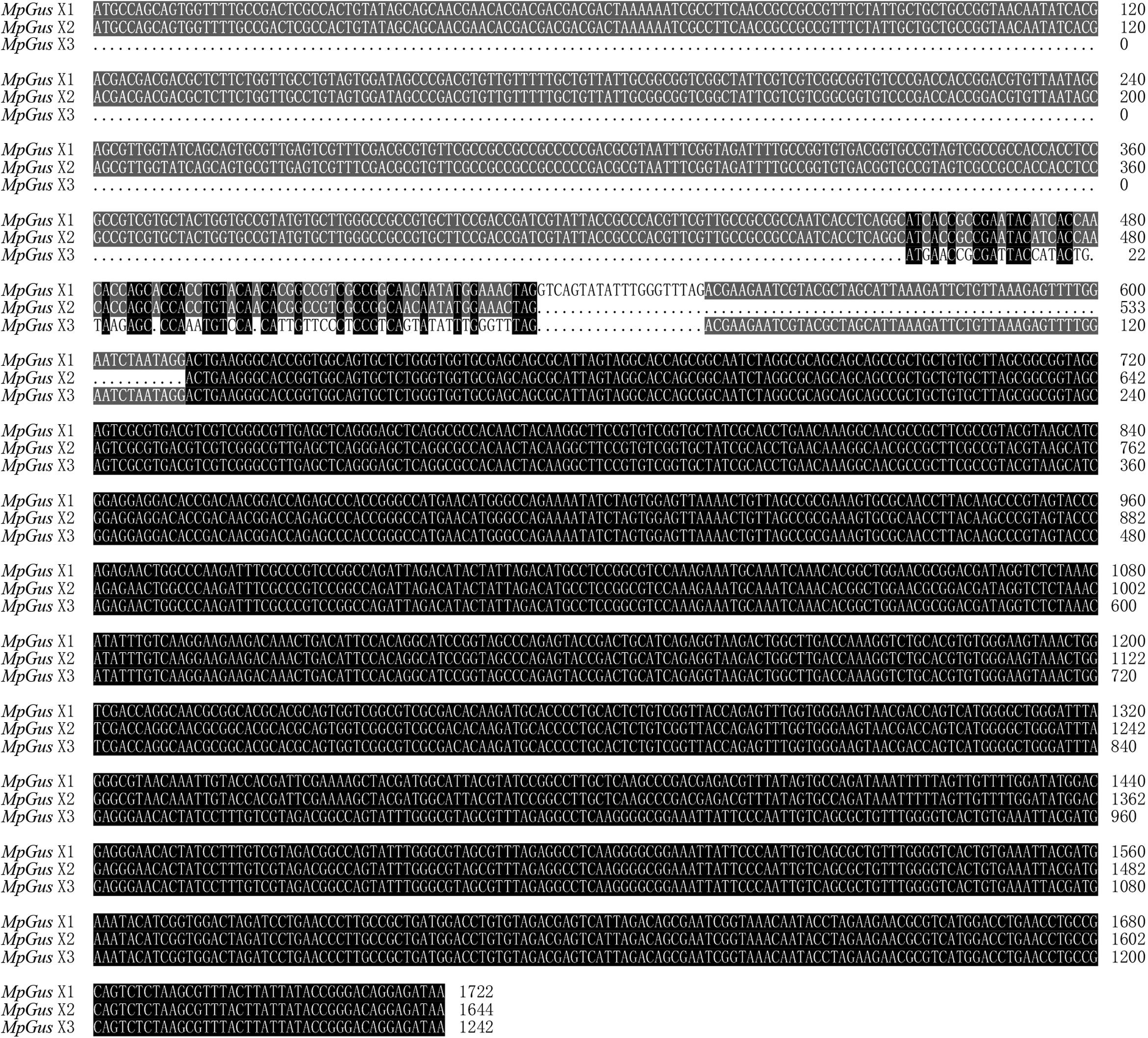
Figure 2. Sequence alignment of three transcriptional variants. Gray indicates that two sequences are consistent; black indicates that three sequences are consistent.
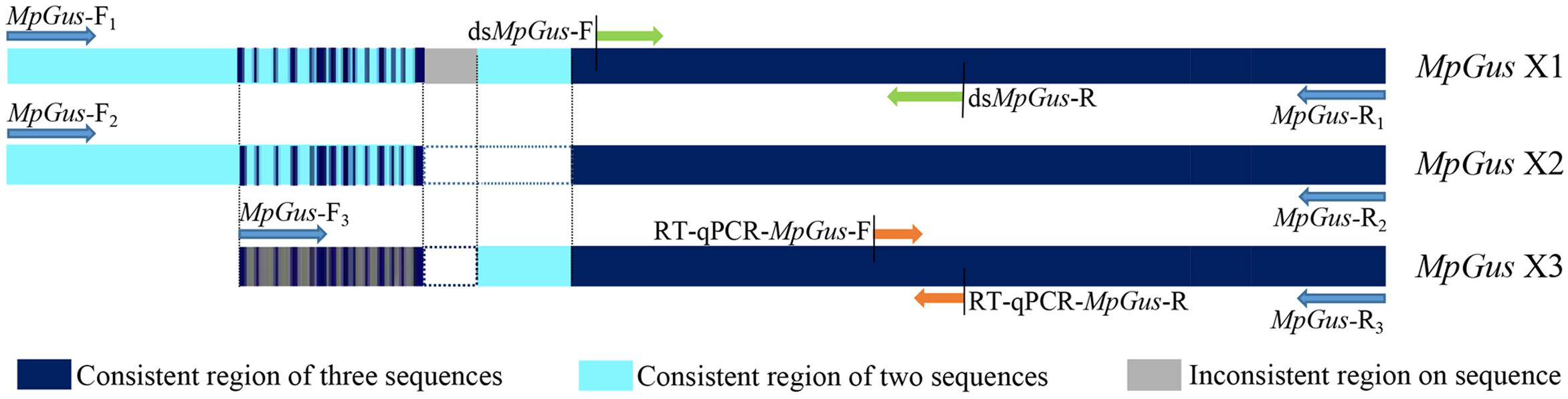
Figure 3. Position information for the primers used in the experiment in the MpGus sequence. MpGus-F1–3/R1–3 were amplification primers for the full length sequence of MpGus X1, MpGus X2, and MpGus X3, respectively. dsMpGus-F/R were used to prepare dsMpGus. RT-qPCR-MpGus-F/R were used to detect the expression levels of MpGus.
The Gus Protein Shows High Levels of Conservation Across a Diverse Range of Species
The MpGus protein contains a conserved SPRY domain and the SOCS box that is shared by all proteins in the Spsb protein family. Compared with the Gus orthologs of other arthropods, the amino acid sequence identity ranged from 77.4 to 99.8% (Figure 4). Further phylogenetic analysis showed that the MpGus protein clustered closely with its orthologs in other aphid species, including Aphis gossypii, Rhopalosiphum maidis, Melanaphis sacchari, Diuraphis noxia, and A. pisum. As expected, the MpGus protein was more distantly related to the Gus orthologs of Bemisia tabaci, D. melanogaster, and Apis dorsata (Figure 5). Nevertheless, regions representing the SPRY and SOCS domains were highly conserved across all of the Gus orthologs we compared.
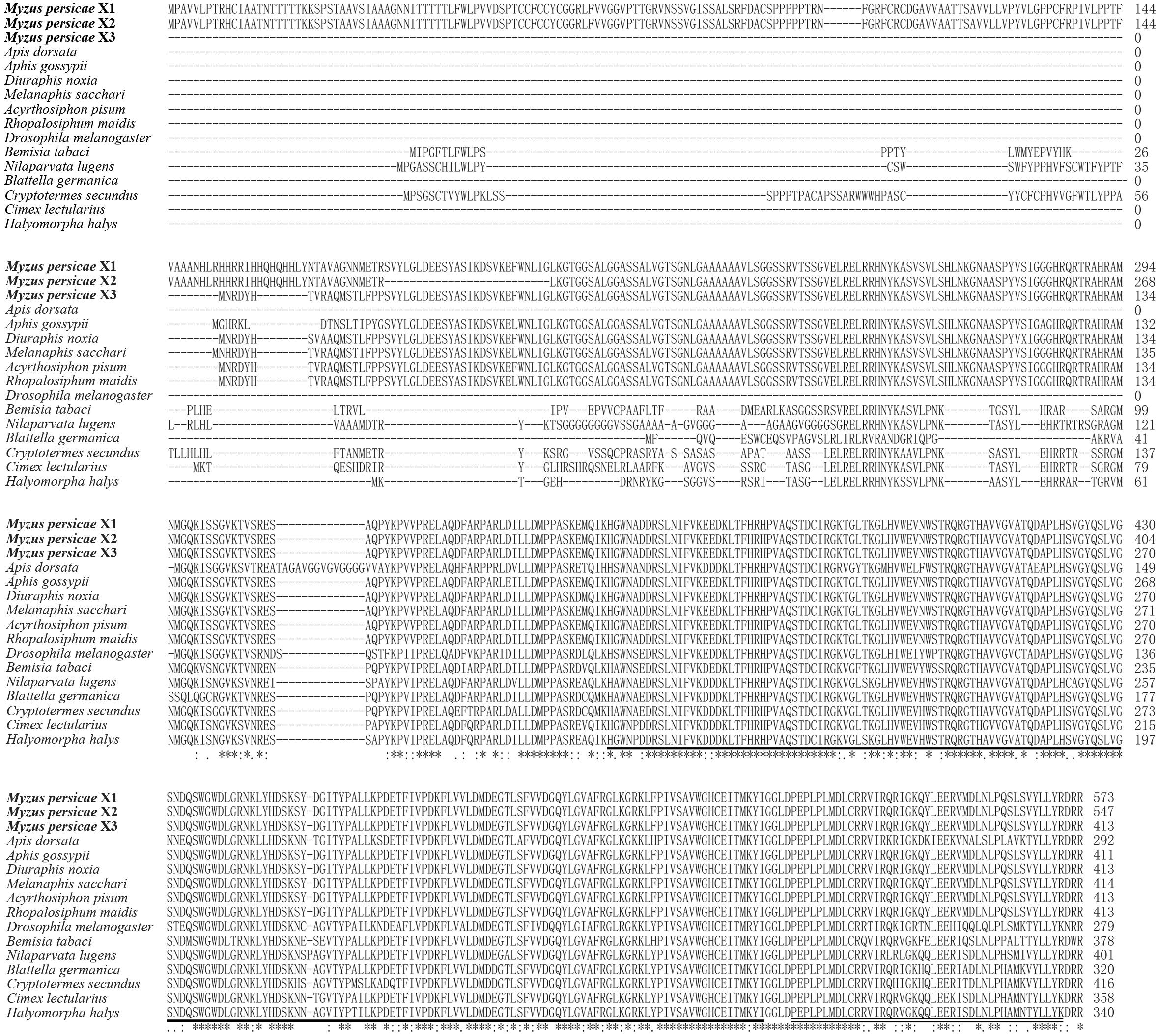
Figure 4. Multiple sequence alignments of MpGus with other 13 Gus orthologs from diverse species. The GenBank accession numbers of these species are Myzus persicae X1 (XP_022169864.1); Myzus persicae X2 (XP_022169865.1); Myzus persicae X3 (XP_022169866.1); Aphis gossypii (XP_027839423.1); Diuraphis noxia (XP_015372875.1); Melanaphis sacchari (XP_025207865.1); Acyrthosiphon pisum (XP_016662132.2); Rhopalosiphum maidis (XP_026820642.1); Apis dorsata (XP_006616315.1); Drosophila melanogaster (sp| A1Z6E0.2|); Bemisia tabaci (XP_018912944.1); Nilaparvata lugens (XP_022187205.1); Blattella germanica (PSN58391.1); Cryptotermes secundus (XP_023706529.1); Cimex lectularius (XP_014262520.1);, and Halyomorpha halys (XP_014272688.1). Highly conserved amino acid residues across all of the species compared are annotated by *. The SPRY domain is underlined, and the SOCS box is marked by double underscores.

Figure 5. A phylogenetic tree of Gus proteins from different organisms based on the comparison of amino acid sequences. The proportion (%) of replicate trees are shown next to the branches. MpGus is shown in bold. Drosophila melanogaster was included as an outgroup.
Expression Levels of MpGus at Different Developmental Stages of Nymphs and in Different Tissues
Next, we used RT-qPCR to investigate changes in the expression of MpGus during the development of different nymph stages. The mRNA levels of MpGus were lowest in first instar nymphs but increased rapidly as nymphs developed from the first to the second instar stage [F(1,4) = 25.878, P < 0.01]. There was no significant change in MpGus expression from the second to the third instar nymph stage (P > 0.05). Subsequently, the expression of MpGus gradually increased from the third to the fourth instar nymph stage (Figure 6A). Therefore, the expression levels of MpGus increased as aphids approached adulthood. The result of MpGus expression levels in cephalothorax, legs, and abdomen of apterous adult aphids showed that among the different tissue types, MpGus expression was significantly higher in abdomen tissue than in cephalothorax or leg tissue (Figure 6B).
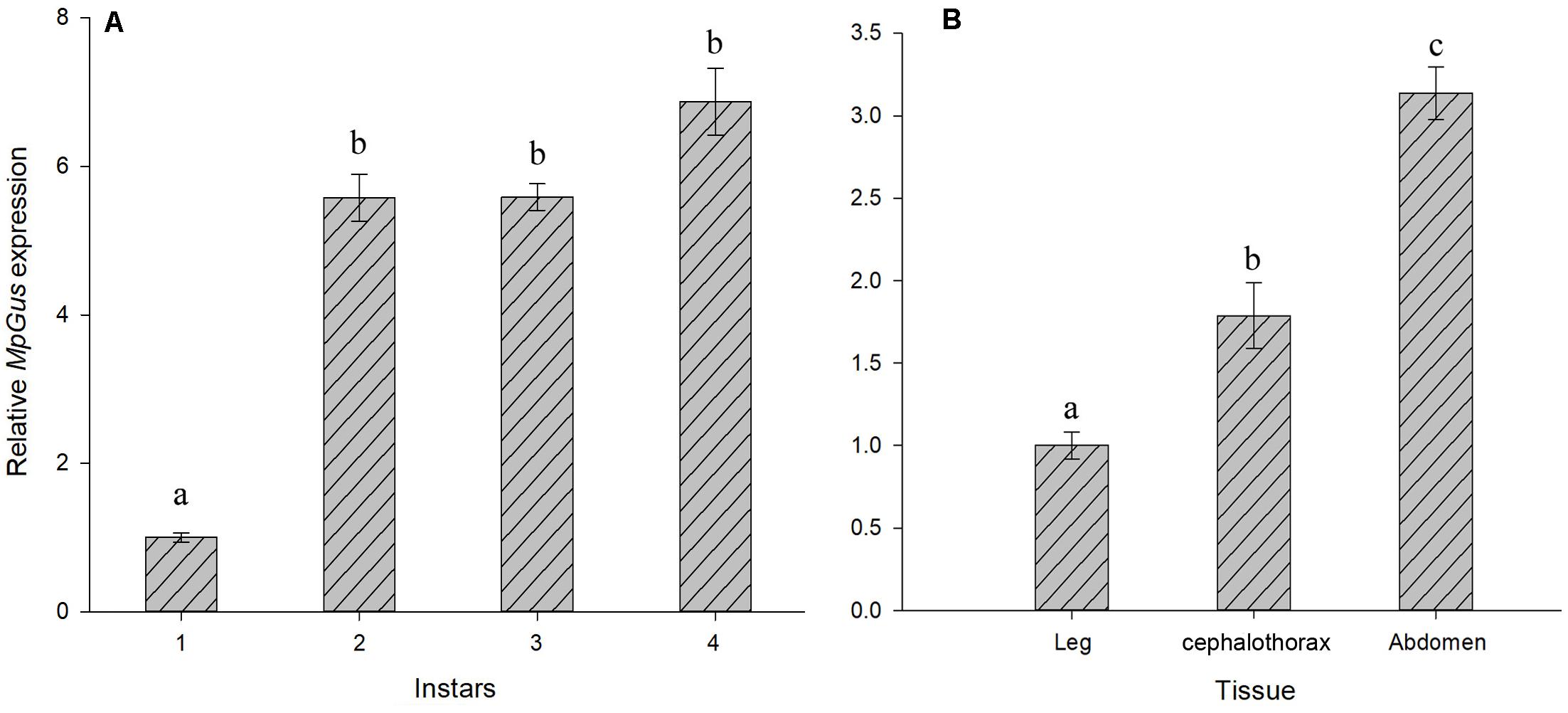
Figure 6. Temporospatial expression profiles of MpGus. (A) Relative expression of MpGus in different instars, as measured by RT-qPCR. (B) Relative expression of MpGus in different instars, as measured by RT-qPCR. Each data point represents the mean of three independent samples, with three repetitions per sample. mRNA expression values were normalized to reference genes (β-actin and GAPDH). Data represent mean ± standard error (SE), n = 3, and different letters indicate statistically significant differences at P < 0.05.
Expression Levels of MpGus Expression Were Down-Regulated by RNAi
In order to determine whether MpGus expression levels could be manipulated with RNAi, and whether such manipulation affected the fecundity of M. persicae fecundity, we added dsRNA targeting MpGus (dsMpGus) to an artificial diet for M. persicae. The control M. persicae group was fed with a non-specific dsRNA based on the green fluorescent protein coding sequence (dsGFP). After 24 h of feeding on the artificial diet containing dsMpGus, we found that the relative expression levels of MpGus did not change in M. persicae fed on dsMpGus at 250 ng/μL [F(1,4) = 0.052, P = 0.83 > 0.05]. The expression levels of MpGus in adult M. persicae were downregulated by 27.9% [F(1,4) = 108.708, P < 0.05] and 50.9% [F(1,4) = 120.661, P < 0.05] following treatment with 500 and 750 ng/μL of dsMpGus, respectively. However, there was no significant difference between the 500 and 750 ng/μL (Figure 7A). Therefore, our data showed that oral intake of dsMpGus was sufficient to cause a significant downregulation of MpGus expression in M. persicae. According to the experimental results, we selected 500 ng/μL dsRNA as the effective RNAi concentration and further determined the time when MpGus expression was significantly downregulated by this concentration. As shown in Figure 7B, the relative expression levels of MpGus were downregulated at 12 h but not significantly [F(1,4) = 0.502, P = 0.518 > 0.05]. MpGus expression was significantly downregulated at 24 h [F(1,4) = 453.933, P < 0.05]. The mortality of M. persicae was shown in Supplementary Figure S3. There is no significant difference between the control group and the dsRNA treatment group.
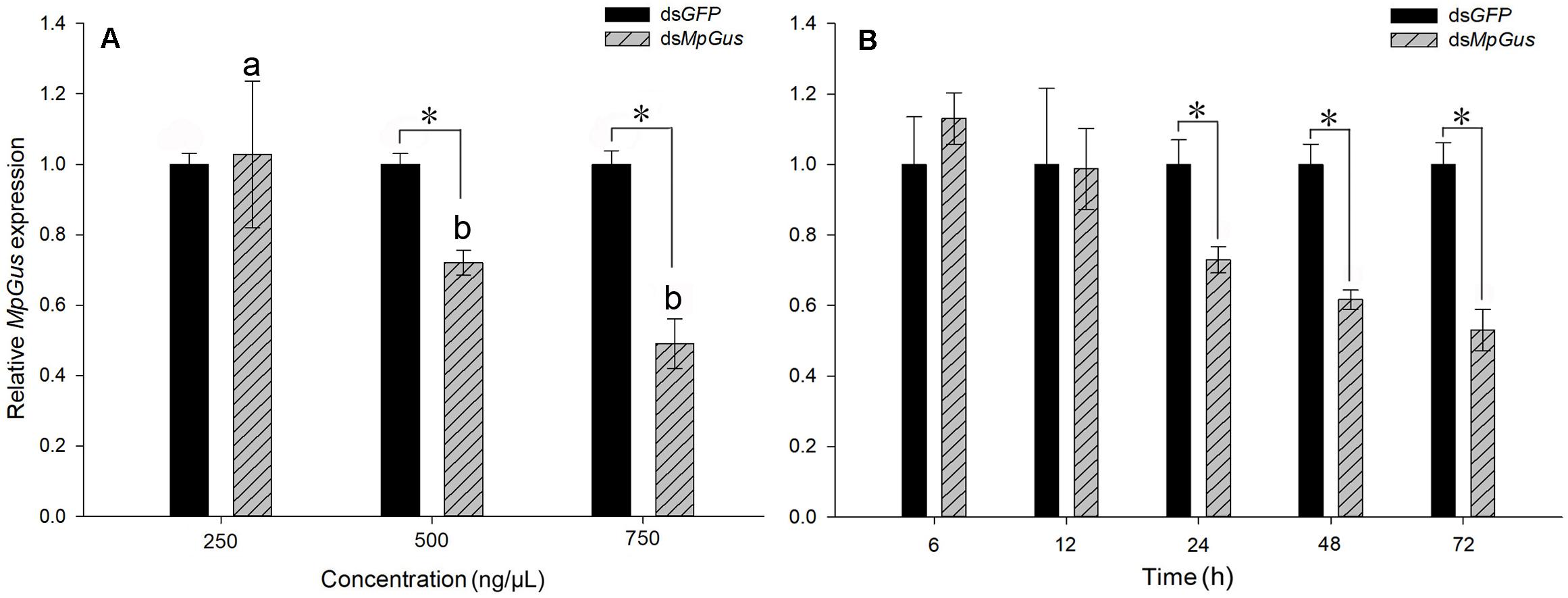
Figure 7. Interference of MpGus expression by dsRNA. (A) Relative expression levels of MpGus fed on different concentrations of dsMpGus for 24 h, as measured by RT-qPCR. (B) Relative expression levels of MpGus fed on 500 ng/μL dsMpGus (final concentration) for 6, 12, 24, 48, and 72 h, as measured by RT-qPCR. Each bar represents the mean of three independent samples, each with three repetitions. β-actin and GAPDH from M. persicae were used as reference genes. Values represent mean ± SE, n = 3. An asterisk indicates a significant difference (P < 0.05) between the expression levels of dsGFP and dsMpGus at the same concentration. Different letters (a b) above each bar denote significant differences (P < 0.05).
RNAi-Mediated Knockdown of MpGus Lowers M. persicae Fecundity
Next, we investigated how the knockdown of MpGus might affect the fecundity of M. persicae. As shown in Figure 7A, an artificial diet containing 500 ng/μL dsMpGus was sufficient to cause significant knockdown of MpGus mRNA levels in M. persicae adults. Next, we investigated M. persicae adults with regard to the number of newborn nymphs they produced while fed on 500 ng/μL dsMpGus, with those fed with 500 ng/μL of dsGFP as controls. As shown in Figure 8, compared with the controls, adults with MpGus knockdown produced significantly fewer newborn nymphs. Specifically, the rates of reduction were 49.2% [F(1,148) = 267.878, P < 0.01], 54.9% [F(1,148) = 381.684, P < 0.01], 57.4% [F(1,148) = 581.800, P < 0.01], 59.5% [F(1,148) = 661.865, P < 0.01], and 58.0% [F(1,148) = 477.965, P < 0.01] for the first 5 days of MpGus knockdown (Figure 8). Collectively, these results strongly suggest that MpGus is a positive regulator of fecundity in M. persicae and that the expression of MpGus at appropriate levels is essential for the reproductive success of this insect.
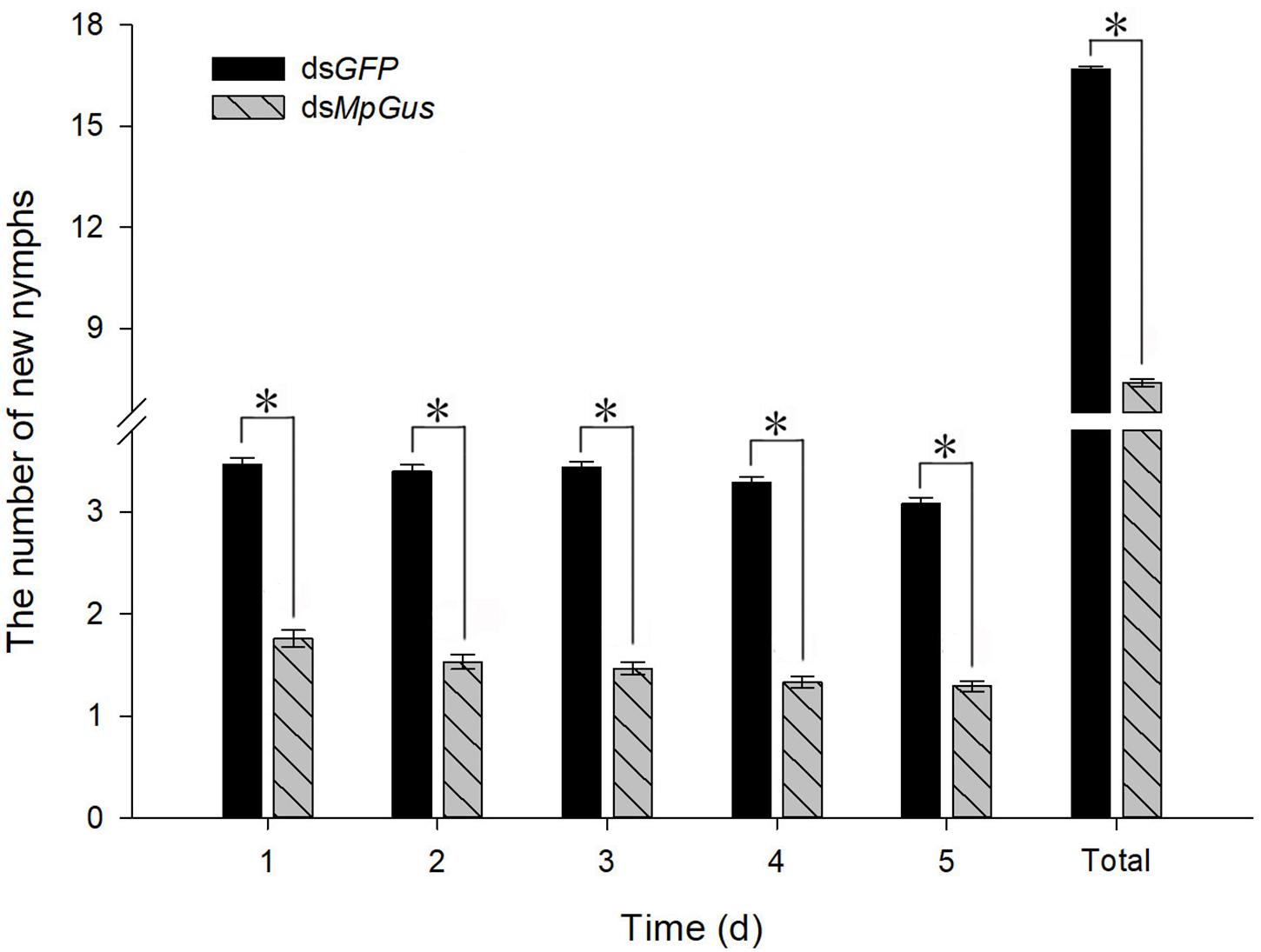
Figure 8. Changes in the fecundity of M. persicae in response to intake of dsRNA. Each bar represents the mean number of newborns (± SD) from 75 mother aphids. Counting was carried out for 5 days, with newborns from the previous day removed after each count. An asterisk indicates a significant difference (P < 0.05) between aphids fed with dsGFP and dsMpGus.
Knockdown of MpGus Reduced the Number of Developing M. persicae Embryos
In the previous experiment, we found that knockdown of MpGus caused M. persicae to produce significantly fewer nymphs. Next, we evaluated whether such knockdown also exerted a negative effect on the number of M. persicae embryos. As it was difficult to spot embryos in the early stages of development, we only counted developing embryos that had distinct red compound eyes. The results are shown in Figure 9. Compared with the control adults treated with dsGFP (Figure 9B), those treated with dsMpGus for 24 h showed a notable reduction in the number of embryos produced (Figure 9C). Further quantification using multiple adult aphids indicated that the mean number of developing embryos per aphid treated with dsMpGus (5.07) was significantly lower (50.6% reduction, [F(1,28) = 59.232, P < 0.01] compared with controls (10.27) (Figure 9A). Therefore, MpGus appears to have a role that ensures the production of healthy embryos.
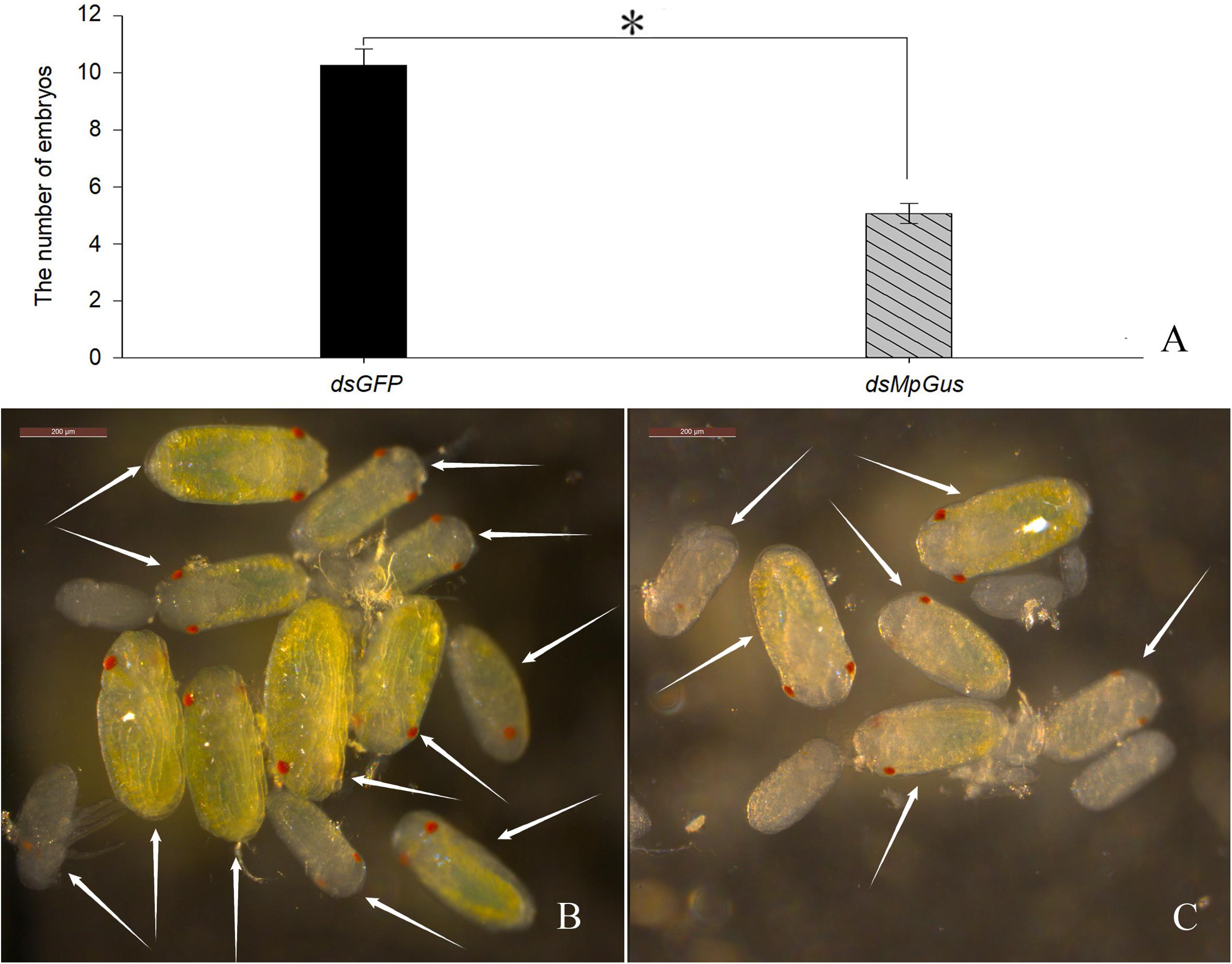
Figure 9. Effect of dsMpGus treatment on the number of developing embryos. Counting was carried out on female aphids treated with dsMpGus for 24 h. (A) Quantitative analysis. Data represent mean ± SD, n = 15. Asterisk indicates a significant difference (P < 0.05) between dsGFP and dsMpGus. (B) Developing embryos produced by adults treated with dsGFP. (C) Developing embryos produced by adults treated with dsMpGus. Arrows in (A,B) indicate embryos with red compound eyes. Scale bars = 200 μm.
The mRNA Levels of MpGus Were Significantly Lower in M. persicae Fed on PVYN-Infected Tobacco Plants
M. persicae is a potent transmission vector for PVY, and the fecundity of M. persicae is known to be affected by the acquisition of PVY (Srinivasan and Alvarez, 2007; Ren et al., 2015; Bak et al., 2017). We therefore considered whether feeding M. persicae with PVY-infected plants might also perturb the expression of MpGus. As shown in Figure 10, levels of MpGus mRNA decreased progressively in M. persicae fed on PVYN-infected tobacco plants. Compared with M. persicae fed on mock-inoculated tobacco plants, the relative expression levels of MpGus in M. persicae maintained on PVYN-infected tobacco plants decreased by 26.9% [F(1,4) = 23.129, P < 0.05], 45.9% [F(1,4) = 96.165, P < 0.01], and 52.8% [F(1,4) = 27.348, P < 0.01] after 24, 48, and 72 h of feeding, respectively.
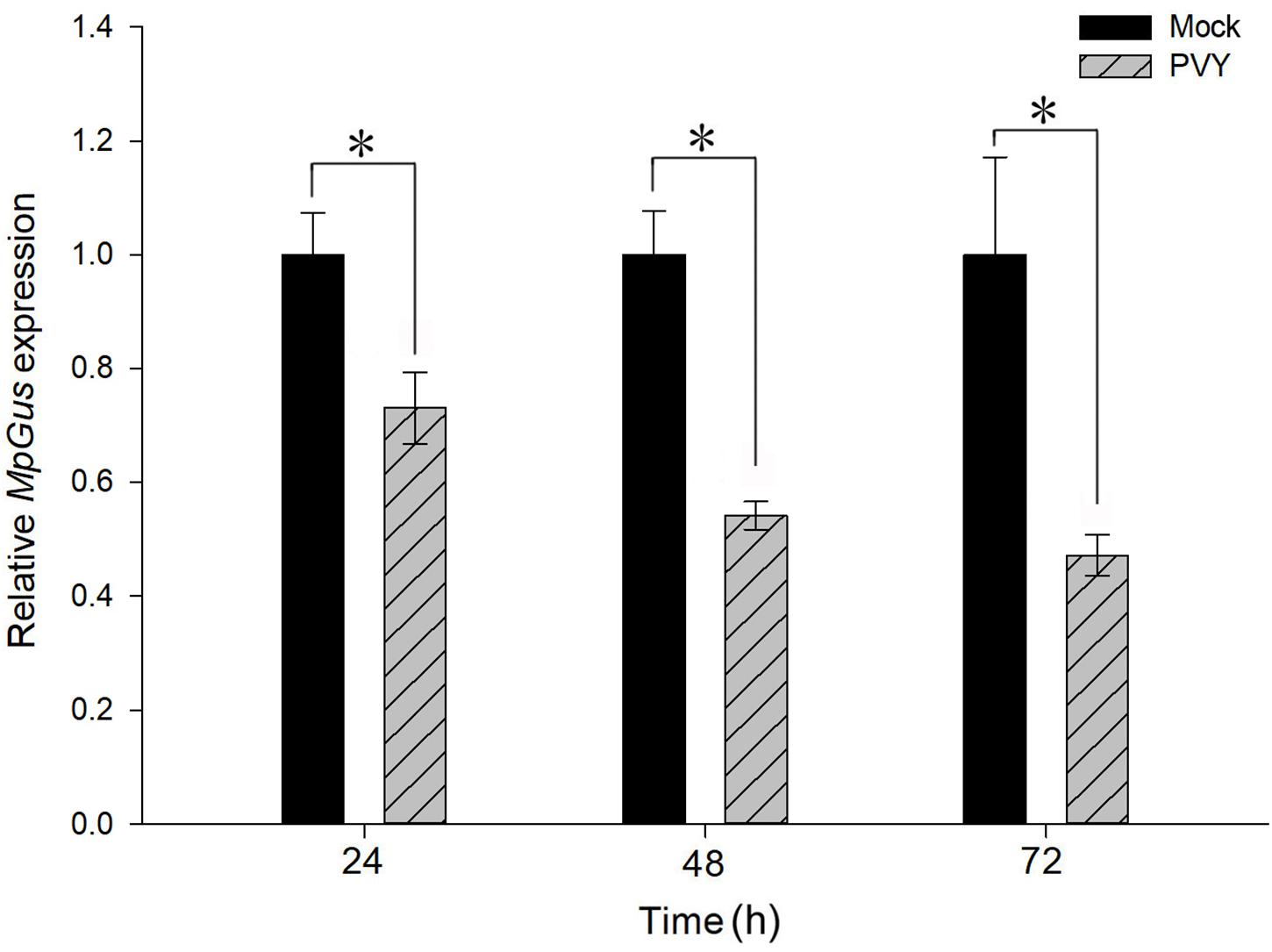
Figure 10. Effects of PVYN-infected tobacco plants on the relative expression of MpGus, as measured by RT-qPCR. Each bar represents the mean of three independent samples, each with three repetitions. β-actin and GAPDH genes from M. persicae were used as references. Values represent mean ± SE, n = 3. Asterisk indicates a significant difference (P < 0.05).
M. persicae Maintained on PVYN-Infected Tobacco Plants Produced a Smaller Number of Nymphs
To determine the effect of PVYN on the fecundity of M. persicae, we counted the numbers of newborn nymphs produced by adult aphids fed on PVYN-infected tobacco plants over a period of 5 days. As shown in Figure 11, both the daily means and the total number of nymphs counted over the 5 days were significantly lower when compared with controls. The mean number of newborn nymphs produced by M. persicae maintained on PVYN-infected tobacco plants decreased by 35.3% [F(1,148) = 94.626, P < 0.01] within 24 h of feeding. However, the level of reduction did not appear to intensify thereafter, as the rate of reduction remained relatively stable at 33.2% [F(1,148) = 91.730, P < 0.01], 33.8% [F(1,148) = 104.113, P < 0.01], 34.8% [F(1,148) = 118.849, P < 0.01], and 33.1% [F(1,148) = 93.663, P < 0.01], over the subsequent 4 days. Collectively, these data suggest that PVYN-infected tobacco plants constituted a hostile environment for M. persicae reproduction, possibly forcing the aphids to migrate to virus-free tobacco, thus facilitating viral transmission. Importantly, the virus-infected plants appear to exert this effect by down-regulating the expression of MpGus, thus highlighting an indispensable role for MpGus in the reproduction of M. persicae.
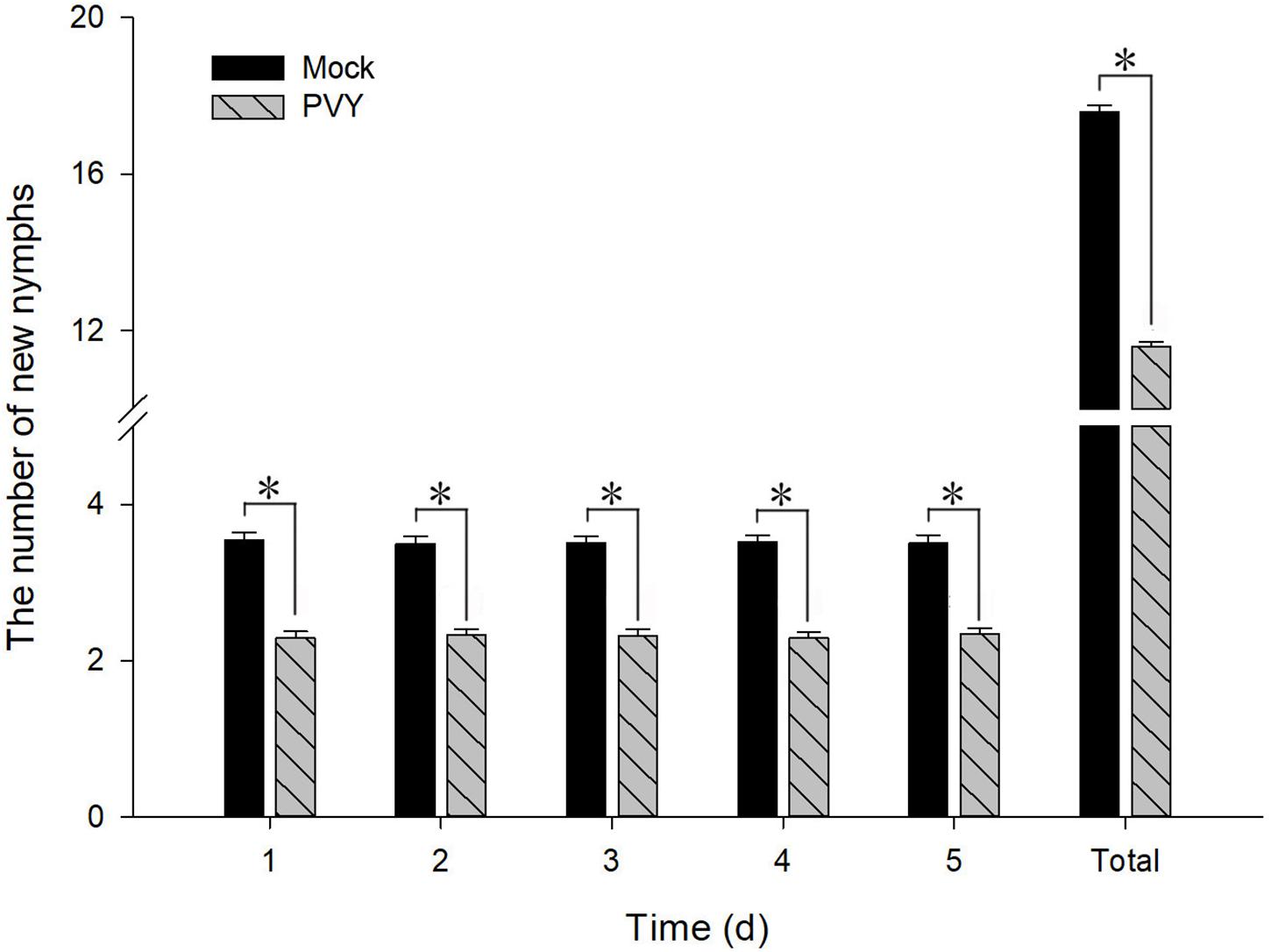
Figure 11. Fecundity of M. persicae fed on PVYN-infected tobacco plants. Newborn nymphs were removed after each count. Values represent mean ± SD. Asterisk indicates a significant difference (P < 0.05).
Discussion
Gustavus is a protein that was first identified in D. melanogaster and found to play an essential part in ensuring the maturation of germline cells (Styhler et al., 2002). A Gus ortholog has since been characterized in a distantly related arthropod (M. nipponense) and was shown to have a strikingly similar reproduction-related role (Zhang et al., 2011). In the present study, we aimed to identify a candidate gene in M. persicae that could be targeted with RNAi to facilitate the control of this agricultural pest. Here, we report the successful identification and preliminary characterization of the Gus ortholog of M. persicae, designated MpGus. This represents the first identification and characterization of a Gus ortholog in an aphid species.
Despite the relatively distant phylogenetic relationship, the MpGus protein shares the highly conserved SPRY domain and SOCS box with its orthologs in D. melanogaster (Styhler et al., 2002) and other insects. These findings were consistent with those reported previously by Xing et al. (2006). Our study showed that about 90 amino acids, located in the N-terminal of the MpGus protein prior to the SPRY domain and SOCS box, were highly conserved in aphid species compared with other species (Figure 4). This species-specific conserved sequence provides us with the possibility to solve the safety problems of RNAi off-target effects on non-target organisms (Roberts et al., 2015). This also suggested that RNAi preparations for MpGus could be used to target a variety of aphids, such as A. gossypii, R. maidis, M. sacchari, D. noxia, and A. pisum. Furthermore, it suggested that MpGus and its ortholog proteins have important biological functions in aphids.
The expression patterns in different tissues demonstrated that MpGus was highly expressed in the abdomens of apterous adult aphids, suggesting a potential role in reproduction. The expression pattern in different instar nymphs showed that the expression of MpGus gradually increased as aphids aged. It has been reported that embryos are present in all developmental stages of aphids, and that mature embryos already existed in nymphs from the fourth instar nymph stage (Xu and Wu, 2020). Styhler et al. (2002) confirmed that the expression of Gus was related to both ovarian and embryonic development in D. melanogaster. Therefore, an increased level of MpGus expression might be related to the development of embryos as well as to reproduction. In addition, the SPRY domain could mediate the release of Ca2+ in the sarcoplasmic reticulum (Ponting, 1997). Zhang et al. (2011) found that the Gus ortholog of M. nipponense is highly expressed in muscle tissue. This suggested that MpGus may also be involved in the development of muscle tissue in nymphs. To further investigate the effect of MpGus on reproduction, we performed RNAi-mediated knockdown experiments on adult aphids.
Our results demonstrated that feeding on 500 ng/μL of dsMpGus for 24 h can significantly reduce the expression of MpGus. We successfully knocked down MpGus expression by feeding adult M. persicae; this resulted in a significant reduction in the number of nymphs produced by these adults. These results further suggest that MpGus regulates the fecundity of M. persicae; these findings are consistent with those of previous studies in D. melanogaster (Styhler et al., 2002; Kugler et al., 2010).
To understand how MpGus affects fecundity, we evaluated the development of embryos in adult M. persicae following the knockdown of MpGus. Similar methods have been used in previous studies to confirm the effects of genes or other factors on the fecundity of aphids (Shang et al., 2017; Xu and Wu, 2020). Our results demonstrated that the RNAi-mediated knockdown of MpGus caused a significant reduction in the number of developing embryos in M. persicae compared with the number of embryos produced by the control group. This reduction in the number of mature embryos explains the decline in fecundity observed in adult aphids affected by dsMpGus. Thus, we speculated that downregulation of MpGus expression affects the processes of embryonic development and maturation, and ultimately reduces the fecundity of M. persicae.
We also found that fecundity was reduced when we fed M. persicae on PVYN-infected tobacco plants. However, our results appear to contradict previous reports by Srinivasan and Alvarez (2007) and Bak et al. (2017), which stated that PVY-infected plants exerted a positive effect on the fecundity of M. persicae. This discrepancy could be attributed to a variety of factors, including viral strains, host plants, M. persicae isolates, and environmental conditions (Ren et al., 2015). Nevertheless, it is notable that the virus-associated reduction in M. persicae fecundity observed in our study was accompanied by lower expression levels of MpGus; this suggests that MpGus plays a role in the regulation of M. persicae fecundity by PVYN-infected plants.
In the experiments described herein, we used home-made mini cages to monitor the daily reproduction of a single aphid maintained on a known plant or artificial diet. This form of investigation has not been described previously. In previous studies, experiments were carried out using a clip cage (2–2.5 cm in diameter) (Srinivasan and Alvarez, 2007; Kersch-Becker and Thaler, 2013; Bak et al., 2017). However, owing to their excessive weight and volume, only 1–2 clip cages can be placed on each leaflet, thus leading to a smaller sample size or the need for a larger number of plants (Srinivasan and Alvarez, 2007; Ren et al., 2015). By contrast, using home-made mini cages, we were able to acquire data from 75 individual aphids on a single tobacco plant in the present study. This strategy helped to reduce statistical error caused by differences between different plants, significantly improved the efficiency of our research, and meant that we could collect data from a larger sample size, thus improving the accuracy of our quantitation. This approach is particularly useful when a large sample size of diverse insects, such as whitefly, thrips, and aphids, is required.
In summary, we identified and characterized the Gus gene in M. persicae (MpGus) and found that MpGus has a critical role in the reproductive cycle of M. persicae. Our results also suggest that MpGus is involved in the effect of PVYN on the fecundity of M. persicae. MpGus may also affect the development and survival of nymphs, but this needs further study. In this study, the downregulation of MpGus inhibited embryo development and reduced fecundity in M. persicae, thus providing evidence that MpGus could be used as a target for the control of M. persicae. Consequently, our findings indicate that MpGus is a new and effective target for the control of M. persicae.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
YL, LD, and DZ designed the research. YG and RR conceived the experiments. YG, JP, and DW analyzed the data. YG drafted the manuscript. XS, LZ, ZZ, CZ, YL, LD, and DZ revised and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the funding from the National Natural Science Foundation of China (Reference: 31801708), Exceptional Talent of Ministry of Agriculture and Rural Affairs of the People’s Republic of China (Reference: 13190200), and the National Key R&D Program of China (Reference: 2017YFD0201103).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.596392/full#supplementary-material
Supplementary Figure 1 | Sequence alignment and electrophoresis detection of PCR product of primer RT-qPCR-MpGus-F/R. (A) Two-sequence alignment between PCR product and MpGus-X3. (B) Agar gelatin electrophoresis result of PCR product.
Supplementary Figure 2 | Amplification specificity and amplification efficiency of primer RT-qPCR-MpGus-F/R in RT-qPCR. (A) Melting curve of RT-qPCR product. (B) Standard curve.
Supplementary Figure 3 | Effect of dsRNA on M. persicae mortality. Each group contained 75 aphids. Counting was carried out for 5 days. Data represent mean ± SD, n = 3. There was no significant difference in mortality among the three groups at each counting point.
Footnotes
- ^ http://www.ncbi.nlm.nih.gov/BLAST/
- ^ https://www.ebi.ac.uk/Tools/msa/clustalo/
- ^ http://www.megasoftware.net/
References
Allen, M. L., and Walker, W. B. (2012). Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J. Insect. Physiol. 58, 391–396. doi: 10.1016/j.jinsphys.2011.12.014
Bak, A., Cheung, A. L., Yang, C., Whitham, S. A., and Casteel, C. L. (2017). A viral protease relocalizes in the presence of the vector to promote vector performance. Nat. Commun. 8:14493. doi: 10.1038/ncomms14493
Bass, C., Puineana, A. M., Zimmera, C. T., Denholm, I., Field, L. M., Foster, S. P., et al. (2014). The evolution of insecticide resistance in the peach potato aphid, Myzus persicae. Insect Biochem. Mol. 51, 41–51. doi: 10.1016/j.ibmb.2014.05.003
Bernays, E. A., and Chapman, R. F. (1994). Host-Plant Selection by Phytophagous Insects. (London: Chapman & Hall), 312. doi: 10.1007/b102508
Bhatia, V., and Bhattacharya, R. (2018). Host-mediated RNA interference targeting a cuticular protein gene impaired fecundity in the green peach aphid Myzus persicae. Pest Manag. Sci. 74, 2059–2068. doi: 10.1002/ps.4900
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Carmo-Sousa, M., Moreno, A., Garzo, E., and Fereres, A. (2014). A non-persistently transmitted-virus induces a pull–push strategy in its aphid vector to optimize transmission and spread. Virus Res. 186, 38–46. doi: 10.1016/j.virusres.2013.12.012
Casteel, C. L., De, A. M., Bak, A., Dong, H. L., Whitham, S. A., and Jander, G. (2015). Disruption of ethylene responses by Turnip mosaic virus mediates suppression of plant defense against the green peach aphid vector. Plant Physiol. 169, 209–218. doi: 10.1104/pp.15.00332
Cervantes, F. A., and Alvarez, J. M. (2011). Within plant distribution of Potato Virus Y in hairy nightshade (Solanum sarrachoides): an inoculum source affecting PVY aphid transmission. Virus Res. 159, 194–200. doi: 10.1016/j.virusres.2011.05.003
Chen, Y. Z., Singh, A., Kaithakottil, G. G., Mathers, T. C., Gravino, M., Mugford, S. T., et al. (2020). An aphid RNA transcript migrates systemically within plants and is a virulence factor. Proc. Natl. Acad. Sci. U.S.A. 117, 12763–12771. doi: 10.1073/pnas.1918410117
Christiaens, O., Niu, J., and Nji Tizi Taning, C. (2020). RNAi in insects: a revolution in fundamental research and pest control applications. Insects 11:415. doi: 10.3390/insects11070415
Dadd, R. H., and Mittler, T. E. (1966). Permanent culture of an aphid on a totally synthetic diet. Experientia 22, 832–833. doi: 10.1007/BF01897447
Gui, S. H., Nji Tizi Taning, C., and Smagghe, G. (2020). Myosuppressin influences fecundity in the Colorado potato beetle, Leptinotarsa decemlineata. Insect Sci. (in press). doi: 10.1111/1744-7917.12855
Gupta, V. (2001). Aphids on the world’s crops. An identification and information guide. Orient. Insects 35:104. doi: 10.1080/00305316.2001.10417292
Gustafson, E. A., Yajima, M., Juliano, C. E., and Wessel, G. M. (2011). Post-translational regulation by gustavus contributes toselective Vasa protein accumulation in multipotent cells during embryogenesis. Dev. Biol. 349, 440–450. doi: 10.1016/j.ydbio.2010.10.031
Hou, Q., Xu, L., Liu, G., Pang, X., Wang, X., Zhang, Y., et al. (2018). Plant-mediated gene silencing of an essential olfactory-related Gqα gene enhances resistance to grain aphid in common wheat in greenhouse and field. Pest Manag. Sci. 75, 1718–1725. doi: 10.1002/ps.5292
Kersch-Becker, M. F., and Thaler, J. S. (2013). Virus strains differentially induce plant susceptibility to aphid vectors and chewing herbivores. Oecologia 174, 883–892. doi: 10.1007/s00442-013-2812-7
Kugler, J. M., Woo, J. S., Oh, B. H., and Lasko, P. (2010). Regulation of Drosophila vasa in vivo through paralogous cullin-RING E3 ligase specificity receptors. Mol. Cell. Biol. 30, 1769–1782. doi: 10.1128/MCB.01100-09
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Liu, F. Z., Yang, B., Zhang, A. H., Ding, D. R., and Wang, G. R. (2019). Plant-mediated RNAi for controlling Apolygus lucorum. Front. Plant. Sci. 10:64. doi: 10.3389/fpls.2019.00064
Liu, X. X., Yang, J. G., Wang, F. L., Qian, Y. M., Shen, L. L., Zhang, S., et al. (2010). Establishment and application of real-time PCR for detection of potato virus Y in tobacco. Chin. Tob. Sci. 31, 9–12. doi: 10.3724/SP.J.1011.2010.01385
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lü, J., Liu, Z. Q., Guo, W., Guo, M. J., Chen, S. M., Yang, C. X., et al. (2020). Oral delivery of dsHvlwr is a feasible method for managing the pest Henosepilachna vigintioctopunctata (Coleoptera: Coccinellidae). Insect Sci. (in press). doi: 10.1111/1744-7917.12784
Mauck, K. E., De Moraes, C. M., and Mescher, M. C. (2014). Biochemical and physiological mechanisms underlying effects of cucumber mosaic virus on host-plant traits that mediate transmission by aphid vectors. Plant Cell Environ. 37, 1427–1439. doi: 10.1111/pce.12249
Ng, J. C. K., and Falk, B. W. (2006). Virus-vector interactions mediating nonpersistent and semipersistent transmission of plant viruses. Annu. Rev. Phytopathol. 44, 183–212. doi: 10.1146/annurev.phyto.44.070505.143325
Ng, J. C. K., and Perry, K. L. (2004). Transmission of plant viruses by aphid vectors. Mol. Plant Pathol. 5, 505–511. doi: 10.1111/j.1364-3703.2004.00240.x
Niu, J., Taning, C. N. T., Christiaens, O., Smagghe, G., and Wang, J. J. (2018). Rethink RNAi in insect pest control: challenges and perspectives. Adv. Insect Physiol. 55, 1–17. doi: 10.1016/bs.aiip.2018.07.003
Pitino, M., Coleman, A. D., Maffei, M. E., Ridout, C. J., and Hogenhout, S. A. (2011). Silencing of aphid genes by dsRNA feeding from plants. PLoS One 6:e25709. doi: 10.1371/journal.pone.0025709
Ponting, C. (1997). SPRY domains in ryanodine receptors (Ca2+-release channels). Trends Biochem. Sci. 22, 193–194. doi: 10.1016/s0968-0004(97)01049-9
Ren, G. W., Wang, X. F., Chen, D., Wang, X. W., Fan, X. J., and Liu, X. D. (2015). Potato virus Y-infected tobacco affects the growth, reproduction, and feeding behavior of a vector aphid, Myzus persicae (Hemiptera: Aphididae). Appl. Entomol. Zool. 50, 239–243. doi: 10.1007/s13355-015-0328-9
Roberts, A. F., Devos, Y., Lemgo, G. N., and Zhou, X. (2015). Biosafety research for non-target organism risk assessment of RNAi-based GE plants. Front. Plant. Sci. 6:958. doi: 10.3389/fpls.2015.00958
Sadeghi, A., Van Damme, E. M., and Smagghe, G. (2009). Evaluation of the susceptibility of the pea aphid, Acyrthosiphon pisum, to a selection of novel biorational insecticides using an artificial diet. J. Insect Sci. 9, 1–8. doi: 10.1673/031.009.6501
Shang, F., Niu, J. Z., Ding, B. Y., Zhang, Q., Ye, C., Zhang, W., et al. (2017). Vitellogenin and its receptor play essential roles in the development and reproduction of the brown citrus aphid, Aphis (Toxoptera) citricidus. Insect Mol. Biol. 27, 221–233. doi: 10.1111/imb.12366
Shrestha, D., Wenninger, E. J., Hutchinson, P. J., Whitworth, J. L., Mondal, S., Eigenbrode, S. D., et al. (2014). Interactions among potato genotypes, growth stages, virus strains, and inoculation methods in the potato virus Y and green peach aphid pathosystem. Environ. Entomol. 43, 662–671. doi: 10.1603/EN13323
Smith, K. M. (1926). A comparative study of the feeding methods of certain Hemiptera and of the resulting effects upon the plant tissue, with special reference to the potato plant. Ann. Appl. Biol. 13, 109–139. doi: 10.1111/j.1744-7348.1926.tb04254.x
Spiller, N. J., Koenders, L., and Tjallingii, W. F. (1990). Xylem ingestion by aphid – a strategy for maintaining water balance. Entomol. Exp. Appl. 55, 101–104. doi: 10.1111/j.1570-7458.1990.tb01352.x
Srinivasan, R., and Alvarez, J. M. (2007). Effect of mixed viral infections (Potato virus Y-potato leafroll Virus) on biology and preference of vectors Myzus Persicae and Macrosiphum euphorbiae (Hemiptera: Aphididae). J. Econ. Entomol. 100, 646–655. doi: 10.1093/jee/100.3.646
Styhler, S., Nakamura, A., and Lasko, P. (2002). VASA localization requires the SPRY-domain and SOCS-box containing protein, GUSTAVUS. Dev. Cell 3, 865–876. doi: 10.1016/S1534-5807(02)00361-1
Tariq, K., Ali, A., Davies, T. G. E., Naz, E., Naz, L., Sohail, S., et al. (2019). RNA interference-mediated knockdown of voltage-gated sodium channel (MpNav) gene causes mortality in peach-potato aphid, Myzus persicae. Sci. Rep. 9:5291. doi: 10.1038/s41598-019-41832-8
Upadhyay, S. K., Chandrashekar, K., Thakur, N., Verma, P. C., Borgio, J. F., Singh, P. K., et al. (2011). RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J. Biosci. 36, 153–161. doi: 10.1007/s12038-011-9009-1
Valkonen, J., Vreugdenhil, D., Bradshaw, J., Gebhardt, C., Govers, F., Mackerron, D. K. L., et al. (2007). “Viruses: economical losses and biotechnological potential,” in Potato Biology and Biotechnology, Advances and Perspectives, ed. D. Vreugdenhil (Amsterdam: Elsevier Science), 619–641. doi: 10.1016/B978-044451018-1/50070-1
van Emden, H. F., Eastop, V. F., Hughes, R. D., and Way, M. J. (1969). The ecology of Myzus persicae. Annu. Rev. Entomol. 14, 197–270. doi: 10.1146/annurev.en.14.010169.001213
van Emden, H. F., and Harrington, R. (eds) (2017). Aphids as Crop Pests, 2nd Edn. (Wallingford: CABI Publishing), 714. doi: 10.1079/9780851998190.0115
Wang, Q., Zhou, J. J., Liu, J. T., Huang, G. Z., Xu, W. Y., Zhang, Q., et al. (2019). Integrative transcriptomic and genomic analysis of odorant binding proteins and chemosensory proteins in aphids. Insect Mol. Biol. 28, 1–22. doi: 10.1111/imb.12513
Weber, G. (1986). Ecological genetics of host plant exploitation in the green peach aphid, Myzus persicae. Entomol. Exp. Appl. 40, 161–168. doi: 10.1111/j.1570-7458.1986.tb00498.x
Xing, Y., Gosden, R., Lasko, P., and Clarke, H. (2006). Murine homologues of the Drosophila gustavus gene are expressed in ovarian granulosa cells. Reproduction 131, 905–915. doi: 10.1530/rep.1.01046
Xu, X. L., and Wu, J. X. (2020). Differences in ovarian and embryonic development between alate and apterous morphs of the soybean aphid, Aphis glycines (Hemiptera: Aphididae). Acta Entomol. Sin. 63, 433–438. doi: 10.16380/j.kcxb.2020.04.006
Yan, S., Qian, J., Cai, C., Ma, Z. Z., Li, J. H., Yin, M. Z., et al. (2019). Spray method application of transdermal dsRNA delivery system for efficient gene silencing and pest control on soybean aphid aphis glycines. J. Pest Sci. 93, 449–459. doi: 10.1007/s10340-019-01157-x
Ye, C., Jiang, Y. D., An, X., Yang, L., Shang, F., Niu, J. Z., et al. (2019). Effects of RNAi-based silencing of chitin synthase gene on moulting and fecundity in pea aphids (Acyrthosiphon pisum). Sci. Rep. 9:3694. doi: 10.1038/s41598-019-39837-4
Yu, N., Christiaens, O., Liu, J., Niu, J., Cappelle, K., Caccia, S., et al. (2013). Delivery of dsRNA for RNAi in insects: an overview and future directions. Insect Sci. 20, 4–14. doi: 10.1111/j.1744-7917.2012.01534.x
Zhan, Y. D., Tian, H. H., Ji, X. L., and Liu, Y. (2020). Myzus persicae (Hemiptera: Aphididae) infestation increases the risk of bacterial contamination and alters nutritional content in storage Chinese cabbage. J. Sci. Food. Agric. 100, 3007–3012. doi: 10.1002/jsfa.10331
Zhang, F., Chen, L., Qin, J., Zhao, W., Wu, P., Yu, N., et al. (2011). cDNA cloning and expression analysis of Gustavus gene in the oriental river prawn Macrobrachium nipponense. PLoS One 6:e17170. doi: 10.1371/journal.pone.0017170
Zhang, J., Khan, S. A., Hasse, C., Ruf, S., Heckel, D. G., and Bock, R. (2015). Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science 34, 991–994. doi: 10.1126/science.1261680
Zhao, Y., Sui, X., Xu, L., Liu, G., Lu, L., You, M., et al. (2018). Plant-mediated RNAi of grain aphid CHS1 gene confers common wheat resistance against aphids. Pest Manag. Sci. 74, 2754–2760. doi: 10.1002/ps.5062
Keywords: Gustavus gene, Myzus persicae (Sulzer), fecundity, RNA interference, potato virus Y
Citation: Gao Y, Ren R, Peng J, Wang D, Shi X, Zheng L, Zhang Z, Zhu C, Liu Y, Dai L and Zhang D (2021) The Gustavus Gene Can Regulate the Fecundity of the Green Peach Aphid, Myzus persicae (Sulzer). Front. Physiol. 11:596392. doi: 10.3389/fphys.2020.596392
Received: 19 August 2020; Accepted: 16 December 2020;
Published: 12 January 2021.
Edited by:
Bin Tang, Hangzhou Normal University, ChinaReviewed by:
Jalal Jalali Sendi, University of Guilan, IranHonggang Tian, Northwest A&F University, China
Copyright © 2021 Gao, Ren, Peng, Wang, Shi, Zheng, Zhang, Zhu, Liu, Dai and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangying Dai, daily@hunau.net; Deyong Zhang, dyzhang73@hotmail.com
 Yang Gao
Yang Gao Ruifan Ren3
Ruifan Ren3 Yong Liu
Yong Liu Deyong Zhang
Deyong Zhang