- 1College of Pharmacy, Hubei University of Chinese Medicine, Wuhan, China
- 2Chinese Medicine Testing and Research Center, Hubei Institute for Drug Control, Wuhan, China
Bungarus multicinctus, an important traditional Chinese medicine, possesses remarkable medicinal activities, while lots of adulterants from other species were misused as B. multicinctus for its large demand and resource starvation. In order to accurately identify B. multicinctus and its common adulterants such as Sinonatrix annularis, Xenochrophis flavipunctatus, Deinagkistrodon acutus, and Naja atra, a simultaneous identification method was designed with multiplex ligation-dependent probe amplification (MLPA) analysis. Five species-specific MLPA probe-couples for B. multicinctus and its common adulterants were designed based on the universal primer amplified COI sequences, which can specifically detect the five species with no mutual interference, and sensitivity analysis showed as less as 5% B. multicinctus or 8.75% adulterants in the mixed samples can be identified in a MLPA assay, especially, the relative quantity of the adulterants can be also inferred based on the MLPA peak area values. Moreover, the results of the present study confirmed the effectiveness of this technique in terms of simultaneous identification of B. multicinctus and its common adulterants in an assay, which has great potential for ensuring the safety of this commercially valuable snake species.
Introduction
According to the reptile database, there are 3,701 kinds of snake species in the world, 286 of which are distributed in China (Yanqing et al., 2019) and about the one tenth are frequently used for medicinal and commercial purposes, for example, snake venom and tissues are used to treat diseases, skins are used in the luxury leather goods industry (Singh et al., 2012; Murray-Dickson et al., 2017). In China, 17 snake species were recorded in the world-famous medical book Compendium of Materia Medica (Pipeng et al., 2013), and there are currently still five snake species included in the Chinese Pharmacopoeia (2015 Edition) as statutory provisions for traditional Chinese medicine. B. multicinctus, belonging to the Elapidae family, is a famous statutory medicinal snake species with effect of dispelling wind and removing obstruction in the meridians, and it was also used as a chief ingredient in many polyherbal formulations for the treatment of cancer (Wuguang et al., 2017; Linchun et al., 2018). However, due to the low reproductive capacity of B. multicinctus and the habitat destruction, the survival of this snake species had been threatening, which exerted a detrimental impact on ecosystems and biodiversity. Furthermore, based on its good effect in treating rheumatoid arthritis, epilepsy, urticaria, and cervical spondylosis (Jinting, 2008), this officinal snake is in huge demand, then, artificially propagated B. multicinctus and its counterfeits were gradually appeared in the medicine market (Tingping, 2001). For the purpose of protecting of species diversity and monitoring the safety of the medical market, it is necessary to develop methods for distinguishing this species.
For resource protection and drug safety, it is generally believed that the first step is to accurately identify species. The morphological characteristics of B. multicinctus and some other processed snakes (Chao et al., 2015), such as D. acutus, N. naja (Yiquan et al., 2000), are very similar. So, morphological examination can´t successfully be employed without visually recognizable features or has been otherwise altered in those species (Yanqing et al., 2019; Yi et al., 2017). In this regard, molecular identification techniques have been proven to be an effective tool for species identification. Although some interesting molecular biology techniques, such as sequence characterized amplified region (SCAR) (Yau et al., 2002) and polymerase chain reaction (PCR) (Jingxue et al., 2010) have been applied to snake identification, these methods can distinguish the authenticity of snake species but not perform simultaneous identification and relative quantitative analysis. In the world, one fifth of snakes are poisonous, most of them belong to Viperidae, Elapidae, Colubridae, and Atractaspididae families (Chi-Hsin et al., 2016). In clinical practice where the snakes are often swallowed by powder, misuse or mixed use of these poisonous snakes will bring hidden dangers to patients.
The multiplex ligation-dependent probe amplification (MLPA) (Schouten et al., 2002) as an emerging molecular diagnostic technique can detect the difference in nucleic acid sequences with a small amount of DNA and possess capability of detecting up to 50 genomic DNA sequence difference in one reaction relied on DNA denaturation, hybridization, ligation, and PCR reaction (Os and Schouten, 2011; Samelak-Czajka et al., 2017). Recently, the technology has been applied to the clinical diagnosis of diseases, such as Duchenne muscular dystrophy (Guertler et al., 2014), Gaucher disease (Schouten et al., 2002), Acute leukemia (Reyes-Núñez et al., 2017), and it also has been received wide attentions in the field of allergen detection (López-Calleja et al., 2016), genetically modified species identification (Holck et al., 2009), and herb identification (Bo et al., 2018).
In the present study, B. multicinctus and the four species usually acted as its adulterants were studied. Five species-specific MLPA probe-couples were designed based on the mitochondrial amplification sequences of the five snake species. The specificity of the probes was verified by hybridization of single probe or probemix to the DNA target, and the proportion of the adulterants in B. multicinctus was estimated by the MLPA peak areas obtained from the capillary electrophoresis analysis. The current study provided a method for simultaneous identification and relative quantification of the adulterants in this important valuable snake, and also provided a reference for adulterants identification in other medicinal species.
Materials and Methods
Sample Material and DNA Extracted
These samples, including B. multicinctus, S. annularis, X. flavipunctatus, D. acutus, and N. atra were collected Guangxi, Yunnan, Hubei, Hunan, Jiangxi, and Zhejiang Province, and identified by Dr. Bo Wang who works in the area of species authenticity and certification in Hubei Institute for Drug Control. The voucher specimens were deposited at the Chinese medicine specimen museum of Hubei Institute for Drug Control and Institute of Chinese Materia Medica China Academy of Chinese Medical Sciences, the sample numbers in two institutions were: HBYJ2016101- HBYJ2016125, and 1040412001-1040412025 respectively. The species identification was confirmed using the COI universal barcode sequence. The total DNA of these samples were extracted using animal tissue genomic DNA kit (Zoman Biotechnology Co., Ltd., Beijing, China) as described by the manufacturer’s instruction. The concentration of DNAs was estimated using ND-2000 spectrometer (Nanodrop Technologies, Wilmington, DE, USA). DNAs were stored at −20°C for further analysis. It is worth mentioning that the collection of all samples studied in this experiment was in compliance with relevant legal provisions, and we required only a small tissue sample, which does not affect snake resources. The protocol was approved by Institutional Animal Care and Use Committee affiliated with the Hubei Provincial Academy of Preventive Medicine & Hubei Provincial Center for Disease Control and Prevention.
Hybrid Sequence Screening and Phylogenetic Reconstruction
A pair of universal primers (Forward primer: 5′-TATTCTCAACTAACCACAA AGA-3′; Reverse primer: 5′-ACTTCTGGTTGACCAAAGAATCA-3′) was screened to amplify the DNA fragments from the five species. PCR amplification was performed in a total volume of 25 µl containing 21 µl of PCR Mix (TsingKe, Beijing, China), 1 µl of forward primer (10 µM, Sangon), 1 µl of reverse primer (10 µM, Sangon), 2 µl of total DNA. Thermal cycling was performed in a Bio-Rad thermocycler (Bio-Rad, California, CA, USA) using a PCR process: 94°C for 3 min; 40 cycles of 94°C for 40 s, 48.5°C for 30 s, and 72°C for 60 s; 72°C for 7 min, and subsequent storage at 4°C. The amplified fragments were sequenced by the ABI3730XL sequencer (Applied Biosystems Co., Shanghai, China). Sequencing data were processed by the CodonCode Aligner V3.7.1 (CodonCode Co., Centerville, MA, USA). Phylogenetic relationship was analyzed by the neighbor-joining (NJ) method in the MEGA7 program (National Institutes of Health, Bethesda, MD, USA).
MLPA Probe Design
The species-specific MLPA probes for B. multicinctus, S. annularis, X. flavipunctatus, D. acutus, and N. atra were designed base on the COI sequences amplified by the universal primers, and were referenced to the suggestions of MRC-Holland (Amsterdam, Netherlands; http://www.mrc-holland.com). In order to find the suitable regions for designing the species- specific probes, the sequences from the five species were aligned and analyzed in the NCBI database (http://www.ncbi.nlm.nih.gov). The LPO and RPO folding and ΔG were tested by mfold Web Server (http://unafold.rna.albany.edu/?q=mfold) to choose the optimal probes. In order to avoid interfering cross-reactivities, Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to evaluate the specificity of five species-specific MLPA probe-couples.
MLPA Assay
All steps of the MLPA reactions were performed in thermocycler (Bio-Rad, California, CA, USA) using the SALSA MLPA EK1 reagent kit (MRC-Holland, Amsterdam, Netherlands) according to manufacturer’s instruction. All components were included in the SALSA MLPA EK1 reagent kit, except DNA samples, ddH2O, probes, and TE buffer. Routinely, 5 µl of DNA sample (50–100 ng) was denatured at 98°C for 5 min. A non-template control with 5 µl of TE buffer was included to reveal possible reagent contaminations. After all samples were cooled to 25°C, removed from the thermocycler to add 3 µl hybridization mix (1.5 µl SALSA MLPA buffer and 1.5 µl probes) and heated at 95°C for 1 min, 60°C for 16 h. For the ligation reaction, 32 µl ligase mix (25 µl ddH2O, 3 µl ligase buffer A, 3 µl ligase buffer B, and 1 µl Ligase-65 enzyme) were added and incubated for 15 min at 54°C. Subsequently, the enzyme was inactivated by heated at 98°C for 5 min. Amplification of the ligation products was carried out with the universal SALSA PCR primers by adding 10 µl polymerase mix (7.5 µl ddH2O, 2 µl SALSA PCR primer mix, and 0.5 µl SALSA polymerase) and implemented the PCR reaction at 35 cycles of 95°C 30 s, 60°C 30 s, 72°C 1 min; 72°C 20 min. PCR products were stored in the dark at 4°C until further analysis. To assess MLPA reproducibility, three replicates of each DNA extract were analyzed in each experiment.
Capillary Electrophoresis
For each reaction, 1 µl PCR product, 9 µl HiDi formamide, and 0.5 µl Genescan 500 LIZ size standard were mixed. The samples were denatured at 95°C for 5 min and then cooled on ice. Collected fragment data were performed 3130xl Genetic analyser (Applied Biosystems) using the following settings: run temperature: 60 °C; injection voltage: 1.2 kV; injection time: 16 s; run voltage: 15 kV, run current: 5 µA, and run time: 1800 s. The size, peak height, and peak area of fluorescent PCR fragments were extracted by GeneMaeker (SoftGenetics, Pennsylvania, PA, USA) and used for quantitative analysis.
Specificity and Sensitivity of MLPA
The sample of B. multicinctus and its common adulterants were used to assess the specificity of the MLPA test, which we tested by DNA hybridized with single probe and probemix, respectively. The sensitivity of MLPA was assessed by change the amount of DNA from 200ng to 2.5ng.
Detection of Mixed Samples
To detect B. multicinctus and its common adulterants in a mixed sample, we mixed B. multicinctus and the four common adulterants into different proportions as composite samples. The ratio of B. multicinctus was 65%, 50%, 30% and 5%, respectively. After capillary electrophoresis analysis, we can visually observe the actual proportion of the five species in the composite sample.
Results
Identification Sequence Analysis
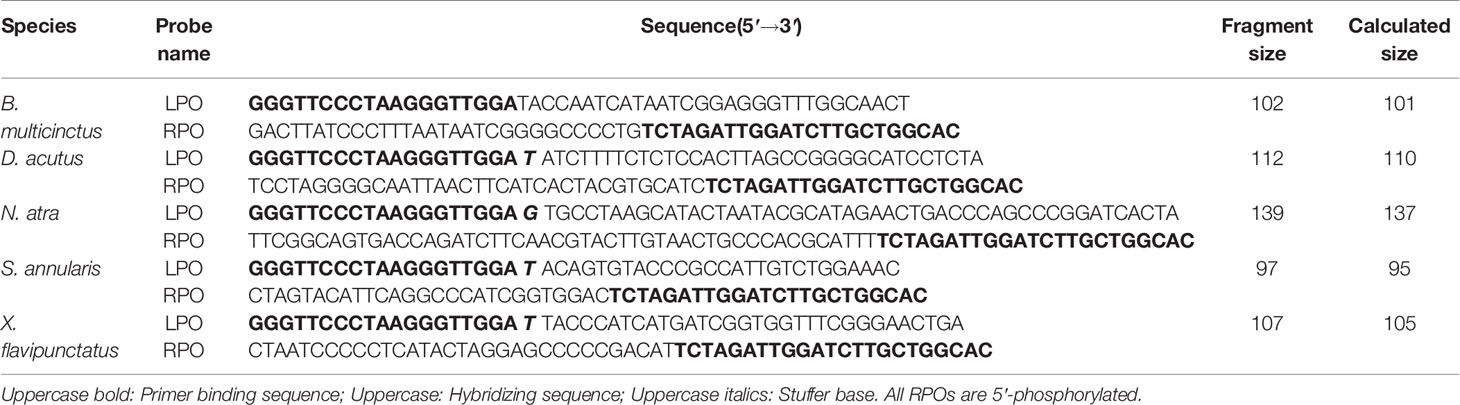
In order to ensure the accuracy and reproducibility of the assay, a NJ tree of the five species was constructed using the snake sequences amplified by the universal primers (Limpiti et al., 2014) (Figure 1). As can be seen from the NJ tree, each species was clustered into one branch, and the support rate was 100%. It was strongly supported that the position of S. annularis and X. flavipunctatus as a sister of the close related species in the Colubridae. For the same reason, both B. multicinctus and N. atra belonging to Elapidae were clustered together. This result indicated that the sequences amplified by the universal primers can be used to distinguish five species and design the species-specific probes for next MLPA analysis. Based on this, the species-specific MLPA probes toward the five snake species were designed, and were shown in Table 1.
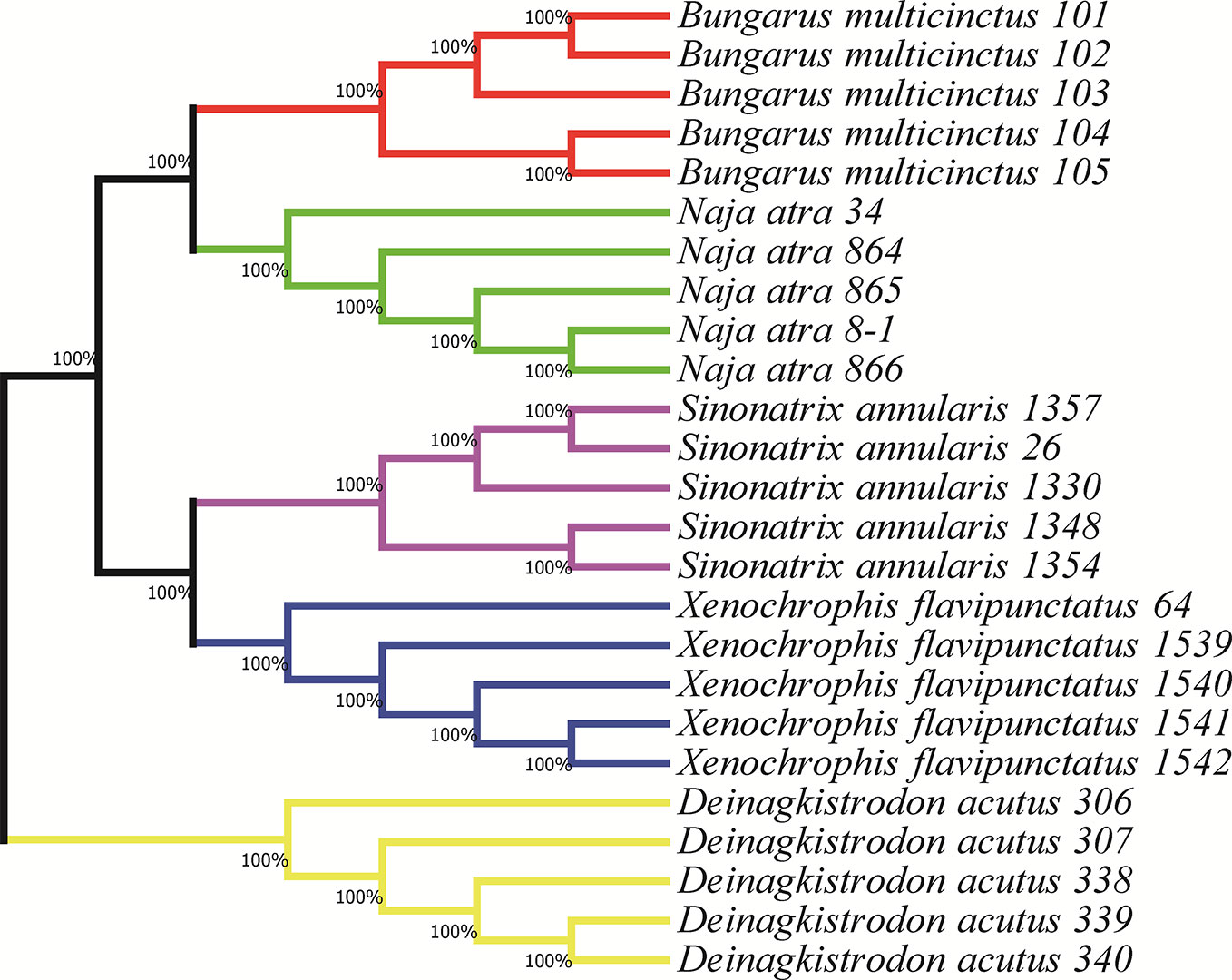
Figure 1 Phylogenetic tree constructed using neighbor joining (NJ), based on the sequences amplified by universal primers from five species.
Evaluation of MLPA Specificity
The complete hybridization sequence (left hybridizing sequence and right hybridizing sequence) for each probe was aligned with the Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and each probe was only matched to a unique species, which theoretically excluding cross-contamination. To assess the specificity of the probes, the single probe and probemix were hybridized to the DNA samples for 16 h, respectively. The size of amplification fragments which hybridization of the DNA target and the single probe were in line with the expected size, S. annularis 95 nt, B. multicinctus 101 nt, X. flavipunctatus 105 nt, D. acutus 110 nt, and N. atra 137 nt (Figure 2). The DNA target of the five species were hybridized with the single probe, the actual length of amplicons were differed from the designed length by 1 to 2 nt, which was due to the different mobility of DNA and/or the distinct labeling of the standard and the amplified fragments (García-García et al., 2017).
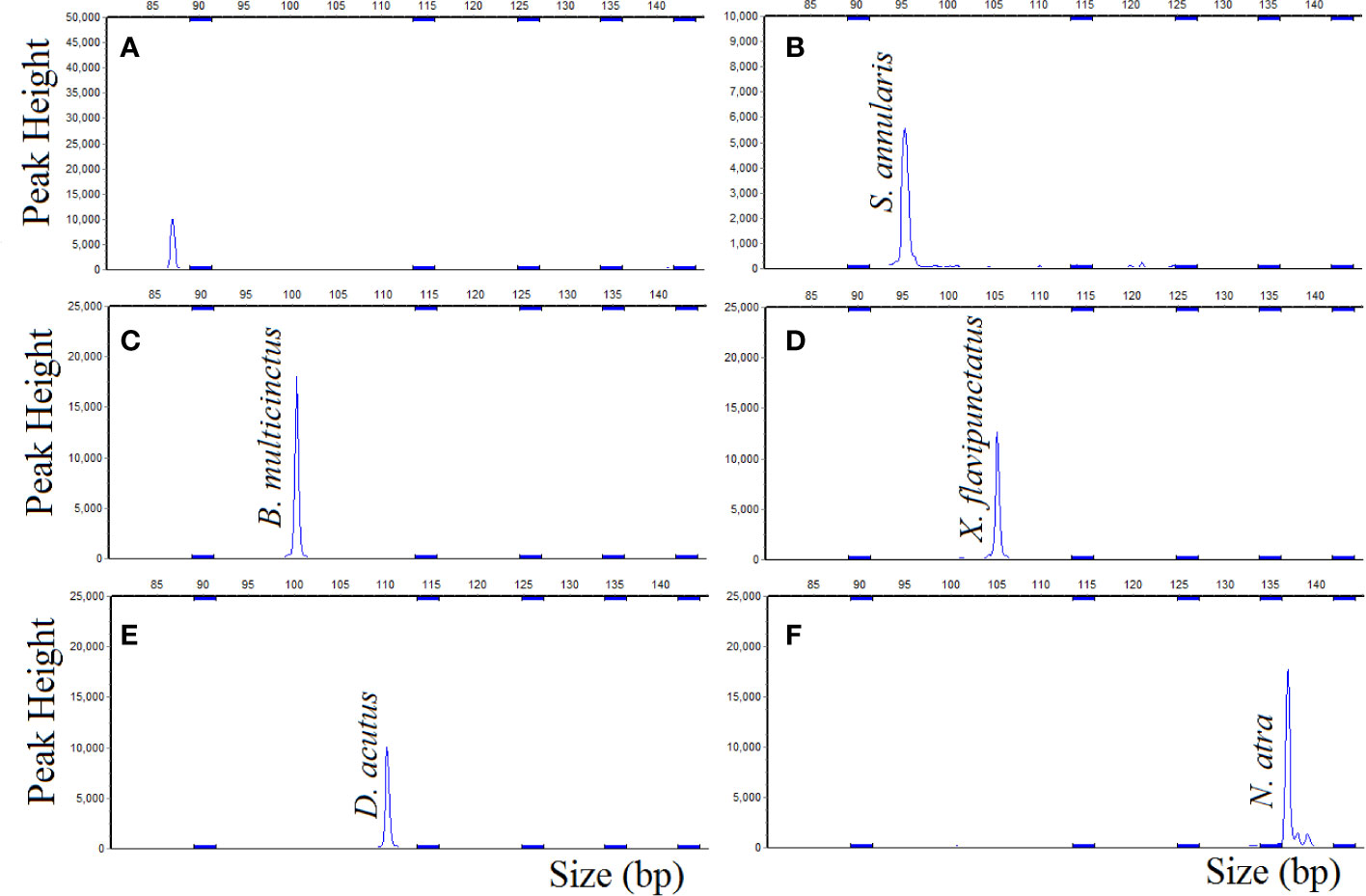
Figure 2 MLPA single probe specificity, DNA target hybridized with single probe. (A) negative control; (B) S. annularis; (C) B. multicinctus; (D) X. flavipunctatus; (E) D. acutus; (F) N. atra.
The results of DNA target hybridized with the probemix (Figure 3) were similar to the results that DNA target hybridized with the single probe. The corresponding probes gave positive signal for S. annularis, B. multicinctus, X. flavipunctatus, D. acutus, and N. atra, respectively, as expected, which was also stable in gradually increased hybridization temperature from 60°C to 68°C with a gradient of 1°C. The results showed that the MLPA probes exhibit strong specificity and reproducibility toward in five species.
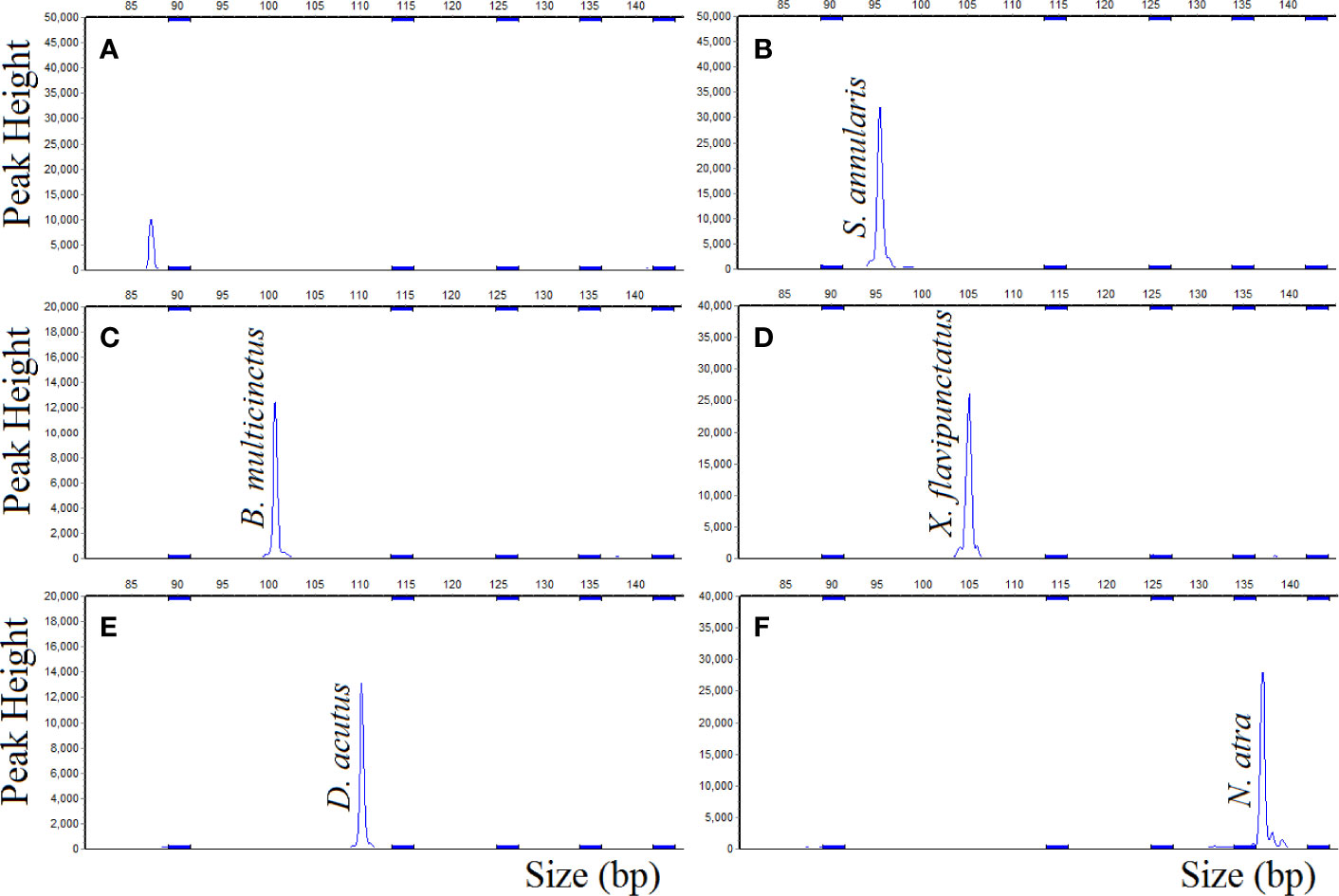
Figure 3 MLPA probemix specificity, DNA sample hybridized with probemix. (A) negative control; (B) S. annularis; (C) B. multicinctus; (D) X. flavipunctatus; (E) D. acutus; (F) N. atra.
Evaluation of MLPA Sensitivity
To verify the ability of MLPA method, five species of snakes were detected in order to promote the application of this technology in traditional Chinese medicine. The DNA concentration of the B. multicinctus and its common adulterants was adjusted to 20 ng/µl before evaluating the sensitivity. Five targets were hybridized with probemix, changing the amount of total DNA samples from 200 ng to 50 ng, all signal peaks were with the expected size, which indicated that the five species could be detected simultaneously even if change the amount of DNA samples. The amount of B. multicinctus DNA sample (100 ng, 80 ng, 60 ng, 32.5 ng, 25 ng, 15 ng, 10 ng, 5 ng, 2.5 ng) and the amount of adulterant DNA sample (80 ng, 47.5 ng, 35 ng, 25 ng, 23.75 ng, 17.5 ng, 11.785 ng, 8.75 ng, 6.25 ng, 4.375 ng) were changed until the amount of B. multicinctus DNA and adulterant DNA were reduced to 2.5 ng and 4.375 ng, respectively, the signal peak of the sample could be detected. The DNA extraction method in this experiment can obtain 10 to 30 µg total DNA from 30 mg sample tissue. In order to obtain 2.5 ng total DNA, only 0.0025 ~0.0075 mg samples were required, indicating that the method requires very little sample. Thus the MLPA method can be used to identify B. multicinctus and its common adulterants in the traditional Chinese medicine market.
Quantitative Adulterations of B. multicinctus Using MLPA Analysis
After MLPA detected the composite samples, the sizes and peak areas of the amplified fragments from the five species were directly obtained from capillary electrophoresis, indicating that five species could be simultaneously identified, differentiated, and relatively quantified by MLPA. The ratio of B. multicinctus in the composite sample was 65%, 50%, 30%, 5%, and the peak area ratio of B. multicinctus was 31.8%, 25.4%, 14.8% and 14.6%, indicated that with the decrease of B. multicinctus DNA target, the peak area of B. multicinctus was decrement. Comparing the ratio of B. multicinctus in the composite samples (theoretical ratio) with the peak area ratio of B. multicinctus (actual ratio), it could be found that when the theoretical ratio of B. multicinctus was more than 30%, the theoretical ratio was nearly twice the actual ratio (Figure 4). The concentration of the DNA samples has been adjusted before the experiment, however, the actual ratio was different from the theoretical ratio possibly due to the different copy number of the five species. When B. multicinctus accounted for 65% of the total DNAs and the four adulterants for 8.75%, five species could be detected simultaneously by MLPA method (Figure 4A). After changing the amount of B. multicinctus to 5%, signal peak of B. multicinctus was still detected (Figure 4D). This means that when B. multicinctus accounted for more than 30%, the proportion of the adulterants can be inferred and when the ratio of B. multicinctus was less than 30%, it can be determined whether the adulterants exist. As a result, the developed MLPA method for B. multicinctus and its common adulterants detection is prompt and reliable for situations where B. multicinctus may be contaminated by its common adulterants.
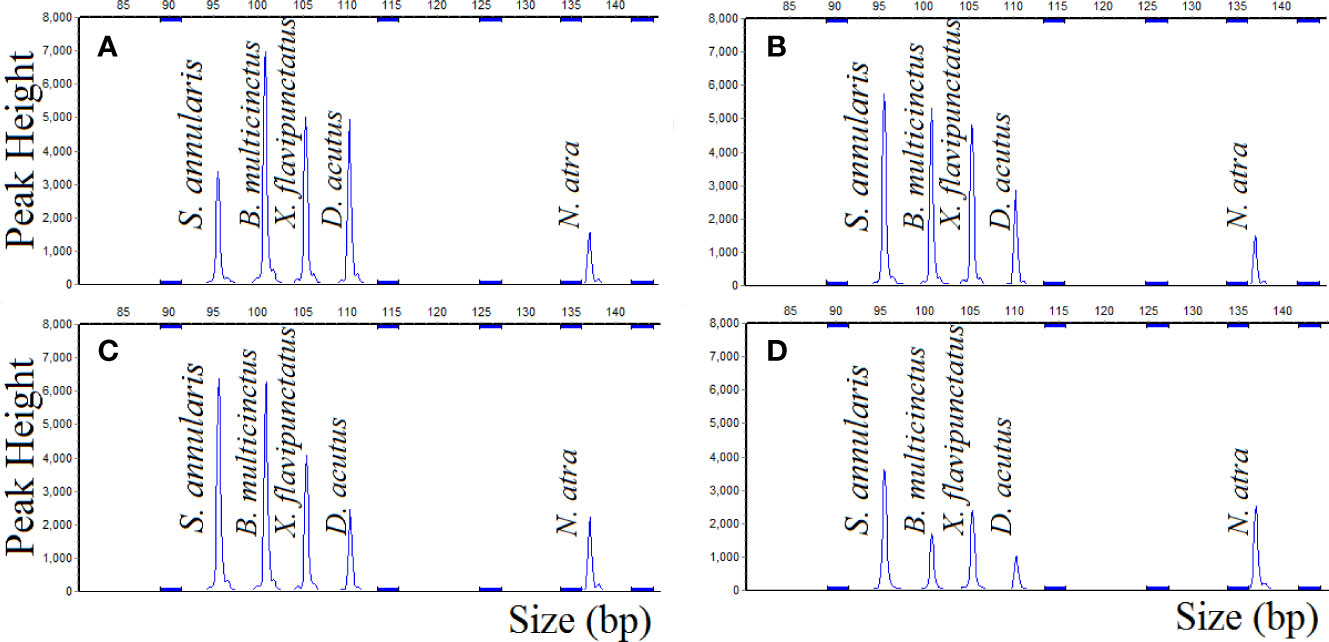
Figure 4 MLPA method simultaneously identifies, differentiates, and relatively quantifies five species (A) B. multicinctus accounts for 65%; (B) B. multicinctus accounts for 50%; (C) B. multicinctus DNA sample accounts for 30%; (D) B. multicinctus DNA sample accounts for 5%.
Discussion
In this study, we investigated the usefulness of the MLPA method as a prompt and effective way to detect S. annularis, X. flavipunctatus, D. acutus, and N. atra, as common adulterants of B. multicinctus. Previous studies have shown that when morphological identification and PCR-based assay are used to authenticate B. multicinctus and its common adulterants (Chengyong, 2002), the results either subjective, inconspicuous, relies on experience, or expensive, time and resource consuming. MLPA, combine the characteristics of DNA probe hybridization and PCR technology, is an easy-to-learn, fast, high throughput, reliable, and low-cost technology. MLPA assay can be performed with thermocycler and capillary electrophoresis system. The MLPA probes are very simple, the base at the left probe 3′-terminal base and ligase-65 enzyme ensure the specificity of the ligation reaction, the right probe 5′-terminal base phosphorylation promote the ligation reaction (Cornelia and Suvi, 2012). Both left probe and right probe can be quickly obtained by synthesis, greatly reducing the test period (Stern et al., 2004). Probe hybridization sequences are short (50~70 nt) (Schouten et al., 2002). It is possible that this method can be used to detect highly fragmented DNA sample. With this advantage, MLPA can be used to detect samples that have been stored for many years, processed and DNA-degraded. The amount of DNA is small, such as research congenital adrenal hyperplasia (CAH) by MLPA, DNA concentration down to 0.3 ng/µl (1.6 ng total) (Karina Meden et al., 2008). In MLPA, a pair of primers amplified all species sequences, and therefore, the amplification efficiency of the sequences was same or similar. Despite the many advantages of the MLPA reaction, this method does have several non-fatal drawbacks. First, we recognized that the MLPA method avoids complicated steps, such as cell culture in medical diagnosis and significantly shortens the diagnosis period, but the long hybridization time is a disadvantage in identification of traditional Chinese medicine. In this work, we referred previous study to shorten the hybridization time to 3 h when evaluating the specificity of MLPA probes (Guertler et al., 2014), and each assay was repeated three times with expected and stable results. When analyzing the relative quantification of the five species in the composite samples, the hybridization time was extended to 16 hours in order to ensure sufficient hybridization of the DNA target and the probemix. However, it has been verified that shortening hybridization time to 30 min can still ensure the stability of the experiment, and the results can be obtained within 6 hours (Saxena et al., 2015). Second, it is important to select proper barcode marker and design the probe. Proper barcode marker should have the feature that effectively distinguish the species involved, and are based on common barcode markers like ITS2, COI, etc.
There are various forms of counterfeit in traditional Chinese medicine. Any medicine that is in large demand and high price has the possibility of adulteration. Common adulterations species were similar in genetic relationships or morphological characteristics. Some adulterants were mixed into traditional Chinese medicine in the form of power. How to accurately identify and quantify adulteration products has become a major problem. DNA-based identification technology was more stable than other material-based identification technologies for its higher stability, cell uniformity, and polymorphism (Sultana et al., 2018). Mitochondria are autonomous organelles in eukaryotes, mtDNA has high rate of evolution, stable amplification, and repeatability. It is rich in genetic differences among different species and is suitable for species identification among order, family, genus, and species (Meng et al., 2018). In the present study, the COI sequence, a gene in mtDNA, was selected to differentiate the five species. After we obtained two sets of species-specific probe from five species, we tested the specificity and determined that only one set could be used for this study.
Compared with identification methods in the previous studies, the highlights of MLPA are the advantages in both simultaneous identification of B. multicinctus and its common adulterants that are S. annularis, X. flavipunctatus, D. acutus, and N. atra, and relative quantitative analysis of the sample. Comparing the method used to identify B. multicinctus indicated that MLPA is a superior process because of easy-to-learn, fast, high throughput, reliable, and low-cost. With the improvement of material living standards, people´s attention on the safety of traditional Chinese medicine is increasing, and the governmental agencies must take effective measures to regulate the safety of traditional Chinese medicine. Our study results indicate that the MLPA method can accurately identify the authenticity of traditional Chinese medicine and ensure the safety of consumers.
Conclusions
Our findings showed that the species-specific MLPA probes can distinguish the snakes including B. multicinctus and its common adulterants that is S. annularis, X. flavipunctatus, D. acutus, and N. atra. A MLPA analysis method was proposed for the identification, differentiation, and relative quantification of these snake species, which proved to be a very sensitive tool for the simultaneous identification of snake genotypes. This method was sensitive enough to detect as less as 5% B. multicinctus or 8.75% adulterant in mixed samples, revealing its adequacy as a simple and fast approach for species simultaneous identification. Thus, this technology has potential for market detection and relative quantification of the adulterants in this important valuable snake, and also provided a reference for adulterants identification in other medicinal species.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Ethics Statement
The animal study was reviewed and approved by the Hubei Provincial Academy of Preventive Medicine & Hubei Provincial Center for Disease Control and Prevention.
Author Contributions
Formal analysis: YZ, SY. Methodology: ZH, BW. Project administration: JN, BW. Writing – original draft: YZ. Writing – review and editing: BW. All authors read and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (81603246), and the Foundation of Hubei Food and Drug Administration (201801001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the Institute of Chinese Materia Medica China Academy of Chinese Medical Sciences and the Institute of Medicinal Plant Development for their assistance in collecting research samples.
References
Bo, W., Yuxin, Z., Gui, Q., Jing, N. (2018). Identification of adulteration in Fritillariae Cirrhosea Bulbus using multiplex ligation-dependent probe amplification technology. Chin. J. Pharm. Anal. 38 (12), 68–73. doi: 10.16155/j.0254-1793.2018.12.07
Chao, J., Yuan, Y., Libing, L., Jingyi, H., Yan, J., Luqi, H. (2015). Homogeneous fluorescent specific PCR for the authentication of medicinal snakes using cationic conjugated polymers. Sci. Rep. 5, 16260. doi: 10.1038/srep16260
Chengyong, H. (2002). Survey of studies on identification of Bungarus multicinctus. Chin. Tradit. Herb. Drugs 38 (12), 68–73. doi: CNKI:SUN:ZCYO.0.2002-04-044
Chi-Hsin, L., Yu-Ching, L., Sy-Jye, L., Liang-Tzung, L., Jen-Ron, C., Wei-Jane, H., et al. (2016). Production and Characterization of Neutralizing Antibodies against Bungarus multicinctus Snake Venom. Appl. Environ. Microbiol. 82 (23), AEM.01876–01816. doi: 10.1128/AEM.01876-16
Cornelia, H. M. H. L., Suvi, S. (2012). Multiplex ligation-dependent probe amplification (MLPA) in tumor diagnostics and prognostics. Diagn. Mol. Pathol. Am. J. Surg. Pathol. Part B 21 (4), 189–206. doi: 10.1097/PDM.0b013e3182595516
García-García, A., Madrid, R., García, T., Martín, R., González, I. (2017). Use of multiplex ligation-dependent probe amplification (MLPA) for screening of wheat, barley, rye and oats in foods. Food Control 84, 268–277. doi: 10.1016/j.foodcont.2017.07.037
Guertler, P., Goerlich, O., Busch, U. (2014). Development of a Multiplex Ligation Dependent Probe Amplification (MLPA) Module for Simultaneous Detection of Five Genetically Modified Rapeseed Events. Agric. Sci. 5 (6), 530–539. doi: 10.4236/as.2014.56055
Holck, A. L., Drømtorp, S. M., Heir, E. (2009). Quantitative, multiplex ligation-dependent probe amplification for the determination of eight genetically modified maize events. Eur. Food Res. Technol. 230 (2), 185–194. doi: 10.1007/s00217-009-1155-4
Jingxue, Z., Guanghong, C., Mintong, X., Shihuan, T. (2010). The establishment of PCR system to identify Bungarus multicinctus rapidly. Acta Pharm. Sin. 10 (10), 1327–1332. doi: 10.16438/j.0513-4870.2010.10.009.
Jinting, H. (2008). Pharmacological action and clinical application of Bungarus multicinctus. J. Modern Med. Health 24 (14), 2132–2133. doi: CNKI:SUN:XYWS.0.2008-14-054
Karina Meden, S. R., Paal Skytt, A., Lars Allan, L., Marianne, S., Schouten, J. P., Nygren, A. O. H. (2008). Multiplex ligation-dependent probe amplification technique for copy number analysis on small amounts of DNA material. Anal. Chem. 80 (23), 9363–9368. doi: 10.1021/ac801688c
López-Calleja, I. M., García, A., Madrid, R., García, T., Martín, R., González, I. (2016). Multiplex ligation-dependent probe amplification (MLPA) for simultaneous detection of DNA from sunflower, poppy, flaxseed, sesame and soy allergenic ingredients in commercial food products. Food Control. 71, 301–310. doi: 10.1016/j.foodcont.2016.06.014
Limpiti, T., Amornbunchornvej, C., Intarapanich, A., Assawamakin, A., Tongsima, S. (2014). iNJclust: Iterative Neighbor-Joining Tree Clustering Framework for Inferring Population Structure. IEEE/ACM Trans. Comput. Biol. Bioinf. 11 (5), 903–914. doi: 10.1109/TCBB.2014.2322372
Linchun, S., Xianming, T., Zhigang, H., Chunying, Z., Lixiao, F., Jinxin, L. (2018). Investigation of medicinal sources for Bungarus Parvus by DNA barcoding technology. Chin. J. Exp. Tradit. Med. Formulae 24 (18), 16–23. doi: 10.13422 /j.cnki.syfjx.20181706
Meng, C., Yun, F., RuBin, C., Zhen, H., Guangji, Z., Jianzhen, C. (2018). Application progress on mitochondrial DNA markers in identification of animal medicinal materials. Chin. Tradit. Herb. Drugs 49 (13), 3134–3142. doi: 10.7501/j.issn.0253-2670.2018.13.027
Murray-Dickson, G., Ghazali, M., Ogden, R., Brown, R., Auliya, M. (2017). Phylogeography of the reticulated python (Malayopython reticulatus ssp.): Conservation implications for the worlds’ most traded snake species. PloS One 12 (8), e0182049. doi: 10.1371/journal.pone.0182049
Os, E. V., Schouten, J. P. (2011). Multiplex Ligation-dependent Probe Amplification (MLPA®) for the detection of copy number variation in genomic sequences. Methods Mol. Biol. 688, 97. doi: 10.1007/978-1-60761-947-5_8
Pipeng, L., Weisheng, W., Xiaoping, L. (2013). Overview of Chinese Snake Protection and Utilization: History, Current Situation and Future. J. Shenyang Normal Univ. (Nat. Sci.) 31 (2), 0129–0135. doi: 10.3969/j.issn.1673-5862.2013.02.001
Reyes-Núñez, V., Galo-Hooker, E., Pérez-Romano, B., Duque, R. E., Ruiz-Arguelles, A., Garcés-Eisele, J. (2017). Simultaneous use of Multiplex Ligation-Dependent Probe Amplification Assay and Flow Cytometric DNA Ploidy Analysis in Patients with Acute Leukemia. Cytom. Part B Clin. Cytom.. 94 (1), 172–181. doi: 10.1002/cyto.b.21523
Samelak-Czajka, A., Marszalek-Zenczak, M., Marcinkowska-Swojak, M., Kozlowski, P., Figlerowicz, M., Zmienko, A. (2017). MLPA-Based Analysis of Copy Number Variation in Plant Populations. Front. Plant Sci. 8 (261), 222. doi: 10.3389/fpls.2017.00222
Saxena, S., Gowdhaman, K., Kkani, P., Vennapusa, B., Subramanian, C. R., Kumar, S. G., et al. (2015). Improved Multiplex Ligation-dependent Probe Amplification (i -MLPA) for rapid copy number variant (CNV) detection. Clin. Chim. Acta 450, 19–24. doi: 10.1016/j.cca.2015.07.028
Schouten, J. P., Mcelgunn, C. J., Raymond, W., Danny, Z., Filip, D., Gerard, P. (2002). Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 30 (12), e57. doi: 10.1093/nar/gnf056
Singh, C. S., Gaur, A., Sreenivas, A., Singh, L. (2012). Species identification from dried snake venom. J. Forensic Sci. 57 (3), 826–828. doi: 10.1111/j.1556-4029.2011.02049.x
Stern, R. F., Roberts, R. G., Mann, K., Yau, S. C., Berg, J., Ogilvie, C. M. (2004). Multiplex ligation-dependent probe amplification using a completely synthetic probe set. Biotechniques 37 (3), 399. doi: 10.2144/04373ST04
Sultana, S., Ali, E., Hossain, M., Asing Naquiah, N., Zaidul (2018). Universal mini COI barcode for the identification of fish species in processed products. Food Res. Int. 105, 19–28. doi: 10.1016/j.foodres.2017.10.065
Tingping, F. (2001). Study on feature of Bungarus multicinctus and its confudes species. Shi Zhen Guo Yi Guo Yao 12 (4), 334–335. doi: 10.3969/j.issn.1008-0805.2001.04.032
Wuguang, L., Lingling, H., Jie, Y., Xiaoyan, S., Huaijiang, Y., Jinman, L., et al. (2017). Isolation and pharmacological characterization of a new cytotoxic L-amino acid oxidase from Bungarus multicinctus snake venom. J. Ethnopharmacol. 213, 311–320. doi: 10.1016/j.jep.2017.11.026
Yanqing, C., Bo, W., Ping, W., Bisheng, H., Hegang, L., Chao, X., et al. (2019). Rapid identification of common medicinal snakes and their adulterants using the Bar-HRM analysis method. Mitochondr. DNA Part A. 30 (2), 367–374. doi: 10.1080/24701394.2018.1532417
Yau, F. C. F., Wong, K. L., Shaw, P. C., But, P. P. H., Wang, J. (2002). Authentication of snakes used in Chinese medicine by sequence characterized amplified region (SCAR). Biodivers. Conserv. 11 (9), 1653–1662. doi: 10.1023/A:1016836017903
Yi, X., Yan, L., Haiping, W., Ying, B. (2017). Progress in molecular identification of snake drugs. Chin. J. Chin. Meteria Med. 42 (15), 2930–2933. doi: 10.19540/j.cnki.cjcmm.20170714.006
Keywords: Bungarus multicinctus, multiplex ligation-dependent probe amplification, adulterant identification, simultaneous identification method, traditional Chinese medicine
Citation: Zhou Y, Nie J, Yu S, Hu Z and Wang B (2020) Multiplex Ligation-Dependent Probe Amplification for Simultaneous Identification of Bungarus multicinctus and Its Common Adulterants in a Single Assay. Front. Pharmacol. 11:501. doi: 10.3389/fphar.2020.00501
Received: 27 August 2019; Accepted: 30 March 2020;
Published: 21 April 2020.
Edited by:
Monique S. J. Simmonds, Royal Botanic Gardens, Kew, United KingdomCopyright © 2020 Zhou, Nie, Yu, Hu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhigang Hu, zghu0608@163.com; Bo Wang, wang_bo1986@hotmail.com
 Yuxin Zhou
Yuxin Zhou Jing Nie
Jing Nie Shiqi Yu
Shiqi Yu Zhigang Hu
Zhigang Hu Bo Wang
Bo Wang