- 1Research Unit Cardiac Epigenetics, Department of Cardiology, University of Heidelberg, Heidelberg, Germany
- 2German Center for Cardiovascular Research (DZHK), Partner Site Heidelberg/Mannheim, Germany
CaMKII has been shown to be activated during different cardiac pathological processes, and CaMKII-dependent mechanisms contribute to pathological cardiac remodeling, cardiac arrhythmias, and contractile dysfunction during heart failure. Activation of CaMKII during cardiac stress results in a broad number of biological effects such as, on the one hand, acute effects due to phosphorylation of distinct cellular proteins as ion channels and calcium handling proteins and, on the other hand, integrative mechanisms by changing gene expression. This review focuses on transcriptional and epigenetic effects of CaMKII activation during chronic cardiac remodeling. Multiple mechanisms have been described how CaMKII mediates changes in cardiac gene expression. CaMKII has been shown to directly phosphorylate components of the cardiac gene regulation machinery. CaMKII phosphorylates several transcription factors such as CREB that induces the activation of specific gene programs. CaMKII activates transcriptional regulators also indirectly by phosphorylating histone deacetylases, especially HDAC4, which in turn inhibits transcription factors that drive cardiac hypertrophy, fibrosis, and dysfunction. Recent studies demonstrate that CaMKII also phosphorylate directly histones, which may contribute to changes in gene expression. These findings of CaMKII-dependent gene regulation during cardiac remodeling processes suggest novel strategies for CaMKII-dependent “transcriptional or epigenetic therapies” to control cardiac gene expression and function. Manipulation of CaMKII-dependent signaling pathways in the settings of pathological cardiac growth, remodeling, and heart failure represents an auspicious therapeutic approach.
Introduction
Heart failure is the leading cause of death in developed countries and is characterized by adverse cardiac remodeling upon pathological stress situations such as arterial hypertension, ischemic injuries or due to genetic causes. Adverse left ventricular remodeling is usually described by a combination of myocardial hypertrophy, cell death, interstitial fibrosis and an activation of a so-called fetal gene program (Koitabashi and Kass, 2012). Calcium-dependent signaling pathways including Calcium/Calmodulin-dependent kinase II (CaMKII) signaling play pivotal roles in adverse cardiac remodeling (Heineke and Molkentin, 2006; Bers, 2008; Backs et al., 2009; Ling et al., 2009). Activation of CaMKII during cardiac stress results in a broad number of biological effects. On the one hand, CaMKII mediates immediate effects due to phosphorylation of cellular proteins such as ion channels and calcium handling proteins (Anderson et al., 2011). On the other hand, CaMKII affects structural features of the cardiac phenotype due to phosphorylation of proteins of the transcriptional machinery (Anderson et al., 2011). This review focuses on CaMKII-dependent transcriptional and epigenetic mechanisms that occur in cardiomyocytes during pathological and physiological processes. First, we will review the different CaMKII genes and splice variants that localize to different subcellular compartments.
CaMKII Isoforms and Splice Variants
In 2003, Colomer and colleagues observed an increased activity of Calcium/Calmodulin-dependent kinases upon pathological pressure overload due to transverse aortic constriction (TAC), and they described the expression patterns of the Calcium/Calmodulin-dependent kinases CaMKI, CaMKII, and CaMKIV (Colomer et al., 2003). They found CaMKI to be expressed in left ventricular tissue, but not up-regulated upon TAC. Whereas artificial overexpression of CaMKIV in a transgenic model was sufficient to induce cardiac hypertrophy in another study (Passier et al., 2000), in the model of Colomer, left ventricular CaMKIV was not detectable, and mice lacking CaMKIV did not display an altered response to TAC, indicating that CaMKIV is not required for cardiac hypertrophy. They convincingly demonstrated that CaMKII is the only multifunctional CaMK that is not only up-regulated on the expression level but also activated after TAC.
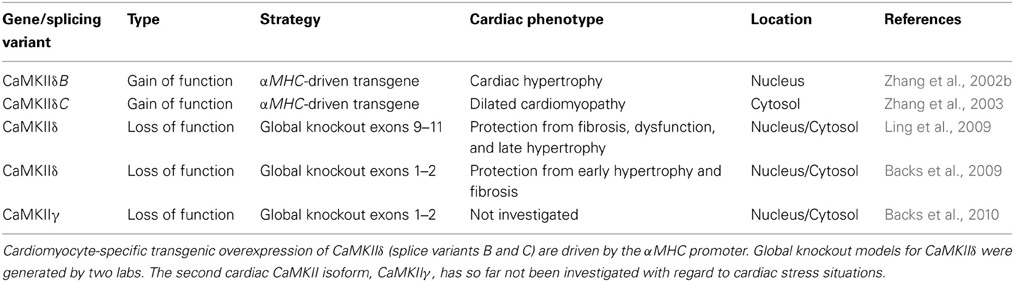
CaMKII consists of four different isoforms with distinct expression patterns. CaMKIIα and CaMKIIβ are enriched in neuronal tissue, and CaMKIIδ, and CaMKIIγ are expressed ubiquitously (Hudmon and Schulman, 2002). CaMKIIδ is the most abundant cardiac CaMKII isoform but CaMKIIγ is also expressed in the heart (Hoch et al., 1999; Colomer et al., 2003). The first in vivo studies establishing CaMKII as a potential target for cardiac arrhythmias and structural heart disease were conducted by the use of a pharmacological inhibitor such as KN-62 or KN-93 and a CaMKII inhibitory peptide (Zhang et al., 2005; Vila-Petroff et al., 2007; Liu et al., 2011). Due to the unclear role of the single CaMKII isoforms and potential unspecific effects of CaMKII inhibitors, isoform-specific genetic loss of function models were generated. Mice with a global deletion of CaMKIIδ were protected against adverse cardiac remodeling (Backs et al., 2009; Ling et al., 2009). CaMKIIδ global knockout mice produced by us were protected from cardiac fibrosis and hypertrophy 3 weeks after TAC surgery. CaMKIIδ global knockout model generated by Ling and colleagues were protected from fibrosis and dysfunction. These mice were not protected from cardiac hypertrophy 2 weeks but only 6 weeks after TAC. These seemingly different phenotypes with regard to cardiac hypertrophy may be explained by different surgery techniques, different genetic backgrounds, or different knockout strategies. With regard to the latter, in the first model, no residual protein was expressed (transcriptional null due to deletion of exon 1 and 2), whereas in the second model the possible existence of a truncated protein encoding a region before exon 8 was not ruled out (exons 9–11 were deleted). The specific role of cardiac CaMKIIγ and a potential redundancy with CaMKIIδ have not been investigated yet. In human and experimental heart failure, enhanced CaMKII activity was mainly attributed to an enhanced expression of the CaMKIIδ splice variants CaMKIIδB and CaMKIIIδC (Edman and Schulman, 1994; Hoch et al., 1999). From transgenic mouse models with artificial overexpression of these splice variants it was concluded that CaMKIIδB (localizes to the nucleus) promotes cardiac hypertrophy and CaMKIIδC (localizes to the cytosol) results in dilated cardiomyopathy, respectively (Zhang et al., 2002b, 2003). Moreover, CaMKIIδ A (localizes to sarcolemmal and nuclear membranes) was implied as another splice variant that is regulated at least in a model of cardiac hypertrophy due to isoproterenol treatment in mice (Xu et al., 2005; Li et al., 2011). However, to our knowledge transgenic models of CaMKIIδ A have not been generated so far. An overview of available genetic mouse models related to cardiac CaMKII is given in Table 1.
CaMKII and Transcriptional Regulation
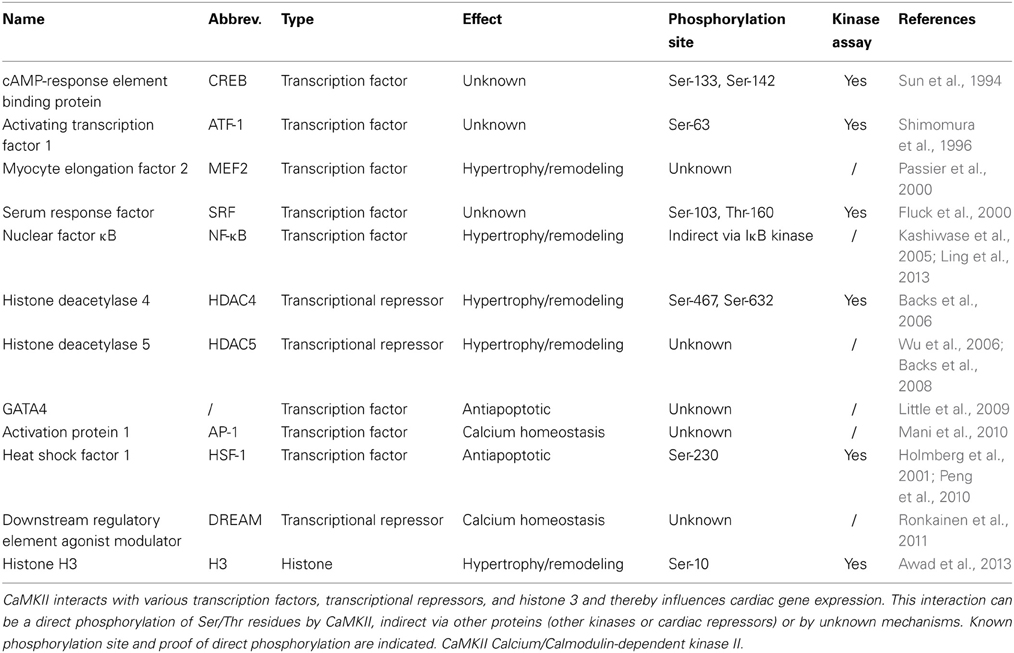
Effects of CaMKII on cardiac gene expression was first reported by the group of Joan Heller Brown when transient expression of CaMKIIδB in neonatal rat ventricular myocytes induced gene expression of atrial natriuretic factor (ANF) and resulted in enhanced transcriptional activation of an ANF-luciferase reporter gene (Ramirez et al., 1997). As we know now, CaMKII is involved in the regulation of many transcription factors such as the activation protein-1 (AP-1) (Antoine et al., 1996), activating transcription factor-1 (ATF-1) (Shimomura et al., 1996), serum response factor (SRF) (Fluck et al., 2000), cAMP-response element binding protein (CREB) (Sun et al., 1994), and myocyte enhancer factor 2 (MEF2). The latter is discussed in the next paragraph. An overview about the identified transcriptional regulators is given in Figure 1.
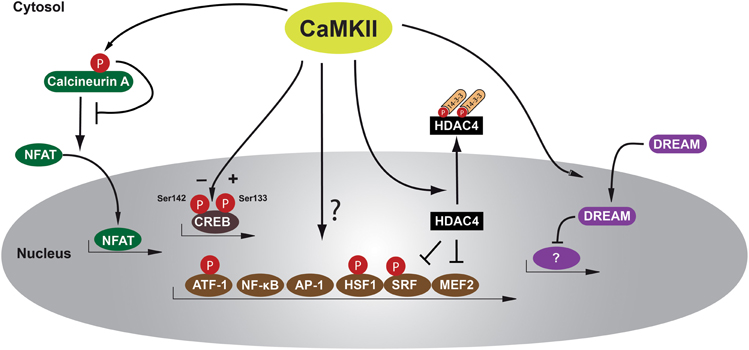
Figure 1. Schematic of transcription factors and transcriptional repressors regulated by CaMKII in cardiomyocytes. CaMKII phosphorylates HDAC4 at Ser-467 and Ser-632, allowing binding of the chaperone protein 14-3-3, leading to nucleo-cytoplasmic shuttling of a phospho-HDAC4/14-3-3 complex out of the nucleus and resulting in derepression of transcription factors such as MEF2 that regulates genes responsible for adverse cardiac remodeling. Other transcription factors such as NF-κB or HSP-1 may play maladaptive or adaptive roles and these factors can be directly or indirectly regulated by CaMKII. Another transcription factor regulated after β-adrenergic stimulation is CREB that can be phosphorylated by CaMKII at two serine residues, Ser-133 and Ser-142, resulting in opposing effects in regard to activation of CREB. However, the functional effects of CREB/CaMKII interaction during cardiac remodeling remain unclear. Another recently recognized mechanism is translocation of the transcriptional repressor DREAM from the cytosol to the nucleus. Calcineurin/NFAT interaction may also be inhibited by direct phosphorylation by CaMKII at Ser-411, leading to decreased NFAT translocation to the nucleus and subsequent reduced transcriptional activity. AP-1 activation protein-1, ATF-1 activating transcription factor-1, CaMKII Calcium/Calmodulin-dependent kinase II, CREB cAMP-response element binding protein, DREAM downstream regulatory element agonist modulator, HDAC4 histone deacetylase 4, HSF-1 heat shock factor 1, MEF2 myocyte elongation factor, NFAT nuclear factor of activated T-cells, NF-κB nuclear factor κB, and SRF serum response factor.
A well-known transcription factor in striated muscle biology is SRF, and Calcium/Calmodulin-dependent regulation of SRF via interaction with CaMKIV and histone deacetylase 4 (HDAC4), but not CaMKII, was demonstrated to be involved in the development of cardiac hypertrophy (Davis et al., 2003). A direct phosphorylation of SRF by CaMKII has been shown in skeletal muscle at Ser-103 and Thr-160 (Fluck et al., 2000), but its relevance in cardiomyocytes remains unclear. However, data from other cell types indicate that SRF-dependent gene transcription might depend on phosphorylation by CaMKII (Ely et al., 2011). As genetic animal models provide evidence for an important role for SRF in the induction and maintenance of the cardiac myogenic program (Lin et al., 1997; Parlakian et al., 2005; Backs et al., 2011), a better understanding of CaMKII-dependent SRF regulation is urgently needed.
AP-1 elements are a group of transcription factors composed of either homodimers of the Jun family (c-Jun, JunB, and JunD) or heterodimers of the Fos and Jun families (c-Fos, Fra-1, Fra-2, and FosB) (Mechta-Grigoriou et al., 2001). There is evidence for CaMKII-dependent transcriptional activity via AP-1 in non-cardiac tissues (Mishra et al., 2005; Zayzafoon et al., 2005). Recently, CaMKII-dependent upregulation of the sodium/calcium exchanger 1 (NCX1) has been shown to depend on AP-1 transcription factors c-Jun and JunB in cardiomyocytes (Mani et al., 2010). However, it remains unclear whether CaMKII phosphorylates AP-1 elements directly or indirectly.
As one of the well characterized transcription factors activated by G-protein coupled receptors, CREB has been shown to be phosphorylated by Calcium/Calmodulin-dependent kinases (Sheng et al., 1991). Several phosphorylation sites in CREB have been identified to regulate transcriptional activity and have been shown to be regulated by CaMKII in a dual way. Phosphorylation of CREB at Ser-133 by PKA and CaMKII is required for CREB activation, whereas phosphorylation of Ser-142 by CaMKII inhibits CREB activity by inhibition of CREB dimerization and protein/protein interactions that are necessary to form an active promoter complex (Wu and McMurray, 2001). In vitro studies using purified CaMKIV and CaMKII have demonstrated that CaMKIV phosphorylates CREB on Ser-133, whereas CaMKII has equal affinity for Ser-133 and Ser-142 (Sun et al., 1994). However, whereas nuclear calcium elevations increase CREB-dependent transcriptional activity (Kobrinsky et al., 2011), it remains unclear whether CaMKII-dependent CREB phosphorylation plays a significant role in cardiac remodeling processes (Li et al., 2006). Another member of the cAMP-responsive transcription factor family is ATF-1. ATF-1 is phosphorylated at Ser-63 by CaMKII, increasing its transcriptional activity (Shimomura et al., 1996), but the relevance of ATF-1 activity in cardiomyocytes is unknown.
Studies using KN-93, a CaMKII-inhibitory chemical compound, postulated CaMKII-dependent activation of transcription factor nuclear factor-κB (NF-κB) leading to cardiomyocyte hypertrophy (Kashiwase et al., 2005). In CaMKIIδ knockout mouse studies, NF-κB-dependent mechanisms and subsequent activation of inflammatory genes were found to play a maladaptive role in myocardial ischemia and ischemia/reperfusion injury in mice (Singh et al., 2009; Ling et al., 2013). Of note, the latter work conducted by the group of Mark Anderson was the first to perform gene expression profiling in a relevant animal model of cardiac stress, after myocardial infarction in mice. In this study, a CaMKII inhibitory peptide was used. Thus, potential off target effects need to be taken into account. For instance, it was shown that the CaMKII inhibitory peptide can also inhibit protein kinase D (PKD) (Backs et al., 2009). Gene expression arrays in genetic loss of function models might provide important additional information.
A possible physiological CaMKII-dependent transcriptional effect mediated by CaMKIIδB is the activation of heat shock factor 1 (HSF-1), a transcription factor responsible for inducible heat shock protein 70 (iHSP70) gene regulation. CaMKII is known to phosphorylate HSF-1 at Ser-230 (Holmberg et al., 2001), and Wei Peng and colleagues provided data suggesting that this might be a CaMKII-dependent antiapoptotic mechanism during cardiac ischemia and reperfusion (Peng et al., 2010). In this context, others could show that CaMKIIδB protects from doxorubicine-induced apoptosis, perhaps through GATA4-related expression of the antiapoptotic bcl-2 (B-cell lymphoma 2) gene (Little et al., 2009), although the mechanism how CaMKII induced GATA4-dependent gene expression was not shown. Taken the central role of GATA4 in cardiac hypertrophy and growth (Oka et al., 2006; Heineke et al., 2007), more data are warranted to clarify the role of GATA4 as a downstream target of CaMKII. Taken together, there are only sparse data available on how CaMKII directly interacts with transcription factors, and for most factors a direct binding and phosphorylation by CaMKII is not yet shown (See also Table 2).
In an interesting recent in vitro study, Jarkko Ronkainen et al. describe how CaMKII potentiates the translocation of the transcriptional repressor DREAM (downstream regulatory element agonist modulator) into the nucleus and thereby promotes DREAM-induced transcriptional repression. In their study, the authors could show that this mechanism is involved in CaMKII-dependent downregulation of the pore-forming α-subunit (Cav1.2) of the L-type calcium channel (LTCC) and postulate this to be a physiological feedback mechanism, which enables cardiomyocytes to adjust calcium influx through the LTCC to calcium-activated CaMKII activity (Ronkainen et al., 2011).
Another “indirect” transcriptional mechanism seems to be mediated by an interaction between CaMKII and calcineurin A. The phosphatase calcineurin A dephosphorylates nuclear factor of activated T-cells (NFAT), resulting in nuclear accumulation of NFAT and consequent activation of NFAT-dependent transcriptional programs and severe cardiac hypertrophy (Molkentin et al., 1998). In an elegant study, it was demonstrated that cytosolic CaMKIIδC phosphorylates calcineurin A within its calmodulin binding domain at Ser-411 and thereby inhibits its activity (MacDonnell et al., 2009). Although the relevance of these findings needs to be proven in vivo, this suggest that CaMKIIδC may act as a negative modulator of calcineurin/NFAT activity.
CaMKII and Epigenetic Regulation
Besides its direct effects on transcriptional regulators, CaMKII regulates gene expression also by phosphorylation of proteins of the epigenetic machinery, especially histone deacetylases (HDACs) and in particular class II HDACs. These interesting mechanisms were initially identified upon the observation that class II HDACs interact with the transcription factor MEF2. MEF2 was introduced to depend on CaMKI and CaMKIV more than 10 years ago by the group of Eric Olson and has been established as a critical transcription factor in cardiac remodeling processes (Passier et al., 2000). MEF2 is a common target for several hypertrophic pathways, although its precise function in cardiac remodeling and the cardiac genes that are modulated by this factor are still under investigation. MEF2 proteins are responsive to calcium-controlled signaling pathways, such as CaMKI, CaMKII, CaMKIV, and Calcineurin (Passier et al., 2000; McKinsey et al., 2002; Zhang et al., 2007). Class II HDACs are expressed in the heart and contain a MEF2 binding domain in the N-terminal region, which is not present in other HDACs. This N-terminal domain binds to the chaperone 14-3-3 and is then exported from the nucleus with the consequent de-repression of MEF2 (Backs and Olson, 2006; McKinsey, 2007; Ling et al., 2013). 14-3-3 binding depends on phosphorylation of HDACs by different kinases. For example, PKD phosphorylates all class II HDAC family members (HDAC4, HDAC5, HDAC7, HDAC9) (Vega et al., 2004; Harrison et al., 2006).
We found that CaMKII selectively signals to HDAC4 via binding to a unique docking site and phosphorylation of Ser-467 and Ser-632 (Backs et al., 2006) (See also Figure 2). These data were confirmed by others and phosphorylation of HDAC4 by CaMKII was suggested as a central mechanism in the development of cardiac hypertrophy and remodeling (Little et al., 2007; Zhang et al., 2007; Backs et al., 2009). HDAC5 does not bind to CaMKII and can therefore only be regulated by CaMKII when it is located in close proximity to HDAC4. When HDAC5 oligomerizes with HDAC4, it can be phosphorylated and exported in a complex with HDAC4 and CaMKII (Backs et al., 2008). Accordingly, HDAC5 has been shown to be regulated by CaMKII under certain conditions. The Bers lab demonstrated that calcium in the nuclear envelope is regulated independently from the global calcium transients that cause contraction at each heartbeat. Interestingly, calcium release from the nuclear envelope activates nuclear CaMKII, which triggers nuclear export of HDAC5 (Wu and Bers, 2006; Wu et al., 2006). Whereas nuclear CaMKIIδB and cytosolic CaMKIIδC exert different effects on the phosphorylation of calcium handling proteins as the ryanodine receptor or phospholamban and on calcium homeostasis (Zhang et al., 2007), both isoforms lead to cytosolic accumulation of HDAC4 and an increase in the activity of the transcription factor MEF2 (Backs et al., 2006). Nuclear CaMKIIδB phosphorylates HDAC4 in the nucleus, leading to nucleo-cytoplasmic shuttling of HDAC4. Activation of cytoplasmic CaMKIIδC phosphorylates HDAC4 in the cytosol and prevents the import of HDAC4 from the cytosol to the nucleus (Backs et al., 2006). Thus, cytoplasmic CaMKII is also capable to regulate transcription in addition to its effects on excitation-contraction coupling. These findings strongly suggested that CaMKII indirectly regulates MEF2 by dissociating HDAC4 and HDAC5. However, HDAC4 binds to many other proteins such as other transcription factors including SRF (Davis et al., 2003), co-repressors as CtBP (C-terminal-binding protein) (Zhang et al., 2001) but also to other chromatin modifying enzymes (Zhang et al., 2002a), opening the possibility that CaMKII exerts via cytosolic accumulation of HDAC4 broader effects than simply activating MEF2 (Lehmann et al., 2013).
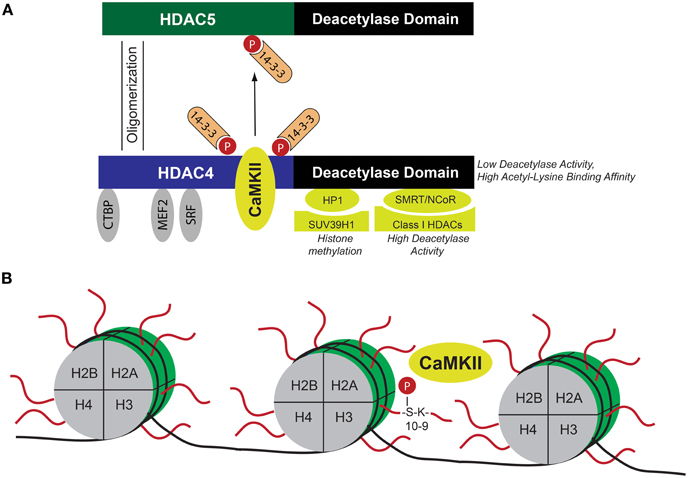
Figure 2. Schematic of CaMKII-dependent epigenetic mechanisms. (A) HDAC4 as a nodal point for CaMKII-dependent epigenetic regulation. CaMKII binds to HDAC4 and phosphorylates HDAC4 at Ser-467 and Ser-632, leading to nucleo-cytoplasmic shuttling of HDAC4. When located in the nucleus, HDAC4 represses transcription factors such as MEF2, SRF, or the co-repressor CtBP. Binding to these transcriptional regulators directs HDAC4 to specific chromatin regions. HDAC4 recruits other chromatin modifying enzymes and direct them to the aforementioned specific chromatin regions. This results in CaMKII/HDAC4-dependent regulation of histone methylation (via interaction with HP1 and histone methyltransferase SUV39H1) and deacetylation (via interaction with SMRT/N-CoR and class I HDACs). Moreover, HDAC4 oligomerizes with HDAC5, and thereby induces 14-3-3 dependent nucleo-cytoplasmic shuttling of the HDAC4/HDAC5 complex, leading to de-repression of HDAC4/HDAC5-dependent transcription factors. Thus HDAC4 integrates CaMKII-dependent signals via epigenetic mechanisms. (B) Histones (H2A, H2B, H3, and H4) assemble with DNA to form nucleosomes. CaMKII directly phosphorylates Ser-10 in the N-terminal region of histone 3, which is located next to Lys-9, a major site for acetylation, and methylation. Phosphorylation at Ser-10 was suggested to result in cardiomyocyte hypertrophy and increased chromatin binding of CaMKII at specific gene loci reactivated during cardiac hypertrophy. CaMKII Calcium/Calmodulin-dependent kinase II, CtBP C-terminal binding protein, HDAC histone deacetylase, HP1 heterochromatin protein 1, MEF2 myocyte elongation factor, N-CoR nuclear receptor co-repressor, SMRT silencing mediator of retinoic acid and thyroid hormone receptor, and SRF serum response factor.
Histones are major components of chromatin and assemble with DNA to form nucleosomes, (Jenuwein and Allis, 2001). The N-terminal regions of histones are subjected to a variety of post-translational modifications, including acetylation, methylation, ubiquitination, SUMOylation, and phosphorylation (Kouzarides, 2007; Ruthenburg et al., 2007). Figure 2 summarizes important interacting proteins of HDAC4 that direct HDAC4 to specific chromatin regions by binding to transcription factors. Of note, the deacetylase activity of class II HDACs is low but the binding affinity to acetylated lysines is high, suggesting that HDAC4 is mainly recruited to acetylated chromatin regions in close proximity to regions where MEF2 or SRF binds (Lahm et al., 2007). Although its deacetylase activity is low, HDAC4 binds indirectly via the co-repressors SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) and N-CoR (nuclear receptor co-repressor) to class I HDACs with high deacetylase activity (Fischle et al., 2002) and via HP1 (heterochromatin protein 1) to histone methyltransferases (Zhang et al., 2002a). Thus it is tempting to speculate that CaMKII effects besides transcriptional activity of MEF2 and SRF also class I HDAC-dependent histone acetylation and histone methyltransferase-dependent histone methylation. Indeed, together with the Maack lab we could show that HDAC4 controls histone methylation in a CaMKII-dependent manner (Hohl et al., 2013). ANF and brain natriuretic peptide (BNP) expression in failing hearts was accompanied by demethylation of histone 3 at lysine 9 (H3K9) and dissociation of HP1 from the promoter regions of ANF and BNP, and this was controlled by HDAC4, possibly by forming a transcriptional repressor complex with the histone methyltransferase SUV39H1 that was disrupted by CaMKII-induced phosphorylation of HDAC4. The importance of the CaMKII/HDAC4/MEF2-pathway with regard to epigenetic mechanisms in cardiac remodeling was underscored by a recent study from the Condorelli lab (Papait et al., 2013). The authors performed chromatin immunoprecipitation combined with genomic sequencing (ChIP-Seq) and RNA sequencing in isolated cardiomyocytes after TAC surgery and found a specific epigenetic signature that regulated gene expression by governing the activity of promoters and enhancers related to cardiac hypertrophy. Interestingly, they found MEF2 to be the main transcription factor to orchestrate this hypertrophic gene program by regulating the activity of transcriptional enhancers.
With regard to epigenetic mechanisms, in cardiac biology most attention so far was paid to histone acetylation and methylation. Histone phosphorylation is thought to be important for cell cycle regulation and was thus not carefully studied in the adult heart (Walter et al., 2008; Baek, 2011). Recently, it was reported by the group of Coralie Poizat that nuclear CaMKII activates cardiac transcription by direct binding to the chromatin. CaMKII was shown to phosphorylate Ser-10 of histone 3 (H3S10) which is located next to Lys-9, a major site for acetylation and methylation (Awad et al., 2013). Phosphorylation of H3S10 was accompanied by hypertrophy of primary cultured cardiomcyocytes and with increased chromatin binding of CaMKII at specific gene loci reactivated during cardiac hypertrophy under control of the transcription factor MEF2. These findings represent an interesting new epigenetic mechanism governed by CaMKII. The possibility, that ventricular remodeling can be mediated by CaMKII-dependent chromatin modifications opens a new avenue of regulatory mechanisms. Important further studies are warranted. ChIP-Seq studies may identify direct target genes of CaMKII that are important for diseases processes.
Summary and Outlook
CaMKII regulates not only immediate cellular functions but also chronic processes such as ventricular remodeling leading to heart failure. In particular, CaMKII integrates several cellular pathways by inducing gene programs that are not understood in detail. Here, we reviewed the yet known transcriptional and epigenetic mechanisms by which CaMKII regulates cardiac gene expression. However, the relative importance of the different downstream mechanisms still needs to be clarified. Unbiased gene expression analyses and epigenetic profiling are warranted to define the specific gene programs that contribute to phenotypic changes induced by CaMKII.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank David Stanmore for editing the manuscript. Johannes Backs was supported by the DZHK (Deutsches Zentrum für Herz-Kreislauf-Forschung—German Center for Cardiovascular Research) and by the European Commission (FP7-Health-2010; MEDIA-261409). Michael M. Kreusser was supported by a research grant from the Ernst-und-Berta-Grimmke foundation and by the Young Investigator program of the University of Heidelberg.
References
Anderson, M. E., Brown, J. H., and Bers, D. M. (2011). CaMKII in myocardial hypertrophy and heart failure. J. Mol. Cell. Cardiol. 51, 468–473. doi: 10.1016/j.yjmcc.2011.01.012
Antoine, M., Gaiddon, C., and Loeffler, J. P. (1996). Ca2+/calmodulin kinase types II and IV regulate c-fos transcription in the AtT20 corticotroph cell line. Mol. Cell. Endocrinol. 120, 1–8. doi: 10.1016/0303-7207(96)03806-3
Awad, S., Kunhi, M., Little, G. H., Bai, Y., An, W., Bers, D., et al. (2013). Nuclear CaMKII enhances histone H3 phosphorylation and remodels chromatin during cardiac hypertrophy. Nucleic Acids Res. 41, 7656–7672. doi: 10.1093/nar/gkt500
Backs, J., Backs, T., Bezprozvannaya, S., McKinsey, T. A., and Olson, E. N. (2008). Histone deacetylase 5 acquires calcium/calmodulin-dependent kinase II responsiveness by oligomerization with histone deacetylase 4. Mol. Cell. Biol. 28, 3437–3445. doi: 10.1128/MCB.01611-07
Backs, J., Backs, T., Neef, S., Kreusser, M. M., Lehmann, L. H., Patrick, D. M., et al. (2009). The delta isoform of CaM kinase II is required for pathological cardiac hypertrophy and remodeling after pressure overload. Proc. Natl. Acad. Sci. U.S.A. 106, 2342–2347. doi: 10.1073/pnas.0813013106
Backs, J., Stein, P., Backs, T., Duncan, F. E., Grueter, C. E., McAnally, J., et al. (2010). The gamma isoform of cam kinase ii controls mouse egg activation by regulating cell cycle resumption. Proc. Natl. Acad. Sci. U.S.A. 107, 81–86. doi: 10.1073/pnas.0912658106
Backs, J., and Olson, E. N. (2006). Control of cardiac growth by histone acetylation/deacetylation. Circ. Res. 98, 15–24. doi: 10.1161/01.RES.0000197782.21444.8f
Backs, J., Song, K., Bezprozvannaya, S., Chang, S., and Olson, E. N. (2006). CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. J. Clin. Invest. 116, 1853–1864. doi: 10.1172/JCI27438
Backs, J., Worst, B. C., Lehmann, L. H., Patrick, D. M., Jebessa, Z., Kreusser, M. M., et al. (2011). Selective repression of MEF2 activity by PKA-dependent proteolysis of HDAC4. J. Cell Biol. 195, 403–415. doi: 10.1083/jcb.201105063
Baek, S. H. (2011). When signaling kinases meet histones and histone modifiers in the nucleus. Mol. Cell 42, 274–284. doi: 10.1016/j.molcel.2011.03.022
Bers, D. M. (2008). Calcium cycling and signaling in cardiac myocytes. Annu. Rev. Physiol. 70, 23–49. doi: 10.1146/annurev.physiol.70.113006.100455
Colomer, J. M., Mao, L., Rockman, H. A., and Means, A. R. (2003). Pressure overload selectively up-regulates Ca2+/calmodulin-dependent protein kinase II in vivo. Mol. Endocrinol. 17, 183–192. doi: 10.1210/me.2002-0350
Davis, F. J., Gupta, M., Camoretti-Mercado, B., Schwartz, R. J., and Gupta, M. P. (2003). Calcium/calmodulin-dependent protein kinase activates serum response factor transcription activity by its dissociation from histone deacetylase, HDAC4. Implications in cardiac muscle gene regulation during hypertrophy. J. Biol. Chem. 278, 20047–20058. doi: 10.1074/jbc.M209998200
Edman, C. F., and Schulman, H. (1994). Identification and characterization of delta B-CaM kinase and delta C-CaM kinase from rat heart, two new multifunctional Ca2+/calmodulin-dependent protein kinase isoforms. Biochim. Biophys. Acta 1221, 89–101. doi: 10.1016/0167-4889(94)90221-6
Ely, H. A., Mellon, P. L., and Coss, D. (2011). GnRH induces the c-Fos gene via phosphorylation of SRF by the calcium/calmodulin kinase II pathway. Mol. Endocrinol. 25, 669–680. doi: 10.1210/me.2010-0437
Fischle, W., Dequiedt, F., Hendzel, M. J., Guenther, M. G., Lazar, M. A., Voelter, W., et al. (2002). Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9, 45–57. doi: 10.1016/S1097-2765(01)00429-4
Fluck, M., Booth, F. W., and Waxham, M. N. (2000). Skeletal muscle CaMKII enriches in nuclei and phosphorylates myogenic factor SRF at multiple sites. Biochem. Biophys. Res. Commun. 270, 488–494. doi: 10.1006/bbrc.2000.2457
Harrison, B. C., Kim, M. S., van Rooij, E., Plato, C. F., Papst, P. J., Vega, R. B., et al. (2006). Regulation of cardiac stress signaling by protein kinase d1. Mol. Cell. Biol. 26, 3875–3888. doi: 10.1128/MCB.26.10.3875-3888.2006
Heineke, J., Auger-Messier, M., Xu, J., Oka, T., Sargent, M. A., York, A., et al. (2007). Cardiomyocyte GATA4 functions as a stress-responsive regulator of angiogenesis in the murine heart. J. Clin. Invest. 117, 3198–3210. doi: 10.1172/JCI32573
Heineke, J., and Molkentin, J. D. (2006). Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 7, 589–600. doi: 10.1038/nrm1983
Hoch, B., Meyer, R., Hetzer, R., Krause, E. G., and Karczewski, P. (1999). Identification and expression of delta-isoforms of the multifunctional Ca2+/calmodulin-dependent protein kinase in failing and nonfailing human myocardium. Circ. Res. 84, 713–721. doi: 10.1161/01.RES.84.6.713
Hohl, M., Wagner, M., Reil, J. C., Muller, S. A., Tauchnitz, M., Zimmer, A. M., et al. (2013). HDAC4 controls histone methylation in response to elevated cardiac load. J. Clin. Invest. 123, 1359–1370. doi: 10.1172/JCI61084
Holmberg, C. I., Hietakangas, V., Mikhailov, A., Rantanen, J. O., Kallio, M., Meinander, A., et al. (2001). Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 20, 3800–3810. doi: 10.1093/emboj/20.14.3800
Hudmon, A., and Schulman, H. (2002). Neuronal CA2+/calmodulin-dependent protein kinase II: the role of structure and autoregulation in cellular function. Annu. Rev. Biochem. 71, 473–510. doi: 10.1146/annurev.biochem.71.110601.135410
Jenuwein, T., and Allis, C. D. (2001). Translating the histone code. Science 293, 1074–1080. doi: 10.1126/science.1063127
Kashiwase, K., Higuchi, Y., Hirotani, S., Yamaguchi, O., Hikoso, S., Takeda, T., et al. (2005). CaMKII activates ASK1 and NF-kappaB to induce cardiomyocyte hypertrophy. Biochem. Biophys. Res. Commun. 327, 136–142. doi: 10.1016/j.bbrc.2004.12.002
Kobrinsky, E., Duong, S. Q., Sheydina, A., and Soldatov, N. M. (2011). Microdomain organization and frequency-dependence of CREB-dependent transcriptional signaling in heart cells. FASEB J. 25, 1544–1555. doi: 10.1096/fj.10-176198
Koitabashi, N., and Kass, D. A. (2012). Reverse remodeling in heart failure–mechanisms and therapeutic opportunities. Nat. Rev. Cardiol. 9, 147–157. doi: 10.1038/nrcardio.2011.172
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128, 693–705. doi: 10.1016/j.cell.2007.02.005
Lahm, A., Paolini, C., Pallaoro, M., Nardi, M. C., Jones, P., Neddermann, P., et al. (2007). Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc. Natl. Acad. Sci. U.S.A. 104, 17335–17340. doi: 10.1073/pnas.0706487104
Lehmann, L. H., Worst, B. C., Stanmore, D. A., and Backs, J. (2013). Histone deacetylase signaling in cardioprotection. Cell. Mol. Life Sci. doi: 10.1007/s00018-013-1516-9. [Epub ahead of print].
Li, B., Dedman, J. R., and Kaetzel, M. A. (2006). Nuclear Ca2+/calmodulin-dependent protein kinase II in the murine heart. Biochim. Biophys. Acta 1763, 1275–1281. doi: 10.1016/j.bbamcr.2006.09.029
Li, C., Cai, X., Sun, H., Bai, T., Zheng, X., Zhou, X. W., et al. (2011). The deltaA isoform of calmodulin kinase II mediates pathological cardiac hypertrophy by interfering with the HDAC4-MEF2 signaling pathway. Biochem. Biophys. Res. Commun. 409, 125–130. doi: 10.1016/j.bbrc.2011.04.128
Lin, Q., Schwarz, J., Bucana, C., and Olson, E. N. (1997). Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 276, 1404–1407. doi: 10.1126/science.276.5317.1404
Ling, H., Gray, C. B., Zambon, A. C., Grimm, M., Gu, Y., Dalton, N., et al. (2013). Ca2+/Calmodulin-dependent protein kinase II delta mediates myocardial ischemia/reperfusion injury through nuclear factor-kappaB. Circ. Res. 112, 935–944. doi: 10.1161/CIRCRESAHA.112.276915
Ling, H., Zhang, T., Pereira, L., Means, C. K., Cheng, H., Gu, Y., et al. (2009). Requirement for Ca2+/calmodulin-dependent kinase II in the transition from pressure overload-induced cardiac hypertrophy to heart failure in mice. J. Clin. Invest. 119, 1230–1240. doi: 10.1172/JCI38022
Little, G. H., Bai, Y., Williams, T., and Poizat, C. (2007). Nuclear calcium/calmodulin-dependent protein kinase IIdelta preferentially transmits signals to histone deacetylase 4 in cardiac cells. J. Biol. Chem. 282, 7219–7231. doi: 10.1074/jbc.M604281200
Little, G. H., Saw, A., Bai, Y., Dow, J., Marjoram, P., Simkhovich, B., et al. (2009). Critical role of nuclear calcium/calmodulin-dependent protein kinase IIdeltaB in cardiomyocyte survival in cardiomyopathy. J. Biol. Chem. 284, 24857–24868. doi: 10.1074/jbc.M109.003186
Liu, N., Ruan, Y., Denegri, M., Bachetti, T., Li, Y., Colombi, B., et al. (2011). Calmodulin kinase II inhibition prevents arrhythmias in RyR2(R4496C+/-) mice with catecholaminergic polymorphic ventricular tachycardia. J. Mol. Cell. Cardiol. 50, 214–222. doi: 10.1016/j.yjmcc.2010.10.001
MacDonnell, S. M., Weisser-Thomas, J., Kubo, H., Hanscome, M., Liu, Q., Jaleel, N., et al. (2009). CaMKII negatively regulates calcineurin-NFAT signaling in cardiac myocytes. Circ. Res. 105, 316–325. doi: 10.1161/CIRCRESAHA.109.194035
Mani, S. K., Egan, E. A., Addy, B. K., Grimm, M., Kasiganesan, H., Thiyagarajan, T., et al. (2010). beta-Adrenergic receptor stimulated Ncx1 upregulation is mediated via a CaMKII/AP-1 signaling pathway in adult cardiomyocytes. J. Mol. Cell. Cardiol. 48, 342–351. doi: 10.1016/j.yjmcc.2009.11.007
McKinsey, T. A. (2007). Derepression of pathological cardiac genes by members of the CaM kinase superfamily. Cardiovasc. Res. 73, 667–677. doi: 10.1016/j.cardiores.2006.11.036
McKinsey, T. A., Zhang, C. L., and Olson, E. N. (2002). MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27, 40–47. doi: 10.1016/S0968-0004(01)02031-X
Mechta-Grigoriou, F., Gerald, D., and Yaniv, M. (2001). The mammalian Jun proteins: redundancy and specificity. Oncogene 20, 2378–2389. doi: 10.1038/sj.onc.1204381
Mishra, J. P., Mishra, S., Gee, K., and Kumar, A. (2005). Differential involvement of calmodulin-dependent protein kinase II-activated AP-1 and c-Jun N-terminal kinase-activated EGR-1 signaling pathways in tumor necrosis factor-alpha and lipopolysaccharide-induced CD44 expression in human monocytic cells. J. Biol. Chem. 280, 26825–26837. doi: 10.1074/jbc.M500244200
Molkentin, J. D., Lu, J. R., Antos, C. L., Markham, B., Richardson, J., Robbins, J., et al. (1998). A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93, 215–228. doi: 10.1016/S0092-8674(00)81573-1
Oka, T., Maillet, M., Watt, A. J., Schwartz, R. J., Aronow, B. J., Duncan, S. A., et al. (2006). Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ. Res. 98, 837–845. doi: 10.1161/01.RES.0000215985.18538.c4
Papait, R., Cattaneo, P., Kunderfranco, P., Greco, C., Carullo, P., Guffanti, A., et al. (2013). Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc. Natl. Acad. Sci. U.S.A. 110, 20164–20169. doi: 10.1073/pnas.1315155110
Parlakian, A., Charvet, C., Escoubet, B., Mericskay, M., Molkentin, J. D., Gary-Bobo, G., et al. (2005). Temporally controlled onset of dilated cardiomyopathy through disruption of the SRF gene in adult heart. Circulation 112, 2930–2939. doi: 10.1161/CIRCULATIONAHA.105.533778
Passier, R., Zeng, H., Frey, N., Naya, F. J., Nicol, R. L., McKinsey, T. A., et al. (2000). CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J. Clin. Invest. 105, 1395–1406. doi: 10.1172/JCI8551
Peng, W., Zhang, Y., Zheng, M., Cheng, H., Zhu, W., Cao, C. M., et al. (2010). Cardioprotection by CaMKII-deltaB is mediated by phosphorylation of heat shock factor 1 and subsequent expression of inducible heat shock protein 70. Circ. Res. 106, 102–110. doi: 10.1161/CIRCRESAHA.109.210914
Ramirez, M. T., Zhao, X. L., Schulman, H., and Brown, J. H. (1997). The nuclear deltaB isoform of Ca2+/calmodulin-dependent protein kinase II regulates atrial natriuretic factor gene expression in ventricular myocytes. J. Biol. Chem. 272, 31203–31208. doi: 10.1074/jbc.272.49.31203
Ronkainen, J. J., Hanninen, S. L., Korhonen, T., Koivumaki, J. T., Skoumal, R., Rautio, S., et al. (2011). Ca2+-calmodulin-dependent protein kinase II represses cardiac transcription of the L-type calcium channel alpha(1C)-subunit gene (Cacna1c) by DREAM translocation. J. Physiol. 589, 2669–2686. doi: 10.1113/jphysiol.2010.201400
Ruthenburg, A. J., Li, H., Patel, D. J., and Allis, C. D. (2007). Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994. doi: 10.1038/nrm2298
Sheng, M., Thompson, M. A., and Greenberg, M. E. (1991). CREB: a Ca(2+)-regulated transcription factor phosphorylated by calmodulin-dependent kinases. Science 252, 1427–1430. doi: 10.1126/science.1646483
Shimomura, A., Ogawa, Y., Kitani, T., Fujisawa, H., and Hagiwara, M. (1996). Calmodulin-dependent protein kinase II potentiates transcriptional activation through activating transcription factor 1 but not cAMP response element-binding protein. J. Biol. Chem. 271, 17957–17960. doi: 10.1074/jbc.271.30.17957
Singh, M. V., Kapoun, A., Higgins, L., Kutschke, W., Thurman, J. M., Zhang, R., et al. (2009). Ca2+/calmodulin-dependent kinase II triggers cell membrane injury by inducing complement factor B gene expression in the mouse heart. J. Clin. Invest. 119, 986–996. doi: 10.1172/JCI35814
Sun, P., Enslen, H., Myung, P. S., and Maurer, R. A. (1994). Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev. 8, 2527–2539. doi: 10.1101/gad.8.21.2527
Vega, R. B., Harrison, B. C., Meadows, E., Roberts, C. R., Papst, P. J., Olson, E. N., et al. (2004). Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 24, 8374–8385. doi: 10.1128/MCB.24.19.8374-8385.2004
Vila-Petroff, M., Salas, M. A., Said, M., Valverde, C. A., Sapia, L., Portiansky, E., et al. (2007). CaMKII inhibition protects against necrosis and apoptosis in irreversible ischemia-reperfusion injury. Cardiovasc. Res. 73, 689–698. doi: 10.1016/j.cardiores.2006.12.003
Walter, W., Clynes, D., Tang, Y., Marmorstein, R., Mellor, J., and Berger, S. L. (2008). 14-3-3 interaction with histone H3 involves a dual modification pattern of phosphoacetylation. Mol. Cell. Biol. 28, 2840–2849. doi: 10.1128/MCB.01457-07
Wu, X., and Bers, D. M. (2006). Sarcoplasmic reticulum and nuclear envelope are one highly interconnected Ca2+ store throughout cardiac myocyte. Circ. Res. 99, 283–291. doi: 10.1161/01.RES.0000233386.02708.72
Wu, X., and McMurray, C. T. (2001). Calmodulin kinase II attenuation of gene transcription by preventing cAMP response element-binding protein (CREB) dimerization and binding of the CREB-binding protein. J. Biol. Chem. 276, 1735–1741. doi: 10.1074/jbc.M006727200
Wu, X., Zhang, T., Bossuyt, J., Li, X., McKinsey, T. A., Dedman, J. R., et al. (2006). Local InsP3-dependent perinuclear Ca2+ signaling in cardiac myocyte excitation-transcription coupling. J. Clin. Invest. 116, 675–682. doi: 10.1172/JCI27374
Xu, X., Yang, D., Ding, J. H., Wang, W., Chu, P. H., Dalton, N. D., et al. (2005). ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell 120, 59–72. doi: 10.1016/j.cell.2004.11.036
Zayzafoon, M., Fulzele, K., and McDonald, J. M. (2005). Calmodulin and calmodulin-dependent kinase IIalpha regulate osteoblast differentiation by controlling c-fos expression. J. Biol. Chem. 280, 7049–7059. doi: 10.1074/jbc.M412680200
Zhang, C. L., McKinsey, T. A., Lu, J. R., and Olson, E. N. (2001). Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J. Biol. Chem. 276, 35–39. doi: 10.1074/jbc.M007364200
Zhang, C. L., McKinsey, T. A., and Olson, E. N. (2002a). Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol. Cell. Biol. 22, 7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002
Zhang, R., Khoo, M. S., Wu, Y., Yang, Y., Grueter, C. E., Ni, G., et al. (2005). Calmodulin kinase II inhibition protects against structural heart disease. Nat. Med. 11, 409–417. doi: 10.1038/nm1215
Zhang, T., Johnson, E. N., Gu, Y., Morissette, M. R., Sah, V. P., Gigena, M. S., et al. (2002b). The cardiac-specific nuclear delta(B) isoform of Ca2+/calmodulin-dependent protein kinase II induces hypertrophy and dilated cardiomyopathy associated with increased protein phosphatase 2A activity. J. Biol. Chem. 277, 1261–1267. doi: 10.1074/jbc.M108525200
Zhang, T., Kohlhaas, M., Backs, J., Mishra, S., Phillips, W., Dybkova, N., et al. (2007). CaMKIIdelta isoforms differentially affect calcium handling but similarly regulate HDAC/MEF2 transcriptional responses. J. Biol. Chem. 282, 35078–35087. doi: 10.1074/jbc.M707083200
Keywords: CaMKII, epigenetics, transcription factors, HDAC4, remodeling
Citation: Kreusser MM and Backs J (2014) Integrated mechanisms of CaMKII-dependent ventricular remodeling. Front. Pharmacol. 5:36. doi: 10.3389/fphar.2014.00036
Received: 14 January 2014; Accepted: 18 February 2014;
Published online: 12 March 2014.
Edited by:
Donald M. Bers, University of California, Davis, USAReviewed by:
Timothy McKinsey, University of Colorado Denver, USADonald Menick, Medical University of South Carolina, USA
Copyright © 2014 Kreusser and Backs. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Johannes Backs, Research Unit Cardiac Epigenetics, Department of Cardiology, University of Heidelberg and DZHK (German Centre for Cardiovascular Research), Im Neuenheimer Feld 410, 69120 Heidelberg, Germany e-mail: johannes.backs@med.uni-heidelberg.de
 Michael M. Kreusser
Michael M. Kreusser Johannes Backs
Johannes Backs