- 1Ben Towne Center for Childhood Cancer Research, Seattle Children’s Research Institute, Seattle, WA, United States
- 2Department of Pediatrics, University of Washington, Seattle, WA, United States
- 3Cancer and Blood Disorders Center, Seattle Children’s Hospital, Seattle, WA, United States
- 4Department of Laboratories, Seattle Children’s Hospital, Seattle, WA, United States
- 5Department of Laboratory Medicine and Pathology, University of Washington, Seattle, WA, United States
- 6Department of Pediatric Neurosurgery, Seattle Children’s Hospital, Seattle, WA, United States
- 7Department of Neurological Surgery, University of Washington, Seattle, WA, United States
Purpose: Somatic molecular profiling of pediatric brain tumors aids with the diagnosis and treatment of patients with a variety of high- and low-grade central nervous system neoplasms. Here, we report follow-up targeted germline evaluation for patients with possible germline variants following tumor only testing in the initial year in which somatic molecular testing was implemented at a single institution.
Patients and Methods: Somatic testing was completed for all tumors of the central nervous system (CNS) undergoing diagnostic workup at Seattle Children’s Hospital during the study period of November 2015 to November 2016. Sequencing was performed in a College of American Pathologists-accredited, Clinical Laboratory Improvements Amendments-certified laboratory using UW-OncoPlex™ assay (version 5), a DNA-based targeted next generation sequencing panel validated to detect genetic alterations in 262 cancer-related genes. We tracked subsequent clinical evaluation and testing on a subgroup of this cohort found to have potential germline variants of interest.
Results: Molecular sequencing of 88 patients’ tumors identified 31 patients with variants that warranted consideration of germline testing. To date, 19 (61%) patients have been tested. Testing confirmed germline variants for ten patients (31% of those identified for testing), one with two germline variants (NF1 and mosaic TP53). Eight (26%) patients died before germline testing was sent. One patient (13%) has not yet had testing.
Conclusion: Clinically validated molecular profiling of pediatric brain tumors identifies patients who warrant further germline evaluation. Despite this, only a subset of these patients underwent the indicated confirmatory sequencing. Further work is needed to identify barriers and facilitators to this testing, including the role of genetic counseling and consideration of upfront paired somatic-germline testing.
Introduction
Over the prior two decades, large pediatric oncology sequencing studies have demonstrated the feasibility of upfront genomic testing at the time of high-risk diagnosis and relapse of childhood cancer (1, 2). While access to sequencing via research protocols has increased for children with cancer over the prior decade, there is currently no consensus regarding best-practice diagnostics for routine clinical practice.
NGS assays designed to detect single-nucleotide variants, insertions and deletions, copy number changes, and rearrangements in genes selected for their clinical significance in cancer, can identify alterations that clarify diagnosis or suggest molecularly targeted therapeutics. In some cases, somatic sequencing can also result in secondary findings, identifying variants in the tumor sample that appear likely to be present in the patient’s germline, which may or may not be related to the development of the patient’s cancer (3–5).
Our group previously reported the findings from a prospective cohort study where clinically validated molecular profiling of pediatric brain tumors aided in the diagnosis and treatment of patients with a variety of high- and low-grade primary, newly-diagnosed and relapsed brain tumors (6). A subset of the patients undergoing tumor only testing during the first year in which somatic clinical targeted sequencing was performed were found to have possible germline variants (6). We sought to evaluate how these results may have impacted further clinical evaluation and testing in this subgroup of the cohort.
Materials and methods
All patients with tumors of the central nervous system (CNS) undergoing diagnostic evaluation at Seattle Children’s Hospital during the study period from November 2015 to November 2016 had their tumors sequenced using the UW-OncoPlex™ assay (version 5; https://testguide.labmed.uw.edu/view/OPX)? (7). This multiplexed targeted next-generation sequencing panel, designed for the detection of genetic alterations in 262 cancer-related genes, is performed in a College of American Pathologists (CAP)-accredited and Clinical Laboratory Improvements Amendments (CLIA)-certified laboratory. Patients with tumor diagnosis by imaging alone were excluded from this study. The patients in this study consented to tumor banking, biology, and return of results (Seattle Children’s Hospital Institutional Review Board No. 00000506). Data were extracted from the medical record.
The molecular tumor results were discussed at a monthly molecular brain tumor conference, including pediatric neuro-oncology, pediatric pathology, molecular pathology, genetic counseling, and neurosurgery along with a research coordinator (6). Each month, all new patients were systematically reviewed in a presentation format consisting of clinical features and magnetic resonance imaging presented by a neuro-oncologist (S.E.S.L.), histologic findings and photomicrographs by a pediatric pathologist (B.L.C.), molecular results by a molecular pathologist (C.M.L.) followed by group discussion including laboratory genetic counseling (S.M.S.). Variants were identified for potential follow-up constitutional testing if they met any of the following criteria: pathogenic or likely pathogenic variants in genes known to be associated with cancer predisposition; or large indels, structural rearrangements or exonic deletions/duplications in genes known to be associated with cancer predisposition; or variants in a patient with clinical features, family history or tumor type suggestive of a constitutional predisposition related to the involved variant (8). The variant allele frequency (VAF) was not used to exclude variants warranting additional follow-up constitutional testing based on other features. Details of the laboratory workflow and methods have previously been published in a Data Supplement (6).
We tracked subsequent clinical evaluation and testing on a subgroup of this cohort who were found to have variants considered to be potentially germline. Individuals in this cohort (or their parents/guardians) were informed of the results and the recommendation for germline testing by their clinical team. Germline confirmatory testing involved the collection a peripheral blood sample or otherwise available non-tumor tissue. Results available in the electronic medical record were tracked on this subgroup of the cohort.
Results
Molecular sequencing was performed on CNS tumors of 88 patients. At the time of biopsy or surgical resection of their tumor, study participants’ ages ranged from 1 month to 21 years (median 7 years). Sequencing identified 31 patients (35%) whose tumors harbored variants warranting consideration of germline testing (Figure 1). The details of these results were previously published (6). To date, 19 patients (61% of those with recommended germline follow-up) have been tested (Table 1) (6). Thirteen (68%) of these patients who underwent testing met with an oncology genetic counselor who coordinated testing, while the other patients had the test coordinated by an oncologist at the institution (2, 11%), a provider at another institution (3, 16%), or an established genetic counselor given known germline genetic diagnosis (1, 5%). Germline testing was performed on peripheral blood for 18 patients and on non-affected brain tissue for one patient (patient 7) who had died prior to confirmatory testing. Confirmatory germline testing was performed a median of 13 months following initial tumor testing and ranged from 3 to 57 months. The patients’ ages at the time of confirmatory testing ranged from 9 months to 21 years (median 13 years). Of these 19 patients, testing confirmed germline variants for 10 patients (52% of those tested; 11.4% of the study population) as of the last follow-up. The confirmed germline findings were identified in ATM, BAP1, BRIP1, FANCA, MSH2, NF1, NF2, RAD51D, and TP53. One patient was found to have two germline variants (NF1 and mosaic TP53) (Table 1). Two other patients each had two somatic variants warranting testing, with one of the two variants in each patient confirmed to be germline. Of these 11 confirmed germline variants (in 10 patients), three (27%) had a corresponding tumor VAF less than 40%. Of the 10 patients with a confirmed germline variant, 8 patients (80%) received a family referral for genetic counseling and 3 (30%) had at least one relative undergo testing (Table 1).
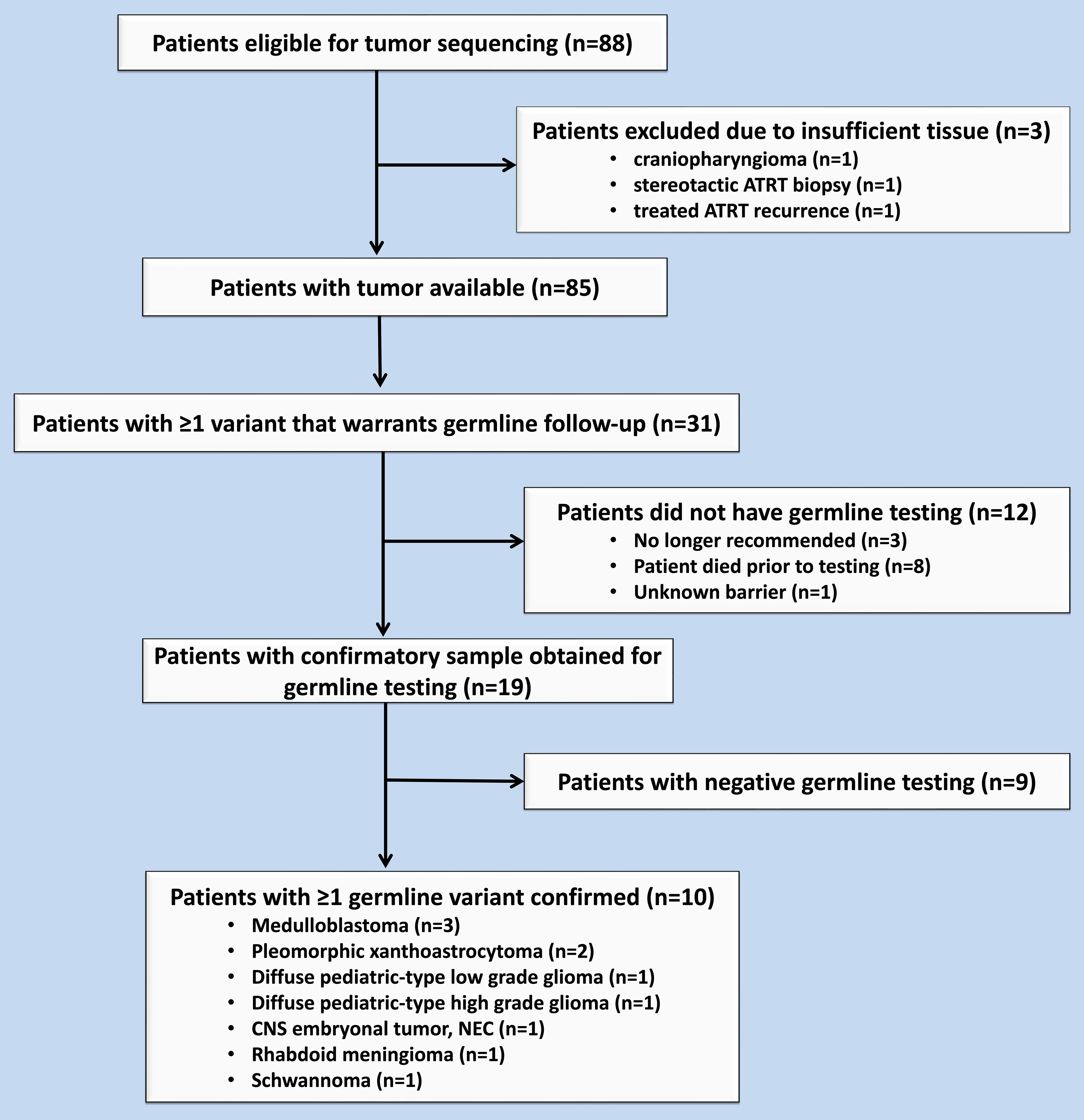
Figure 1 Flow chart of eligible patients during the study period from November 2015 to November 2016.
For some patients, confirmation of a germline variant clarified their diagnosis or was relevant to their treatment plan. Sequencing of one patient’s high grade pleomorphic xanthoastrocytoma (PXA) demonstrated a somatic BRAF variant and a TP53 variant, which was confirmed to be present in the germline (patient 39, Table 1). These findings directly impacted their treatment plan, with the use of a BRAF inhibitor, rather than radiation therapy. Another patient’s posterior fossa tumor was found to have an ATM variant, confirmed on germline testing (patient 61, Table 1). If this variant had been identified prior to initiating therapy, the concern for radiation sensitivity with a germline ATM variant would have likely impacted management (9). Unfortunately, this result was not available prior to treatment, and the patient ultimately experienced radiation necrosis (10). Another patient with medulloblastoma was found to have a germline variant of uncertain significance (VUS) in SUFU (patient 83, Table 1). This prompted cascade testing in her mother who was found to have the same variant. The patient’s maternal grandmother had a basal cell carcinoma diagnosed in her 40s and reported several other clinical features that could be consistent with Gorlin syndrome, including an extra rib, extra teeth, and history of oophorectomy for ruptured ovarian cyst. For this reason, sequencing of her maternal grandmother’s basal cell carcinoma was performed, but the variant was not identified.
Twelve (39%) of the patients with somatic variants indicating a possible germline variant have not had confirmatory testing. Eight (26%) of these patients died before genetic counseling and germline testing coordination. Three (10%) of the patients had variants that were reclassified and ultimately did not have germline testing recommended. The remaining patient (3%) had insurance authorization for testing, which has not yet been performed. No patients who were offered confirmatory testing actively declined.
Discussion
Molecular profiling of pediatric brain tumors is typically pursued with the priority of identifying potentially clinically significant variants. However, somatic sequencing may uncover potential germline variants, as demonstrated by this cohort. These variants can be important for informing a patient’s own cancer risk, can translate into life-saving surveillance and risk reduction interventions for self and family members, and – if identified in time – can sometimes impact therapeutic decisions (3, 4). The cohort on which we report includes several patients whose secondary germline findings could have influenced their treatment plan or surveillance care. Additionally, the majority of the patients with confirmed germline variants had at least one family member referred for genetic counseling and consideration of cascade testing. In one case, resultant cascade testing in family members assisted in diagnostic clarification (patient 83, Table 1). This patient, diagnosed with a medulloblastoma, was found to have a germline VUS in SUFU. Given the patient’s family history of basal cell carcinoma, one might suspect this variant was pathogenic. However, the variant was not detected upon subsequent testing of the grandmother’s basal cell carcinoma. Considering its absence in the grandparent’s tumor, along with the identification of a somatic RELA fusion in the patient’s recurrence, it was determined to be unlikely that the detected SUFU variant played a significant role in the development of this patient’s tumor.
Subsequent tracking of our cohort of patients highlights several challenges that arise with standard incorporation of tumor molecular sequencing into clinical care. In this protocol, the identification of somatic variants warranting germline follow-up testing was determined by the multi-disciplinary tumor board, a determination that might have variability across laboratories. Although the VAF was not used to exclude any variants in our cohort, it was documented for each case. Variants with an allele frequency of approximately 50% are generally assumed to be of germline origin; however, this threshold varies widely in the literature, ranging from 25-70% (8, 11–13) and is not the sole reliable indicator of need for germline testing (8). Tissue or tumor heterogeneity can result in a lower VAF (as can large indels, due to reference allele bias), and in some cases, copy number alterations, such as deletions, can mask germline point variants. It is therefore possible that potential germline variants are missed by laboratories using VAF thresholds to infer germline status or flag variants for germline follow-up. In our cohort, three of the confirmed germline variants (in two patients) had a corresponding tumor VAF less than 40%. One patient with a posterior fossa tumor treated with radiation therapy was found to have a variant in ATM with VAF of 25% (Patient 61, Table 1) (10). Had our standard process of evaluation used VAF alone, this patient would not have met the threshold for germline testing. Unfortunately, given delays in testing this variant was not discovered until after the patient had already received radiation treatment. The patient ultimately developed radiation necrosis – a long term effect noted more frequently in patients with germline ATM variants (9).
Given the challenges of using a somatic sample to identify which patients should undergo germline sequencing – including delays to confirmation of potentially time-sensitive, clinically relevant germline variants – some experts have advocated for upfront paired testing of both tumor and germline samples (14, 15). Paired testing is not without additional challenges. It is worth noting some laboratories intend their test to be only somatic and therefore intentionally restrict reporting likely constitutional variants, which can occur both in paired testing approaches and with bioinformatic filtering of potential germline variants (12). Aside from the technical and laboratory requirements/barriers, it necessitates a more nuanced informed consent process, which is already complicated by the distress of a new diagnosis or relapse of cancer (16, 17). ASCO guidelines recommend that oncology providers communicate the potential for incidental and secondary germline information to patients before they conduct somatic molecular profiling (4, 18). Further investigation is needed to establish more nuanced communication recommendations for oncologists to discuss the potential impacts of molecular sequencing testing and results with their patients and their families. Genetic counselors (GCs) have expertise in this communication, though their involvement may be limited early in the process by the urgency of this testing or at the time of return-of-results by GC availability, particularly at less resourced institutions. At our institution, this sequencing protocol led to institutional support for an integrated GC in our multidisciplinary neuro-oncology clinic. Incorporation of GCs into a multidisciplinary clinic in similar settings, such as adult gynecologic oncology clinic, improves adherence to genetic testing recommendations (19, 20). This and other quality improvement efforts in pediatric oncology clinics can be effective tools to improve uptake of genetic testing recommendations in pediatric patients with increased risk of cancer predisposition syndrome (21).
One identified barrier to patients undergoing appropriate follow-up testing in this cohort was patient death. Difficulty with follow-up testing when performed on a research protocol is further challenged when funding for administrative support does not continue following the completion of the research protocol. One patient who had prior insurance prior authorization for confirmatory germline testing but has not yet undergone the testing did not have a barrier identified in the medical record. It is possible that this might represent the family passively declining the recommendation. It is striking that all other patients who were recommended germline testing ultimately had a germline test performed. This might be related to the timing of this information in relation to their child’s initial diagnosis, with some not receiving the sequencing results and recommendation until after their child’s completion of upfront tumor-directed treatment. While for some families a delay in this discussion may have allowed more “mental space” to consider germline testing, others may have preferred to hear about this information sooner, particularly if the findings of the germline test had the potential to impact their child’s treatment plan. For those patients with confirmed germline variants, uptake of cascade testing in family members was variable. This is not unique to our cohort; in general, cascade testing remains suboptimal due to a variety of barriers at the patient-, provider-, and health systems-level (22–24).
The implications of this study should be considered in the context of a few limitations. First, this study reports on a small cohort of patients with brain tumors at a single institution. A limitation is that we did not consider germline testing of all variants found on somatic sequencing and therefore cannot validate our selection method prior to pursuing testing. Finally, the follow-up details of patient testing were determined by chart review, which may have simplified or misrepresented the extent to which the sequencing impacted the patients’ care or discussions about further testing.
Experience from this cohort highlights important considerations as upfront molecular sequencing becomes ever more frequent in childhood cancer care, both on clinical research protocols, such as the Molecular Characterization Initiative funded by the National Cancer Institute, and as part of the standard clinical care of patients (25). Additional research is needed to better delineate the impact of molecular sequencing both on patients’ clinical care and outcomes, as well as the psychosocial and financial impact on patients and their families.
Data availability statement
The data analyzed in this study is subject to the following licenses/restrictions: available upon request. Requests to access these datasets should be directed to BG, brittany.greene@seattlechildrens.org.
Ethics statement
The studies involving humans were approved by Seattle Children’s Hospital Institutional Review Board No. 00000506. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
BG: Conceptualization, Data curation, Formal analysis, Visualization, Writing – original draft, Writing – review & editing. SS: Conceptualization, Data curation, Writing – original draft, Writing – review & editing. MT: Validation, Writing – review & editing. NW: Conceptualization, Writing – review & editing. BC: Data curation, Writing – review & editing. CL: Data curation, Writing – review & editing. VP: Data curation, Writing – review & editing. JB: Data curation, Writing – review & editing. AL: Writing – review & editing. JO: Writing – review & editing. RE: Writing – review & editing. JS: Data curation, Writing – review & editing. SL: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Supported by the Seattle Children’s Hospital Funding Hope Award and the Pediatric Brain Tumor Research Fund. Also supported in part through the Focus on Kids Laboratory Guild and from a gift to Seattle Children’s Hospital from Kandace and Richard Holley. BG is supported in part by the Rally Foundation for Childhood Cancer Research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Newman S, Nakitandwe J, Kesserwan CA, Azzato EM, Wheeler DA, Rusch M, et al. Genomes for kids: the scope of pathogenic mutations in pediatric cancer revealed by comprehensive DNA and RNA sequencing. Cancer Discovery. (2021) 11:3008–27. doi: 10.1158/2159-8290.Cd-20-1631
2. Vo KT, Parsons DW, Seibel NL. Precision medicine in pediatric oncology. Surg Oncol Clin N Am. (2020) 29:63–72. doi: 10.1016/j.soc.2019.08.005
3. Alba-Pavón P, Alaña L, Gutierrez-Jimeno M, García-Obregón S, Imízcoz T, Panizo E, et al. Identification of germline cancer predisposition variants in pediatric sarcoma patients from somatic tumor testing. Sci Rep. (2023) 13. doi: 10.1038/s41598-023-29982-2
4. Raymond VM, Gray SW, Roychowdhury S, Joffe S, Chinnaiyan AM, Parsons DW, et al. Germline findings in tumor-only sequencing: points to consider for clinicians and laboratories. J Natl Cancer Inst. (2016) 108. doi: 10.1093/jnci/djv351
5. Deleonardis K, Hogan L, Cannistra SA, Rangachari D, Tung N. When should tumor genomic profiling prompt consideration of germline testing? J Oncol Pract. (2019) 15:465–73. doi: 10.1200/jop.19.00201
6. Cole BL, Lockwood CM, Stasi S, Stevens J, Lee A, Ojemann JG, et al. Year 1 in the molecular era of pediatric brain tumor diagnosis: application of universal clinical targeted sequencing in an unselected cohort of children. JCO Precis Oncol. (2018) 2):1–13. doi: 10.1200/po.17.00151
7. Kuo AJ, Paulson VA, Hempelmann JA, Beightol M, Todhunter S, Colbert BG, et al. Validation and implementation of a modular targeted capture assay for the detection of clinically significant molecular oncology alterations. Pract Lab Med. (2020) 19:e00153. doi: 10.1016/j.plabm.2020.e00153
8. MacFarland SP, Zelley K, Surrey LF, Gallo D, Luo M, Raman P, et al. Pediatric somatic tumor sequencing identifies underlying cancer predisposition. JCO Precis Oncol. (2019) 3. doi: 10.1200/po.19.00062
9. Rothblum-Oviatt C, Wright J, Lefton-Greif MA, McGrath-Morrow SA, Crawford TO, Lederman HM. Ataxia telangiectasia: a review. Orphanet J Rare Dis. (2016) 11:159. doi: 10.1186/s13023-016-0543-7
10. Tam L, Cole B, Stasi S, Paulson V, Wright J, Hoeppner C, et al. Somatic versus Germline— A Case Series of Three Children with ATM- mutated Medulloblastoma. JCO Precis Oncol. (2023).
11. Li MM, Chao E, Esplin ED, Miller DT, Nathanson KL, Plon SE, et al. Points to consider for reporting of germline variation in patients undergoing tumor testing: a statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med. (2020) 22:1142–8. doi: 10.1038/s41436-020-0783-8
12. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the association for molecular pathology, American society of clinical oncology, and college of American pathologists. J Mol Diagn. (2017) 19:4–23. doi: 10.1016/j.jmoldx.2016.10.002
13. Mandelker D, Donoghue M, Talukdar S, Bandlamudi C, Srinivasan P, Vivek M, et al. Germline-focussed analysis of tumour-only sequencing: recommendations from the ESMO Precision Medicine Working Group. Ann Oncol. (2019) 30:1221–31. doi: 10.1093/annonc/mdz136
14. Schrader KA, Cheng DT, Joseph V, Prasad M, Walsh M, Zehir A, et al. Germline variants in targeted tumor sequencing using matched normal DNA. JAMA Oncol. (2016) 2:104–11. doi: 10.1001/jamaoncol.2015.5208
15. Forman A, Sotelo J. Tumor-based genetic testing and familial cancer risk. Cold Spring Harb Perspect Med. (2020) 10. doi: 10.1101/cshperspect.a036590
16. Mandrell BN, Gattuso JS, Pritchard M, Caples M, Howard Sharp KM, Harrison L, et al. Knowledge is power: benefits, risks, hopes, and decision-making reported by parents consenting to next-generation sequencing for children and adolescents with cancer. Semin Oncol Nurs. (2021) 37:151167. doi: 10.1016/j.soncn.2021.151167
17. Mandrell BN, Johnson LM, Caples M, Gattuso J, Maciaszek JL, Mostafavi R, et al. Parental preferences surrounding timing and content of consent conversations for clinical germline genetic testing following a child’s new cancer diagnosis. JCO Precis Oncol. (2022) 6:e2200323. doi: 10.1200/po.22.00323
18. Robson ME, Bradbury AR, Arun B, Domchek SM, Ford JM, Hampel HL, et al. American society of clinical oncology policy statement update: genetic and genomic testing for cancer susceptibility. J Clin Oncol. (2015) 33:3660–7. doi: 10.1200/JCO.2015.63.0996
19. Kentwell M, Dow E, Antill Y, Wrede CD, McNally O, Higgs E, et al. Mainstreaming cancer genetics: A model integrating germline BRCA testing into routine ovarian cancer clinics. Gynecol Oncol. (2017) 145:130–6. doi: 10.1016/j.ygyno.2017.01.030
20. Senter L, O’Malley DM, Backes FJ, Copeland LJ, Fowler JM, Salani R, et al. Genetic consultation embedded in a gynecologic oncology clinic improves compliance with guideline-based care. Gynecol Oncol. (2017) 147:110–4. doi: 10.1016/j.ygyno.2017.07.141
21. Knoll J, Li A, Smith CH, Schratz K, Cooper SL, Meah T, et al. Improving detection of cancer predisposition syndromes in pediatric oncology. J Pediatr Hematol Oncol. (2021) 43:e891–e6. doi: 10.1097/mph.0000000000001987
22. Whitaker KD, Obeid E, Daly MB, Hall MJ. Cascade genetic testing for hereditary cancer risk: an underutilized tool for cancer prevention. JCO Precis Oncol. (2021) 5:1387–96. doi: 10.1200/po.21.00163
23. Frey MK, Ahsan MD, Bergeron H, Lin J, Li X, Fowlkes RK, et al. Cascade testing for hereditary cancer syndromes: should we move toward direct relative contact? A systematic review and meta-analysis. J Clin Oncol. (2022) 40:4129–43. doi: 10.1200/jco.22.00303
24. Griffin NE, Buchanan TR, Smith SH, Leon AA, Meyer MF, Liu J, et al. Low rates of cascade genetic testing among families with hereditary gynecologic cancer: An opportunity to improve cancer prevention. Gynecol Oncol. (2020) 156:140–6. doi: 10.1016/j.ygyno.2019.11.005
Keywords: childhood cancer, brain tumor, molecular testing, next-generation sequencing, germline
Citation: Greene BL, Stasi SM, Ting MA, Waligorski N, Cole BL, Lockwood CM, Paulson VA, Buchan JG, Lee A, Ojemann JG, Ellenbogen RG, Stevens J and Leary SES (2024) Looking beyond year 1 in the molecular era of pediatric brain tumor diagnosis: confirmatory testing of germline variants found on tumor sequencing. Front. Oncol. 14:1338022. doi: 10.3389/fonc.2024.1338022
Received: 14 November 2023; Accepted: 20 February 2024;
Published: 06 March 2024.
Edited by:
Angela Mastronuzzi, Bambino Gesù Children’s Hospital (IRCCS), ItalyReviewed by:
Fern Tsien, Louisiana State University Health Sciences Center, United StatesLuigi Boccuto, Clemson University, United States
Copyright © 2024 Greene, Stasi, Ting, Waligorski, Cole, Lockwood, Paulson, Buchan, Lee, Ojemann, Ellenbogen, Stevens and Leary. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brittany L. Greene, brittany.greene@seattlechildrens.org
 Brittany L. Greene
Brittany L. Greene Shannon M. Stasi
Shannon M. Stasi Michelle A. Ting
Michelle A. Ting Natalie Waligorski3
Natalie Waligorski3 Christina M. Lockwood
Christina M. Lockwood Jeffrey Stevens
Jeffrey Stevens Sarah E. S. Leary
Sarah E. S. Leary